Abstract
Lower jaw development is a complex process in which multiple signaling cascades establish a proximal-distal organization. These cascades are regulated both spatially and temporally and are constantly refined through both induction of normal signals and inhibition of inappropriate signals. The connective tissue of the tongue arises from cranial neural crest cell-derived ectomesenchyme within the mandibular portion of the first pharyngeal arch and is likely to be impacted by this signaling. Although the developmental mechanisms behind later aspects of tongue development, including innervation and taste acquisition, have been elucidated, the early patterning signals driving ectomesenchyme into a tongue lineage are largely unknown. We show here that the basic helix-loop-helix transcription factor Hand2 plays key roles in establishing the proximal-distal patterning of the mouse lower jaw, in part through establishing a negative-feedback loop in which Hand2 represses Dlx5 and Dlx6 expression in the distal arch ectomesenchyme following Dlx5- and Dlx6-mediated induction of Hand2 expression in the same region. Failure to repress distal Dlx5 and Dlx6 expression results in upregulation of Runx2 expression in the mandibular arch and the subsequent formation of aberrant bone in the lower jaw along with proximal-distal duplications. In addition, there is an absence of lateral lingual swelling expansion, from which the tongue arises, resulting in aglossia. Hand2 thus appears to establish a distal mandibular arch domain that is conducive for lower jaw development, including the initiation of tongue mesenchyme morphogenesis.
Keywords: Aglossia, bHLH, Craniofacial, Hinge and caps, Homeosis, Neural crest cell, Mouse
INTRODUCTION
Most of the craniofacial skeleton arises from neural crest cells (NCCs), including the bone, cartilage and connective tissue of the face, neck and tongue (Hall, 1982). Arising along the neural folds of vertebrates before migrating ventrally into the pharyngeal arches (Bronner-Fraser, 1995; Couly et al., 1996; Kontges and Lumsden, 1996; Le Douarin, 1982), NCCs are patterned by numerous environmental cues (Chai and Maxson, 2006). These signals, organized into hierarchical cascades, initiate NCC patterning and differentiation, thus determining both their identity and fate (Clouthier and Schilling, 2004; Depew and Simpson, 2006).
In contrast to our knowledge of the signals regulating bone and cartilage development in the mandibular portion of the first pharyngeal arch, the early signals initiating tongue morphogenesis are poorly understood. Development of the tongue involves NCCs both from three pharyngeal arches and from somitic myoblasts (Noden and Francis-West, 2006). Although the mouse mandibular arch is a complex structure composed of multiple tissue and gene expression domains at embryonic day (E) 10.5 (Depew and Simpson, 2006; Clouthier and Schilling, 2004), a noticeable tongue bud is not evident. However, by E12.5, a recognizable tongue is present and includes somitic myoblasts that have migrated through the hypoglossal duct and that will give rise to the tongue musculature (Noden and Francis-West, 2006). Besides transforming growth factor-beta receptor 2 (Tgfbr2) signaling (Hosokawa et al., 2010), the identity of other factors regulating this early aspect of tongue morphogenesis is not known.
One key pathway responsible for establishing the proximal-distal organization of the pharyngeal arches is initiated by signaling from the endothelin-A receptor (Ednra) located on NCCs (Clouthier et al., 1998; Yanagisawa et al., 1998). Ednra signaling is induced soon after NCCs reach the pharyngeal arches by its ligand, endothelin-1 (Edn1) (Clouthier et al., 1998; Maemura et al., 1996; Miller et al., 2000; Yanagisawa et al., 1998). This initiates one or more signaling cascades responsible for NCC identity and fate within the mandibular pharyngeal arch (Clouthier et al., 2010; Clouthier and Schilling, 2004). Two induced factors, Dlx5 and Dlx6, contribute to the ‘Dlx code’ hypothesized to establish proximal-distal identity within the arches (Depew et al., 2002; Depew and Simpson, 2006; Depew et al., 2005; Jeong et al., 2008). Loss of Ednra, Edn1, or Dlx5 and Dlx6 results in homeotic transformation of mandibular arch-derived bone and soft tissue structures into more maxillary-like derivates (Beverdam et al., 2002; Depew et al., 2002; Ozeki et al., 2004; Ruest et al., 2004). However, aglossia is not observed in any mouse mutants in the Ednra pathway (Ednra, Edn1, Ece1, Mef2c, Dlx5, Dlx6) (Clouthier et al., 2010).
One direct transcriptional target of Dlx5 and Dlx6 is the gene encoding the basic helix-loop-helix transcription (bHLH) factor Hand2 (Charité et al., 2001). Although Hand2 mutant mice die by E10.5 owing to vascular defects (Thomas et al., 1998; Yamagishi et al., 2000), fate mapping Hand2 daughter cells using the Hand2 arch-specific enhancer (Charité et al., 2001) illustrated that these cells were found throughout most of the lower jaw (Ruest et al., 2003a). In addition, ventral arch cartilage derivatives in hand2 mutant zebrafish (hans6, which survive longer than mouse mutants), are severely affected (Miller et al., 2003) and early gene expression is disrupted. In mice, targeted deletion of the Hand2 arch-specific enhancer (Hand2BA/BA) results in only partial loss of the Hand2 expression domain in the mandibular arch and limited developmental changes (Yanagisawa et al., 2003). However, NCC-specific deletion of Hand1 (the other Hand gene expressed in the pharyngeal arches) (Clouthier et al., 2000; Cserjesi et al., 1995) on the hypomorphic Hand2BA/BA background resulted in cleft palate and a small, thickened mandible (Funato et al., 2009). Based on in vitro findings, the changes in the mandible were hypothesized to result from a loss of Hand2-Runx2 interaction, leading to changes in bone ossification. However, the full function of Hand2 in facial morphogenesis is not clear, as Hand2 was not completely inactivated in these mice.
To address problems associated with global Hand2 inactivation, two groups have created Hand2 conditional knockout mice. Conditional ablation of Hand2 in NCCs has revealed a crucial role for Hand2 in the development of sympathetic ganglion neurons (Hendershot et al., 2008; Morikawa et al., 2007), the enteric nervous system (Hendershot et al., 2007; D'Autreaux et al., 2007) and the cardiac outflow tract (Holler et al., 2010). In addition, conditional inactivation of Hand2 in the developing palatal epithelium has uncovered a role for Hand2 in palatal shelf fusion (Xiong et al., 2009). However, the role of Hand2 in NCC patterning and differentiation during craniofacial development has yet to be examined. In this report, we show that inactivating Hand2 specifically in NCCs leads to changes in jaw development, with the most striking being aglossia. These changes are preceded by an earlier failure to downregulate Dlx5 and Dlx6 expression in the distal mandibular arch, indicating that Hand2-mediated repression of Dlx5 and Dlx6 expression is crucial for establishing a domain in the mandibular arch that is conducive for tongue mesenchymal morphogenesis.
MATERIALS AND METHODS
Mice
Hand2flox/flox (Hand2fl/fl) (Hendershot et al., 2007; Hendershot et al., 2008), Hand2fl/− (Hendershot et al., 2008), Wnt1-Cre (Danielian et al., 1998) and R26R (Soriano, 1999) mice have been described previously.
Breeding and genotyping
Hand2fl/fl mice were bred with Wnt1-Cre transgenic mice to generate Hand2fl/+;Wnt1-Cre animals. Hand2fl/+;Wnt1-Cre mice were bred with Hand2fl/fl mice to generate Hand2fl/fl;Wnt1-Cre embryos (referred to as Hand2cko). For fate-mapping experiments, Hand2fl/fl mice were bred with R26R mice (Soriano, 1999) to homozygosity. These mice were then bred with Hand2fl/+;Wnt1-Cre animals to create Hand2fl/fl;Wnt1-Cre;R26R+/− embryos. Hand2fl, Cre and lacZ genotyping were performed as described previously (Ruest and Clouthier, 2009; Soriano, 1999; Hendershot et al., 2008).
Skeleton staining
Skeleton (Ruest and Clouthier, 2009) and cartilage (Clouthier et al., 1998) staining was performed as described previously. Stained bone and cartilage preparations were analyzed and photographed using an Olympus SZX12 stereomicroscope fitted with a DP11 digital camera.
Whole-mount β-galactosidase staining
β-galactosidase staining was performed as described previously (Ruest et al., 2003a).
Histology
For histological analysis, E18.5 embryos were fixed, embedded in paraffin, sectioned and stained as described previously (Ruest et al., 2004). For immunostaining, sections were incubated with monoclonal anti-skeletal myosin (1:400; MY-32; Sigma, St Louis, MO, USA), anti-TROMO-1 (1:25; Developmental Studies Hybridoma Bank, Iowa City, IA, USA), and/or Alexa Fluor 568 phalloidin (Invitrogen) as described previously (Clouthier et al., 1997). Trichrome staining was performed as described previously (Clouthier et al., 1997). After staining, all sections were examined and photographed with an Olympus BX50 compound microscope fitted with an Olympus DP71 digital camera.
In situ hybridization
Whole-mount in situ hybridization (ISH) analysis was performed as described previously (Clouthier et al., 1998) using digoxigenin (DIG)-labeled antisense cRNA riboprobes against Dlx1, Dlx2, Dlx3, Dlx5, Dlx6, FoxF1 (Foxf1a – Mouse Genome Informatics), goosecoid homeobox (Gsc – Mouse Genome Informatics), Hand1, Hand2, Msx1, Msx2, Pitx1, Pitx2, Runx2, sonic hedgehog and Twist1. Embryos were photographed using an Olympus SZX12 microscope as described above.
Real-time PCR
Mandibular pharyngeal arches were collected and stored in RNA Later (Ambion) until genotyped. RNA was collected from arches using the QIAShredder and RNeasy kits (Qiagen). cDNA was prepared from total RNA using the Quantitect cDNA Synthesis Kit (Qiagen). Real-time quantitative PCR was performed using 5 ng of cDNA and the Quantitect SYBR Green PCR Kit (Qiagen), including Quantitect Assay primers (Qiagen). RT-PCR and data analysis was performed using a MyiQ2 machine (BioRad).
Volume measurements for the Runx2 expression domain
Volume measurements for Runx2 expression following ISH were conducted using ImageJ software (http://rsb.info.nih.gov/ij/) (Abramoff et al., 2004). Briefly, the scale (pixels/micron) was calculated using a micrometer and the ImageJ ruler tool, then was set globally in ImageJ. A region of interest (ROI) was specified using the polygon tool. The lower jaw ROI was specified as the entire lower jaw. We included the tongue in control embryos to ensure that our area measurements between control and Hand2cko embryos were equivalent (as Hand2cko embryos lack a tongue). The Runx2 ROI was specified as the entire region exhibiting DIG reactivity. Once the ROIs were outlined, the total area contained in the outlined region was calculated in ImageJ using the Measure tool. The percentage of expression was calculated by dividing the Runx2 ROI area by the lower jaw ROI area. Measurements were conducted on three embryos of each genotype and values averaged.
Analysis of cellular proliferation and apoptosis
Three 7 μm sections through the rostral, middle and caudal aspects of the mandibular arch, each separated by 60 μm, were used. Analysis of cell death at E10.5 was performed using the DeadEnd Labeling Kit (Promega) as described previously (Abe et al., 2007). Detection of cell death at E11.5 was performed using the In Situ Cell Death Detection Kit (Roche). Proliferating cells at both ages were detected using 5-ethynyl-2′-deoxyuridine (EdU) incorporation and the Invitrogen Click-It EdU Kit (Promega). At E10.5, these assays were conducted on consecutive sections; at E11.5, assays were performed on the same section. After staining, slides were counterstained with 4′,6-diamidino-2-phenlylindole dihydrochloride (DAPI; Sigma, 2.0 μg/ml) for 10 minutes, washed and coverslipped using ProLong Gold Antifade Reagent (Invitrogen). Images were captured using the DP71 digital camera, with reconstruction of the entire arch region accomplished using TrakEM2, a component of Fiji (http://pacific.mpi-cbg.de). To quantify cell death and proliferation in specific arch regions, a grid was manually placed over the arch image using Adobe Photoshop. Vertical grid lines were placed on the midline and the outermost regions of the left and right lateral arch halves. The midline of the left or right arch half was then calculated by dividing the distance between lateral outer edge and arch midline in half; this established the boundary between the anterior/posterior arch and the lateral arch. Horizontal grid lines were calculated by setting the upper and lower limits at the point where the arch midline reaches the ectoderm. The length between upper and lower horizontal lines was then divided in two to create the horizontal midline of the arch, separating the anterior and posterior arch. Cell counts within the left and right lateral regions and posterior and anterior regions were then manually counted using the Cell Counter plugin for ImageJ. The final incidence of proliferation and cell death was calculated as the number of proliferating or TUNEL-positive cells as a percentage of total cells.
Cell culture
MC3T3 cells (ATCC, Manassas, VA, USA) were maintained in MEM alpha (Invitrogen) supplemented with 20% fetal bovine serum (Sigma), penicillin/streptomycin (Invitrogen), and fungizone (Invitrogen) at 37°C in a humidified chamber with 5% CO2.
Luciferase assays
MC3T3 cells seeded on 12-well plates were transfected with 0.05 μg pRL-TK (Promega), 0.5 μg pGL3-I56i-luc [generated by placing the I56i enhancer both 5′ and 3′ of the firefly luciferase cDNA in the pGL3 (Promega) vector] or pGL3-I56iΔEbox-luc [created using the QuikChange Lightning Site-Directed Mutagenesis Kit (Stratagene); primers 5′-CAAATTGGATGGCACTGAAGCTGGAGGCTTTGTTC-3′ and 5′-GAACAAAGCCTCCAGCTTCAGTGCCATCCATTG-3′ were used to change the putative E-box site CAGCTG to AAGCTG within the pGL2-I56i-luc template vector], and 0.8 μg of each expression vector using Fugene 6 (Roche). Expression vectors included: pCS2+MT-Dlx5, pCS2+MT-Hand2, pCS2+MT-Hand1 (gift from Hiromi Yanagisawa, University of Texas Southwestern Medical Center, Dallas, TX, USA) and pcDNA3.1+MT-ΔN(1-90)-Hand2 (gift from Anthony Firulli, Indiana University School of Medicine, Indianapolis, IN, USA). Experiments were performed in triplicate and collected 24 hours after transfection. Lysate was prepared and firefly (experimental reporter) and renilla (normalizing reporter) luciferases were measured simultaneously using the Dual-Luciferase Reporter Assay System (Promega) and a Luminoskan Ascent 2.4 machine (Thermo Scientific). Activity of the experimental reporter was calculated by dividing the firefly luciferase value by the renilla luciferase value. Fold change was then calculated by dividing the expression vector activity by the empty expression vector (mock) activity.
Statistical analysis
All statistical analysis of results was performed using an unpaired two-tailed t-test with Prism software (GraphPad).
RESULTS
Changes in facial development in Hand2fl/fl;Wnt1-Cre embryos
Mice homozygous for the conditional allele (Hand2fl/fl) were viable, healthy and fertile (Hendershot et al., 2007; Hendershot et al., 2008) and showed no alterations in skull morphology (data not shown). We next crossed Hand2fl/fl mice with the Wnt1-Cre transgenic strain to target the Hand2 gene deletion to NCCs (Chai et al., 2000). Grossly, E18.5 Hand2fl/fl;Wnt1-Cre (Hand2cko) embryos had retrognathia and low-set pinnae (data not shown) along with mystacial vibrissae (the whisker-like sensory organs normally confined to the snout) on the lower jaw epithelium (data not shown).
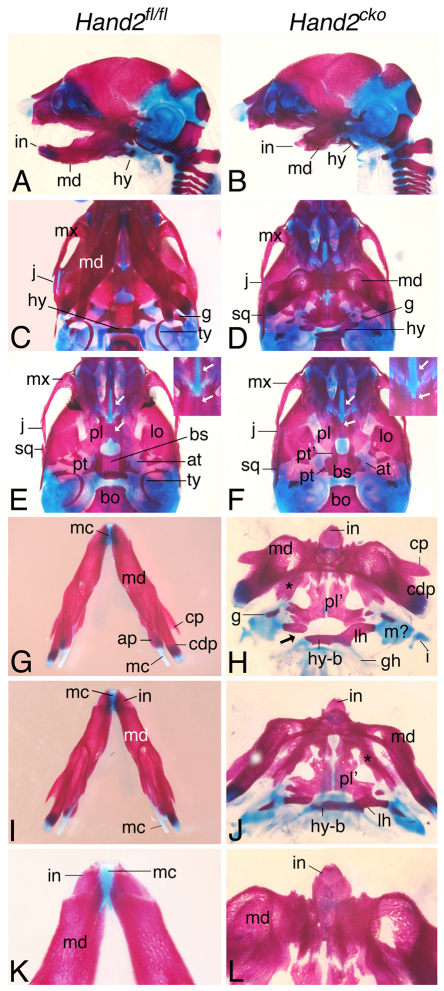
Alizarin Red- and Alcian Blue-stained E18.5 Hand2cko skeletons revealed a loss of both the angular processes of the mandible and the tympanic rings, significant retrognathia and an almost complete loss of Meckel's cartilage (Fig. 1B,D,H,J,L). Bone structures resembling duplicated pterygoid bones were fused to the actual pterygoid bones (Fig. 1F), indicating a role for Hand2 in establishing or maintaining NCC identity. Also, in contrast to Hand2BA/BA embryos (Barbosa et al., 2007; Funato et al., 2009; Yanagisawa et al., 2003), gross cleft palate was not observed (Fig. 1F), indicating that Hand2 does not play a direct role in NCC-mediated palatal shelf elevation. In the lower jaw, the mandible was dysmorphic, resembling the mandible seen in Hand2BA/BA;Hand1fl/fl;Wnt1-Cre embryos (Barbosa et al., 2007; Funato et al., 2009). A second bilateral membranous bone ran medial to the mandible (asterisks in Fig. 1H,J), though its identity was not clear. Other changes included aberrantly ossified body and lesser horns of the hyoid bone, with the latter fused, either unilaterally or bilaterally, to duplicated palatine bones (Fig. 1H,J). In addition, both lesser horns also formed articulations with the malformed cartilage anlage of the mallei (Fig. 1H,J; data not shown), whereas the greater horns articulated with the styloid process (data not shown). Finally, only a single midline incisor was observed in most embryos (Fig. 1H,J,L), though two closely abutting lower incisors were also observed in some embryos (data not shown). Because skull defects in E18.5 Hand2fl/−;Wnt1-Cre embryos were identical to those observed in Hand2fl/fl;Wnt1-Cre embryos (data not shown), the remainder of our analysis was performed using Hand2fl/fl;Wnt1-Cre embryos. Taken together, these changes suggest that Hand2 plays crucial roles in both NCC patterning and establishment of NCC identity in the mandibular arch.
Fig. 1.
Analysis of Hand2cko embryo skulls. Alizarin Red (bone) and Alcian Blue (cartilage)-stained E18.5 Hand2fl/fl (left column) and Hand2cko (right column) mouse embryos. (A,B) Lateral view illustrates retrognathia. (C-F) Ventral view shows loss of tympanic rings (ty), absence of cleft palate (white arrows, magnified in insets) and duplicated pterygoid bones (pt′) in Hand2cko embryos (D,F). (G,H) Ventral view of the Hand2cko mandible complex shows duplicated palatine bones (pl′), loss of the angular processes (ap) of the mandible (md) and Meckel's cartilage (mc), and unilateral fusion of the hyoid bone to middle ear cartilage (H). (I,J) Oral view of a Hand2cko mandible shows the presence of aberrant bone medial to the mandible (asterisks in H,J). (K,L) Incisors (in) in Hand2cko embryos are fused. at, ala temporalis; bo, basioccipital; bs, basisphenoid; cdp, condylar process; cp, coronoid process; g, gonial; gh, greater horns of the hyoid; hy, hyoid; hy-b, body of hyoid; i, incus; j, jugal; lh, lesser horns of the hyoid; lo, lamina obturans; mx, maxilla; sq, squamosal.
Histological analysis of skull structures reveals aglossia
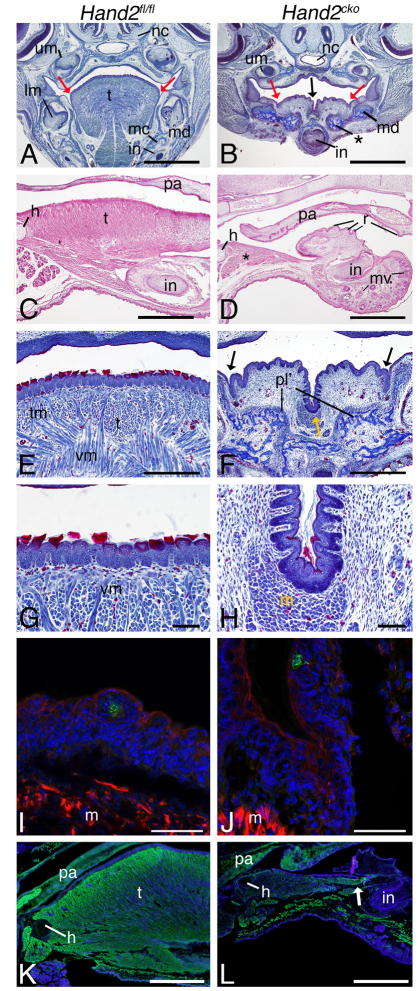
In serial frontal sections through the head of E18.5 control and Hand2cko embryos, aberrant bone was observed running along the medial mandible (Fig. 2B). Only a single incisor was present along the midline, whereas upper and lower molar development appeared normal (Fig. 2B; data not shown). Mystacial vibrissae on the lower jaw epithelium were also observed (Fig. 2D), and rugae, raised epithelial ridges normally confined to the roof of the mouth, were duplicated on the lower oral cavity surface (Fig. 2D). That duplicated vibrissae and rugae were also observed in Edn1, Ednra and Dlx5/Dlx6 mutant embryos (Depew et al., 2002; Ozeki et al., 2004; Ruest et al., 2004) suggest that loss of Hand2 might contribute to these mandibular to maxillary transformations.
Fig. 2.
Histological analysis of Hand2cko embryo skulls. Frontal (A,B,E-J) and sagittal (C,D,K,L) sections from E18.5 Hand2fl/fl (left column) and Hand2cko (right column) mouse embryos. (A,B) Trichrome-stained sections show the mandible (md), aberrant bone (asterisk) and absent tongue (t; black arrow in B). Red arrows distinguish the junction between non-keratinized and keratinized stratified squamous epithelium. (C,D) Hematoxylin and eosin-stained sagittal sections illustrate aglossia, limited muscle (asterisk), rugae (r) in the upper and lower oral ectoderm and mystacial vibrissae (mv) on the lower jaw ectoderm (D). (E-H) Trichrome staining illustrates the non-keratinized to keratinized stratified squamous epithelium transition (black arrows in F) and the presence of keratin (red) in both control (E,G) and mutant (F,H) tongue epithelium. Limited muscle is present in the mutants (yellow arrow in F; m in H). (I,J) Immunofluorescent staining [nuclei, blue (DAPI); F-actin, red (phalloidin); immature taste buds, green (cytokeratin-8)] detects taste buds in the tongue epithelium in both control and mutant embryos. (K,L) Using an anti-myosin antibody (green), muscle is detected in the anterior oral cavity (white arrow). Counterstained with DAPI. h, hyoid; in, incisor; mc, Meckel's cartilage; nc, nasal capsule; pa, palate; pl, palatine bones; tm, transversus muscle; vm, verticalis muscle; lm, lower molars; um, upper molars.
The most striking finding in Hand2cko embryos was the absence of a tongue (Fig. 2B,D,F,H,L). In frontal and sagittal sections, a cleft existed where the tongue should normally reside, though a small amount of muscle was present at the cleft base (asterisk in Fig. 2D; m in Fig. 2H,J). Although the tongue was absent, the tongue epithelium was present. In both control and Hand2cko embryos, the delineation between the non-keratinized and keratinized stratified squamous epithelium of tongue was readily apparent (red arrows in Fig. 2A,B; black arrows in Fig. 2F). Squamous cells overlying the keratinized epithelium were also present in control and Hand2cko embryos, though fewer were observed in Hand2cko embryos (Fig. 2E-H). We also identified taste buds in both control (Fig. 2I) and Hand2cko (Fig. 2J) embryos using an antibody against cytokeratin-8 (CK8, Troma-1; Krt8 – Mouse Genome Informatics), a taste bud marker (Thirumangalathu et al., 2009). Taken together, our results indicate that although a failure of NCC patterning, proliferation or differentiation leads to a near absence of tongue mesenchyme, partial tongue epithelial development occurs.
Tongue morphogenesis requires the interaction of NCC-derived cells and myoblasts (Hosokawa et al., 2010). Aglossia could, therefore, result from failed somitic myoblast migration to the future tongue domain. However, the muscle below the tongue of Hand2cko embryos was stained by phalloidin (an F-actin marker) (Fig. 2J). Similarly, anti-myosin staining was observed in the anterior lower jaw of Hand2cko embryos (white arrow in Fig. 2L), though it was far more scattered than that observed in control embryos (Fig. 2K). Thus, although we cannot rule out a partial myoblast migration defect, the aglossia does not appear to be due to a complete absence of myoblasts.
Tongue mesenchyme morphogenesis is never initiated
We next examined gross oral morphology in control and Hand2cko embryos between E12.5 and E13.5. As a marker, we used sonic hedgehog (Shh) expression, which is expressed by the incisor buds, palatal rugae and developing taste papilla of the tongue. At E12.5, Shh expression in the upper jaw of control and Hand2cko embryos was observed in the mystacial vibrissae, palatal rugae and incisor fields (Fig. 3A,B). In the lower jaw of control embryos, Shh expression was observed in the incisor domains and fungiform papilla on the tongue (Fig. 3C,E). By contrast, although Shh expression in E12.5 Hand2cko embryos marked the incisor fields, expression was also observed in the anterior soft tissue and aberrant mystacial vibrissae (Fig. 3D,F), but was absent in the posterior soft tissue, consistent with absence of tongue development (asterisk in Fig. 3F). By E13.5, Shh expression in control embryos was still observed in the fungiform papilla (Fig. 3G). In E13.5 Hand2cko embryos, Shh expression in the lower jaw had resolved into lateral lines that resembled developing palatal rugae (black arrows in Fig. 3H). Although a midline cleft was still present, it met two other clefts anteriorly that together formed the appearance of primary and secondary palates (see lines in Fig. 3H′). The primary and secondary palates are complex structures; not all aspects of these structures were duplicated in the lower jaw of Hand2cko embryos. However, it does appear that loss of Hand2 disrupts tongue morphogenesis and leads to partial transformation of NCC fate from a mandibular to maxillary identity.
Fig. 3.

Tongue development of Hand2cko embryos. Shh expression in Hand2fl/fl (left column) and Hand2cko (right column) mouse embryos. (A,B) Oral view of the upper jaw from E12.5 control and Hand2cko embryos shows Shh expression in developing mystacial vibrissae (mv), incisors (in) and rugae (r). (C-F) In E12.5 control embryos, Shh expression is observed in the fungiform papilla of the tongue (t) (C,E). With the tongue absent (asterisk) in Hand2cko embryos, expression is observed diffusely in the anterior oral cavity (F) and in aberrant mystacial vibrissae (mv′) (D). (G-H′) At E13.5, Shh expression in the lower jaw of controls remains in fungiform papilla (arrow in G), whereas expression in the anterior oral cavity in Hand2cko embryos has organized into horizontal stripes resembling rugae of the upper jaw (arrows in H). The aberrant proximal-distal seam has persisted in the lower jaw and joined two other seams in the anterior oral cavity that resemble the fusion point of the primary and secondary palates in the upper jaw (see blue lines in H′). md, mandible.
Changes in proliferation and cell death in Hand2cko embryos
Aglossia could result from defects in NCC or somitic myoblast migration to the pharyngeal arches, or their subsequent proliferation and survival. Because Hand2 expression within the ectomesenchyme is first observed at ~E9.25-9.5 (Clouthier et al., 2000; Thomas et al., 1998), NCC migration defects were not expected (Abe et al., 2007; Chai et al., 2000). To confirm this, we crossed the Hand2 conditional mutant strain into the R26R Cre reporter strain (Soriano, 1999) and found that in E10.5 embryos, the distribution of β-gal-labeled cells was similar in the arches of both Hand2fl/+;R26R;Wnt1-Cre and Hand2cko;R26R;Wnt1-Cre embryos (data not shown). This indicates that NCC migration was unaffected by loss of Hand2.
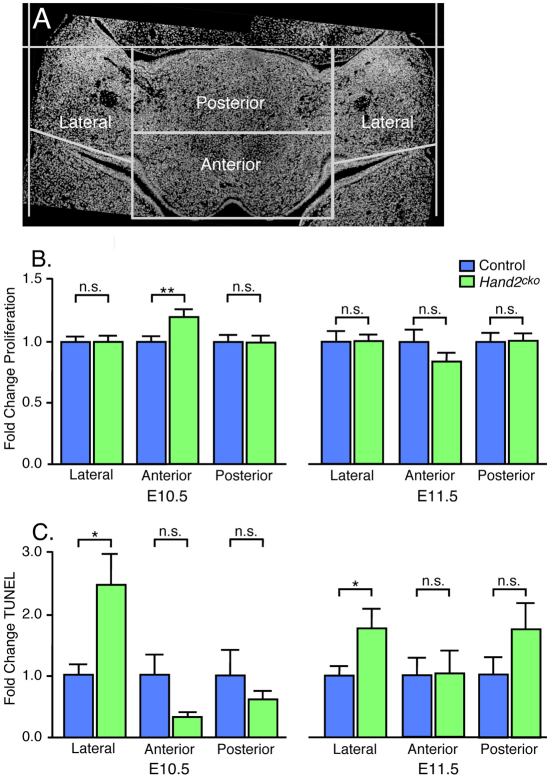
Conventional targeting of Hand2 in mice results in widespread mandibular arch mesenchyme apoptosis by E10.0 (Thomas et al., 1998). Thus, we examined changes in both cellular proliferation and apoptosis in E10.5 and E11.5 control and Hand2cko mandibular arches using a grid pattern that would allow us to determine if and where (lateral, anterior or posterior) changes were occurring (Fig. 4A). Because the tongue arises from the posterior arch, changes in this region could provide a partial mechanism for the observed aglossia. We found that the percentage of proliferating cells was similar between control and Hand2cko embryos at E10.5 in the lateral and posterior arch (Fig. 4B). However, in the anterior arch, EdU incorporation was slightly but significantly elevated. By E11.5, proliferation was statistically similar in control and Hand2cko embryos throughout the mandibular arch (Fig. 4B). Examination of cell death using TUNEL revealed a significant increase in apoptotic nuclei in the lateral mandibular arch of both E10.5 and E11.5 Hand2cko embryos compared with that observed in control embryos (Fig. 4C). Although the incidence of apoptosis trended higher in the posterior arch domain of E11.5 Hand2cko embryos, the change was not statistically significant. These data indicate that decreased proliferation and increased cell death are not major contributors to the observed aglossia.
Fig. 4.
Cell proliferation and death in Hand2cko mouse embryo arches. (A) A representative section showing the grid used for analysis of proliferation and cell death. (B,C) Sections from EdU-treated embryos were analyzed for EdU incorporation (proliferation, B) and TUNEL (apoptosis, C). After counterstaining with DAPI, labeled and total cells were manually counted. The percentage of labeled cells was calculated as the total number of EdU- or TUNEL-positive cells divided by the total number of DAPI positive cells, with this number then used to calculate fold change from control. Error bars represent s.e.m. *P<0.05; **P<0.01; n.s., not significant (P>0.05). All other significance numbers are at least P>0.1 except for the change in anterior arch TUNEL between E10.5 control and Hand2cko embryos (P=0.07).
Gene expression analysis of Hand2cko embryos
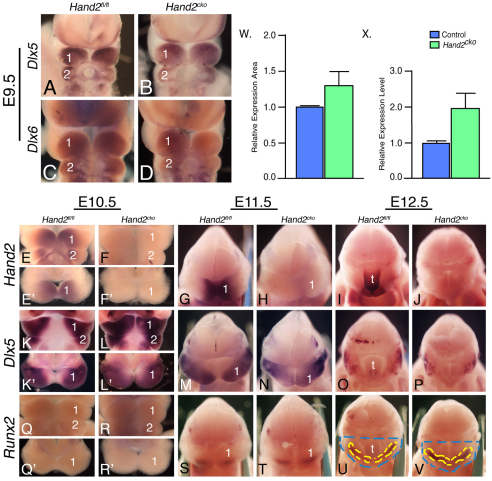
To define the molecular changes that accompany the observed cellular changes, we performed whole-mount ISH analysis on E10.5 control and Hand2cko embryos. As expected, Hand2 expression was absent in the pharyngeal arches of Hand2cko embryos (Fig. 5B). Surprisingly, expression of Hand1 was almost completely downregulated in the mandibular arch (Fig. 5D). This suggests that Hand1 expression in this area is under transcriptional control of another signaling pathway. Gsc, a gene involved in mouse middle ear development (Rivera-Perez et al., 1995; Yamada et al., 1995), is downregulated in the pharyngeal arches of hans6 zebrafish (Miller et al., 2003), a pattern also observed in the pharyngeal arches of Hand2cko embryos (Fig. 5F). By contrast, the zebrafish msx genes msxb and msxe are upregulated in the pharyngeal arches of hans6 embryos (Miller et al., 2003). However, we found that the expression of Msx1 and Msx2 was unchanged in Hand2cko embryos (Fig. 5L,N), as were Pitx1 and Pitx2 (Fig. 5P,R), both involved in lower jaw and incisor development (Gage et al., 1999; Lin et al., 1999; Liu et al., 2003; Mitsiadis and Drouin, 2008). Expression of Twist1 was also examined, as it can interact with Hand2 to influence limb development (Firulli et al., 2005; McFadden et al., 2002) and shows an expanded expression domain that parallels the decreased Hand2 expression domain in Ednra−/− embryos (Ruest et al., 2004). However, Twist1 expression in Hand2cko embryos did not appear grossly different (Fig. 5T). Expression of the distal-less gene family members Dlx2 and Dlx3 was also not noticeably changed (Fig. 5H,J). Finally, FoxF1 expression was examined, as it appears to be involved in distal lower jaw morphogenesis downstream of Shh (Jeong et al., 2004). However, FoxF1 expression was unchanged in Hand2cko embryos (Fig. 5V).
Fig. 5.
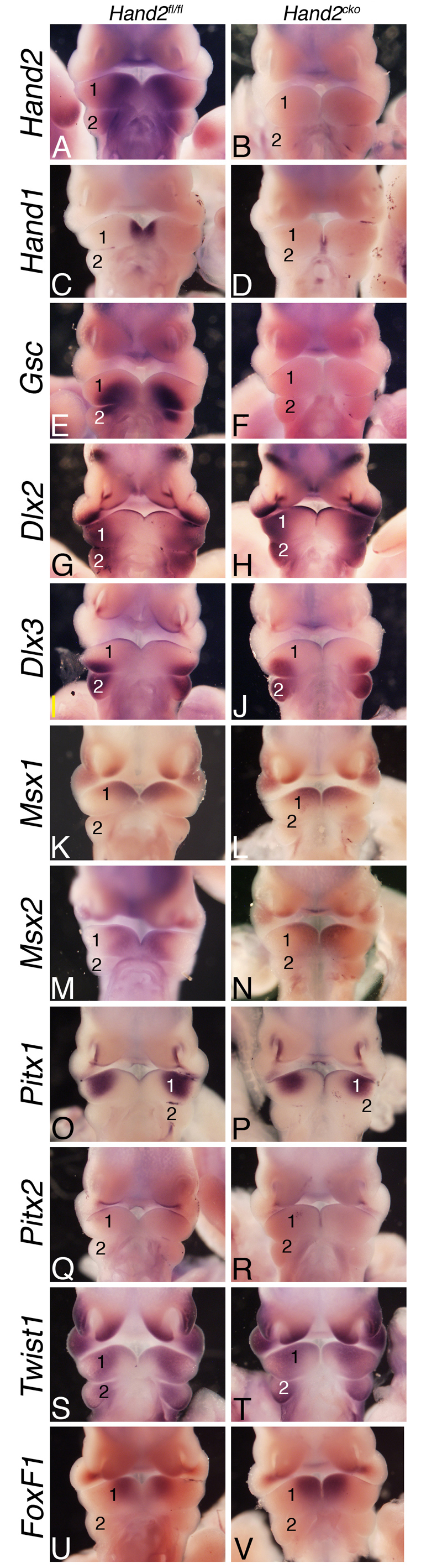
Gene expression analysis in Hand2cko embryos. (A-V) In situ hybridization analysis of the indicated genes in E10.5 Hand2fl/fl (left column) and Hand2cko (right column) mouse embryos. 1, mandibular pharyngeal arch; 2, second pharyngeal arch.
Loss of Hand2 leads to aberrant maintenance of both Dlx5 and Dlx6 expression
We also examined the expression of Dlx5 and Dlx6, two genes that induce arch expression of Hand2 (Charité et al., 2001; Depew et al., 1999; Depew et al., 2002). In E9.5 Hand2cko embryos, Hand2 expression was absent (data not shown), whereas Dlx5 and Dlx6 were both expressed throughout the mandibular arch of control and Hand2cko embryos (Fig. 6A-D). By E10.5, Hand2 expression in control embryos was observed in the disto-oral mandibular arch mesenchyme (Fig. 6E,E′), but was absent in Hand2cko embryos (Fig. 6F,F′). Dlx5 expression in E10.5 control embryos was downregulated in the distal and oral aspects of the mandibular arch but remained in the proximal arch (Fig. 6K,K′). By contrast, Dlx5 expression was present throughout the mandibular arch of E10.5 Hand2cko embryos (Fig. 6L,L′). By E11.5, Hand2 message was present on the disto-oral surface of the mandibular arch in control embryos corresponding to the lateral lingual swelling (Fig. 6G). As observed in E10.5 control embryos, this coincided with a repression of Dlx5 expression in the lateral lingual swelling (Fig. 6M). In E11.5 Hand2cko embryos, Hand2 expression was absent (Fig. 6H), with Dlx5 expression continuing in the disto-oral region (Fig. 6N). By E12.5, Hand2 expression in control embryos was confined to the tongue and distal mandibular arch (Fig. 6I), whereas Dlx5 expression was confined to the tongue and proximal jaw (Fig. 6O). In E12.5 Hand2cko embryos, Hand2 expression was absent (Fig. 6J) whereas Dlx5, owing to the absence of the tongue, was only observed in the proximal jaw (Fig. 6P). The expression pattern of Dlx6 in control and Hand2cko embryos was identical to that of Dlx5 (data not shown). These results suggest that one function of Hand2 is to restrict the expression domains of Dlx5 and Dlx6 from the distal mandibular arch.
Fig. 6.
Hand2, Dlx5 and Runx2 expression analysis in Hand2cko embryo arches. (A-V) In situ hybridization analysis of Dlx5 (A,B,K-P), Dlx6 (C,D), Hand2 (E-J) and Runx2 (Q-V) expression in E9.5-12.5 Hand2fl/fl (left column of each time point) and Hand2cko (right column of each time point) mouse embryos. (A-D) Ventral views of Dlx5 (A,B) and Dlx6 (C,D) expression in E9.5 embryos. (E-F′) Ventral (E,F) and oral (E′,F′) views of Hand2 expression. (G-J) An oral view of Hand2 expression in the mandibular arch at both E11.5 (G,H) and E12.5 (I,J). (K-L′) Ventral (K,L) and oral (K′,L′) views of Dlx5 expression at E10.5. (M-P) Oral views of Dlx5 expression in the mandibular arch at E11.5 (M,N) and E12.5 (O,P). (Q-R′) Ventral (Q,R) and oral (Q′,R′) views of Runx2 expression at E10.5. (S-V) Oral views of Runx2 expression at E11.5 (S,T) and E12.5 (U,V). The area of the arch (blue dashed lines) and Runx2 expression domains (yellow dashed lines) used for the calculation of Runx2 expansion in Fig. 6W are denoted. (W) Quantification of Runx2 expression area in E12.5 control and Hand2cko embryos. Values are expressed relative to control values. (X) Analysis of Runx2 expression in E12.5 mandibular arches. Levels were normalized against β-actin and expressed as fold change versus control levels. Error bars represent s.e.m.
Expression of Dlx5 and Dlx6 is an early event in osteogenesis and acts in part to induce Runx2 expression (Holleville et al., 2007), which is required for normal osteogenesis (Ducy et al., 1997; Komori et al., 1997; Otto et al., 1997). As Hand2 can bind Runx2 in vitro (Funato et al., 2009), we examined whether aberrant Dlx5 and Dlx6 expression in Hand2cko embryos affected Runx2 expression. At E10.5, Runx2 expression was not detectable by ISH in control (Fig. 6Q,Q′) or Hand2cko (Fig. 6R,R′) embryos. Likewise, Runx2 expression was similarly present in the mandibular arches of E11.5 control and Hand2cko embryos (Fig. 6S,T). However, by E12.5, Runx2 expression in Hand2cko embryos appeared expanded (Fig. 6V). These observations were supported by qRT-PCR, which showed an almost twofold increase in Runx2 expression (Fig. 6X), and by area analysis (Fig. 6W). These results indicate that Hand2 loss might lead to expanded initiation of an osteogenic pathway in the mandibular arch.
Hand2 represses Dlx5 and Dlx6 expression through the Dlx5/Dlx6 pharyngeal arch-specific enhancer
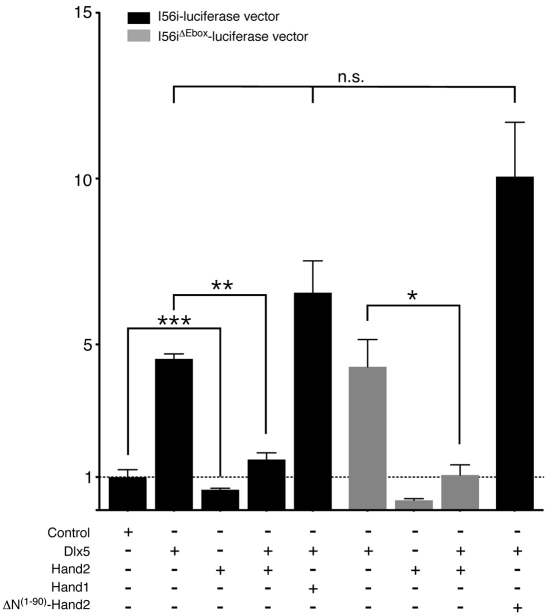
To examine how Hand2 might repress Dlx5 and Dlx6 expression, we transfected MC3T3-E1 cells with a luciferase reporter construct under control of the arch-specific intergenic enhancer from the Dlx5/Dlx6 locus (Zerucha et al., 2000a). This enhancer, referred to as I56i, directs transgene expression to the Dlx5/Dlx6 expression domain within the mandibular arch (Ruest et al., 2003b; Zerucha et al., 2000a). Co-transfection of the I56i-luciferase vector and Dlx5, a known inducer of the I56i enhancer, resulted in an almost fivefold increase in luciferase activity (Fig. 7). By contrast, co-transfection of the I56i-luciferase vector and Hand2 led to a twofold reduction in luciferase activity (Fig. 7). Transfecting the I56i-luciferase vector with both Dlx5 and Hand2 resulted in essentially baseline levels of luciferase activity (Fig. 7), indicating that Hand2 acts as a transcriptional repressor of Dlx5 and Dlx6 expression, with this repression being dominant to the activation effect of Dlx5. Similar results were obtained when Dlx6 was used in place of Dlx5 (data not shown). By contrast, co-transfection of the I56i-luciferase vector with Dlx5 and Hand1 did not block the inductive effect of Dlx5, illustrating that the effect is specific to Hand2.
Fig. 7.
In vitro analysis of Hand2 repression of the Dlx5/6 arch specific enhancer. An I56i-firefly luciferase vector was transfected into MC3T3-E1 cells along with other cDNAs and a renilla reporter construct (efficiency control). All conditions were normalized to corresponding empty vectors of gene expression constructs. Error bars represent s.e.m. Control, representative empty vector; ***P<0.0005; **P<0.005; *P<0.05; n.s., not significant (P>0.1).
As part of a bHLH dimer pair, Hand2 binds to the E-box CANNTG and regulates transcription (Dai and Cserjesi, 2002). We examined the I56i enhancer for E-boxes and found one putative site in the 3′ end of the enhancer. However, when the first nucleotide of this E-box was mutated from a C to an A (a mutation that should inactivate the E-box) (McLellan et al., 2006), the ability of Hand2 to repress both basal enhancer activity and Dlx5-mediated activity was unchanged (Fig. 7), suggesting that Hand2 was not acting through this E-box to regulate I56i activity. As Hand2 has been previously demonstrated to interact with Runx2 in vitro through Hand2's N-terminal domain (Funato et al., 2009), we tested the ability of a mutant version of Hand2 lacking the first 90 amino acids (ΔN(1-90)–Hand2) to inhibit Dlx5-induced enhancer activity. In these experiments, ΔN(1-90)–Hand2 was unable to block Dlx5-induced luciferase activity, suggesting that Hand2's N-terminal domain is crucial to Hand2's ability to repress Dlx5 and Dlx6 expression. Although Dlx5 does not appear to directly associate with Hand2 (data not shown), Hand2 could still act as part of a protein complex. Overall, our data indicates that Hand2 is acting as a transcriptional repressor of Dlx5 and Dlx6 in the distal mandibular arch.
DISCUSSION
Although a ‘Dlx code’ appears to establish the proximal-distal identity of the mandibular arch during craniofacial development (for a review, see Depew and Simpson, 2006), the mechanism by which they act remains only partially elucidated. Here, we have shown that Hand2 temporally limits the action of Dlx5 and Dlx6 during mandibular arch patterning. Loss of this negative-feedback loop leads to aglossia and loss of identity of the mandibular soft tissue. These findings illustrate that Hand2 is likely to be a key mediator of the Dlx code during arch patterning.
Hand2 regulates mandibular arch patterning through negative regulation of Dlx5 and Dlx6
Dlx5 and Dlx6 induce Hand2 expression in the distal half of the mandibular arch (Charité et al., 2001). Here, we have shown that Hand2 then acts in a negative-feedback loop to shut down Dlx5 and Dlx6 expression in the Hand2 domain, acting through the Dlx5 and Dlx6 intergenic arch-specific enhancer (Fig. 8). Although it might be surprising that changes limited to this distal domain can cause such extensive changes in lower jaw development, we have shown through analysis of Hand2 daughter cell fate that the Hand2 domain contributes to the entire lower jaw and much of the middle ear (excluding the incus and stapes) (Ruest et al., 2003a). Further, Bronner-Fraser and colleagues have illustrated in axolotl that the distal half of the mandibular arch gives rise to the lower jaw (mandible) whereas the proximal portion of the mandibular arch gives rise to more proximal hinge structures of the jaw (the palatoquadrate) (Cerny et al., 2004).
Fig. 8.
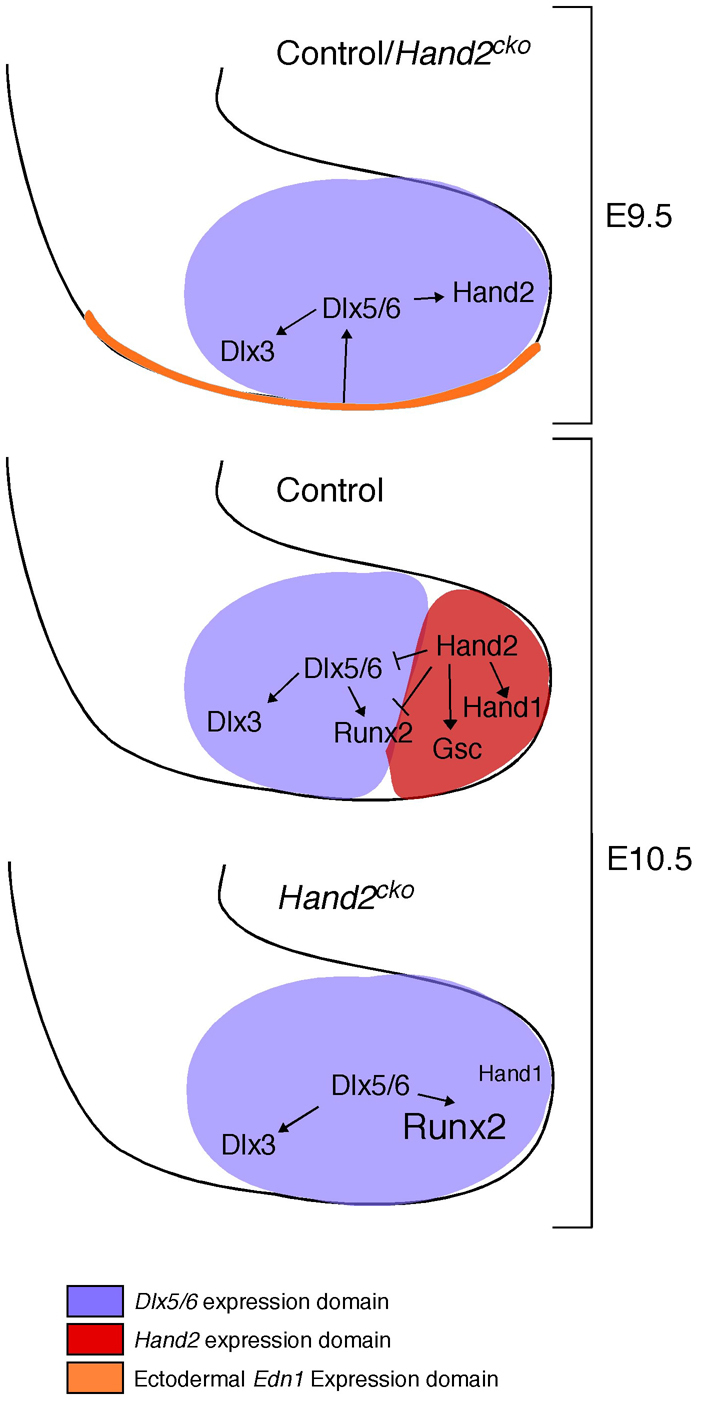
Model of Hand2 activation and regulation of Dlx5 and Dlx6 during mandibular arch development. At E9.5, ectodermal-derived Edn1 activates Ednra signaling in neural crest cells (NCCs), thus inducing Dlx5 and Dlx6 expression broadly within the mandibular arch. Dlx5 and Dlx6 in turn activate Hand2 expression distally and Dlx3 expression proximally. In E10.5 control animals, Dlx5 and Dlx6 expression is restricted from the distal arch by Hand2, which also activates or maintains expression of Hand1 and Gsc. In Hand2cko embryos, absence of Hand2 leads to continued Dlx5 and Dlx6 expression in the distal arch and aberrant expression of Runx2. In addition, Gsc expression is lost and Hand1 expression is greatly reduced.
So why does the maintained presence of Dlx5 and Dlx6 lead to aglossia? Targeted disruption of smoothened (Smo – Mouse Genome Informatics) in the mandibular arch mesenchyme leads to loss of distal arch tissue and subsequent absence of the tongue and most of the lower jaw (Jeong et al., 2004). However, in these mutants, the expression of Fox genes, including FoxF1, was disrupted in the mandibular arch, accompanied by increased Hand2 expression and distal arch cell death. Such changes were not observed in Hand2cko embryos. Based on these findings, it does not appear that aglossia is due to absence of sufficient arch tissue. Rather, it would seem more likely that the distal arch tissue is repatterned owing to the presence of aberrant signals that include Dlx5 and Dlx6, with this disrupting normal developmental interactions with other cell types, including somitic myoblasts. That some tongue muscle is present in near-term embryos probably reflects the complex signaling environment required for proper tongue muscle morphogenesis (Noden and Francis-West, 2006).
The findings from this study support the ‘hinge and caps’ model of jaw development (for a review, see Depew and Simpson, 2006). This model proposes that the polarity and modularity of the upper and lower jaws is driven by the articulation of the first arch prominences (the hinge) and signals located on the distal aspects of the two prominences and from the lamboidal junction (where the maxillary prominence meets the olfactory placode) (the caps). Our findings support this model by illustrating that the loss of a cap signal (Hand2) specifically expressed in the distal mandibular arch mesenchyme disrupts jaw polarity (duplication of maxillary elements in the lower jaw). Further, our findings illustrate the importance of modular signals within the lower jaw, as improper regulation of cap signals (in this case, the failure to downregulate Dlx5 and Dlx6 expression in the distal arch) probably contributes to the observed Hand2cko phenotype in the jaw, including changes in distal jaw shape and fused incisors. It will be important to understand better the communication between Hand2 and the hinge region and to elucidate the molecules through which such communication occurs.
Hand2 function is crucial for osteogenic fates
Mouse embryos hypomorphic for Hand2 have premature ossification of mandibular arch tissue (Barbosa et al., 2007; Funato et al., 2009). Based on both in vitro and in vivo data, Yanagisawa and colleagues proposed that Hand2 could both repress early Runx2 expression and antagonize Runx2 activity via physical association and subsequent sequestration (Funato et al., 2009). Because Runx2 is required for both the maintenance and differentiation of preosteoblastic mesenchyme into osteoblasts and subsequent initiation of osteogenesis (Bialek et al., 2004; Flores et al., 2006; Goldring et al., 2006; Hinoi et al., 2006), loss of either of these functions could lead to premature ossification and thus mandibular hypoplasia. Here, we show a third potential mechanism in which Hand2 functions in a negative-feedback loop to inhibit distal Dlx5 and Dlx6 expression. As Dlx5 can induce Runx2 (Holleville et al., 2007; Miyama et al., 1999), failure to downregulate Dlx5 and Dlx6 expression in the distal arch mesenchyme would probably lead to enhanced Runx2 expression (as shown in the present study) and thus the premature establishment of signaling cascades favorable to bone formation. Enhanced Runx2 expression could also pattern neighboring cells that were competent to respond to Runx2 but do not normally see it and could explain the presence of the aberrant bone in the lower jaw of our Hand2cko embryos. It remains to be seen whether Runx2 binds Hand2 in vivo (Abe et al., 2009), though our model could allow aberrant Runx2 activity without a physical interaction.
Hand gene dosage and bHLH dimer pools
As findings from this and other studies emerge illustrating the importance of Hand family members in developmental processes, one aspect to consider is whether both Hand1 and Hand2 have unique or redundant functions. A neural crest-specific deletion of Hand1 on a hypomorphic Hand2 background leads to jaw defects (Barbosa et al., 2007; Funato et al., 2009), suggesting that Hand proteins act redundantly in far distal arch development (i.e. in the Hand2 domain in which Hand1 is expressed). However, it is not possible to prove this point specifically, as we showed that Hand2 regulates Hand1 expression in the arch. The converse (that loss of Hand1 in the NCC leads to loss of Hand2) is not true (Barbosa et al., 2007). One intriguing question to be addressed is whether the small amount of Hand1 remaining in the distal arch is functionally significant. Answering this will require NCC-specific deletion of both Hand1 and Hand2.
Another aspect to consider is how bHLH dimer pools change in response to loss of specific bHLHs within a cell. Although bHLHs can bind other factors through their 5′ end (Bialek et al., 2004; Funato et al., 2009), they typically form dimer pairs with the ubiquitous class A bHLH molecules (such as E-proteins) or other class B bHLH proteins (Cai and Jabs, 2005; Firulli, 2003). When one dimer partner is removed, the stoichiometry of dimer pools is disrupted, allowing for potential changes in binding partners and subsequent aberrant gene regulation (Cai and Jabs, 2005). Dimer partner choice is known to influence strongly the cellular response to Twist family proteins (Firulli et al., 2003), with changes in Twist1-Hand2 dimerization resulting in Saethre-Chotzen syndrome (Firulli et al., 2005). In addition, conditional disruption of either Hand2 (this study) or Twist1 (Rinon et al., 2007) in NCCs results in defects in craniofacial bone and cartilage structures. To understand fully the role of Hand2 in facial morphogenesis, it will therefore be important to understand how loss of Hand2 affects the function of other Twist family members, including Twist1.
Note added in proof
While this manuscript was in review, Talbot et al. (Talbot et al., 2010) reported that the expression of several Dlx genes expands into the ventral arches of hand2 mutant zebrafish embryos.
Acknowledgements
We thank Nicholas Bennetts and Alicia Navetta for technical help and Hiromi Yanagisawa, Andrew McMahon and James Martin for probes. This work was supported by NIH grants DE018899 (to D.E.C.) and NS040644 and DK067064 (to M.J.H.). Deposited in PMC for release after 12 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
References
- Abe M., Ruest L.-B., Clouthier D. E. (2007). Fate of cranial neural crest cells during craniofacial development in endothelin-A receptor deficient mice. Int. J. Dev. Biol. 51, 97-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe M., Michikami I., Fukushi T., Abe A., Maeda Y., Ooshima T., Wakisaka S. (2009). Hand2 regulates chondrogenesis in vitro and in vivo. Bone 46, 1359-1368 [DOI] [PubMed] [Google Scholar]
- Abramoff M. D., Magelbaes P. J., Ram S. J. (2004). Image Processing with ImageJ. Biophotonics Int. 11, 36-42 [Google Scholar]
- Barbosa A. C., Funato N., Chapman S., McKee M. D., Richardson J. A., Olson E. N., Yanagisawa H. (2007). Hand transcription factors cooperatively regulate development of the distal midline mesenchyme. Dev. Biol. 310, 154-168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beverdam A., Merlo G. R., Paleari L., Mantero S., Genova F., Barbieri O., Janvier P., Levi G. (2002). Jaw transformation with gain of symmetry after Dlx5/Dlx 6 inactivation: mirror of the past. Genesis 34, 221-227 [DOI] [PubMed] [Google Scholar]
- Bialek P., Kern B., Yang X., Schrock M., Sosic D., Hong N., Wu H., Yu K., Ornitz D. M., Olson E. N., et al. (2004). A twist code determines the onset of osteoblast differentiation. Dev. Cell 6, 423-435 [DOI] [PubMed] [Google Scholar]
- Bronner-Fraser M. (1995). Origins and developmental potential of the neural crest. Exp. Cell Res. 218, 405-417 [DOI] [PubMed] [Google Scholar]
- Cai J., Jabs E. W. (2005). A twisted hand: bHLH protein phosphorylation and dimerization regulate limb development. BioEssays 27, 1102-1106 [DOI] [PubMed] [Google Scholar]
- Cerny R., Lwigale P., Ericsson R., Meulemans D., Epperlein H.-H., Bronner-Fraser M. (2004). Developmental origins and evolution of jaws: new interpretations of “maxillary” and “mandibular”. Dev. Biol. 276, 225-236 [DOI] [PubMed] [Google Scholar]
- Chai Y., Maxson J. R. E. (2006). Recent advances in craniofacial morphogenesis. Dev. Dyn. 235, 2353-2375 [DOI] [PubMed] [Google Scholar]
- Chai Y., Jiang X., Ito Y., Bringas P., Jr, Han J., Rowitch D. H., Soriano P., McMahon A. P., Sucov H. M. (2000). Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development 127, 1671-1679 [DOI] [PubMed] [Google Scholar]
- Charité J., McFadden D. G., Merlo G. R., Levi G., Clouthier D. E., Yanagisawa M., Richardson J. A., Olson E. N. (2001). Role of Dlx6 in regulation of an endothelin-1-dependent, dHAND branchial arch enhancer. Genes Dev. 15, 3039-3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouthier D. E., Schilling T. F. (2004). Understanding endothelin-1 function during craniofacial development in the mouse and zebrafish. Birth Defects Res. C 72, 190-199 [DOI] [PubMed] [Google Scholar]
- Clouthier D. E., Comerford S. A., Hammer R. E. (1997). Hepatic fibrosis, glomerulosclerosis, and a lipodystrophy-like syndrome in PEPCK-TGF-β1 transgenic mice. J. Clin. Invest. 100, 2697-2713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouthier D. E., Hosoda K., Richardson J. A., Williams S. C., Yanagisawa H., Kuwaki T., Kumada M., Hammer R. E., Yanagisawa M. (1998). Cranial and cardiac neural crest defects in endothelin-A receptor-deficient mice. Development 125, 813-824 [DOI] [PubMed] [Google Scholar]
- Clouthier D. E., Williams S. C., Yanagisawa H., Wieduwilt M., Richardson J. A., Yanagisawa M. (2000). Signaling pathways crucial for craniofacial development revealed by endothelin-A receptor-deficient mice. Dev. Biol. 217, 10-24 [DOI] [PubMed] [Google Scholar]
- Clouthier D. E., Garcia E., Schilling T. F. (2010). Regulation of facial morphogenesis by endothelin signaling: insights from mouse and fish. Am. J. Med. Genet. A 152, 2962-2973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couly G. F., Grapin-Botton A., Coltey P., Le Douarin N. M. (1996). The regeneration of the cephalic neural crest, a problem revisited: the regenerating cells originate from the contralateral or from the anterior and posterior neural fold. Development 122, 3393-3407 [DOI] [PubMed] [Google Scholar]
- Cserjesi P., Brown D., Lyons G. E., Olson E. N. (1995). Expression of the novel basic helix-loop-helix gene eHAND in neural crest derivatives and extraembryonic membranes during mouse development. Dev. Biol. 170, 664-678 [DOI] [PubMed] [Google Scholar]
- D'Autreaux F., Morikawa Y., Cserjesi P., Gershon M. D. (2007). Hand2 is necessary for terminal differentiation of enteric neurons from crest-derived precursors but not for their migration into the gut or for formation of glia. Development 134, 2237-2249 [DOI] [PubMed] [Google Scholar]
- Dai Y. S., Cserjesi P. (2002). The basic helix-loop-helix factor, HAND2, functions as a transcriptional activator by binding to E-boxes as a heterodimer. J. Biol. Chem. 277, 12604-12612 [DOI] [PubMed] [Google Scholar]
- Danielian P. S., Muccino D., Rowitch D. H., Michael S. K., McMahon A. P. (1998). Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr. Biol. 8, 1323-1326 [DOI] [PubMed] [Google Scholar]
- Depew M. J., Simpson C. A. (2006). 21st century neonatology and the comparative development of the vertebrate skull. Dev. Dyn. 235, 1256-1291 [DOI] [PubMed] [Google Scholar]
- Depew M. J., Liu J. K., Long J. E., Presley R., Meneses J. J., Pedersen R. A., Rubenstein J. L. (1999). Dlx5 regulates regional development of the branchial arches and sensory capsules. Development 126, 3831-3846 [DOI] [PubMed] [Google Scholar]
- Depew M. J., Lufkin T., Rubenstein J. L. (2002). Specification of jaw subdivisions by Dlx genes. Science 298, 381-385 [DOI] [PubMed] [Google Scholar]
- Depew M. J., Simpson C. A., Morasso M., Rubenstein J. L. R. (2005). Reassessing the Dlx code: the genetic regulation of branchial arch skeletal pattern and development. J. Anat. 207, 501-561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducy P., Zhang R., Geoffroy V., Ridall A. L., Karsenty G. (1997). Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89, 747-754 [DOI] [PubMed] [Google Scholar]
- Firulli A. B. (2003). A HANDful of questions: the molecular biology of the heart and neural crest derivatives (HAND)-subclass of basic helix-loop-helix transcription factors. Gene 312, 27-40 [DOI] [PubMed] [Google Scholar]
- Firulli B. A., Howard M. J., McDaid J. R., McIlreavey L., Dionne K. M., Centonze V. E., Cserjesi P., Virshup D. M., Firulli A. B. (2003). PKA, PKC, and the protein phosphatase 2A influence HAND factor function: a mechanism for tissue-specific transcriptional regulation. Mol. Cell 12, 1225-1237 [DOI] [PubMed] [Google Scholar]
- Firulli B. A., Krawchuk D., Centonze V. E., Vargesson N., Virshup D. M., Conway S. J., Cserjesi P., Laufer E., Firulli A. B. (2005). Altered Twist1 and Hand2 dimerization is associated with Saethre-Chotzen syndrome and limb abnormalities. Nat. Genet. 37, 373-381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores M. V., Lam E. Y., Crosier P., Crosier K. (2006). A hierarchy of Runx transcription factors modulate the onset of chondrogenesis in craniofacial endochondral bones in zebrafish. Dev. Dyn. 235, 3166-3176 [DOI] [PubMed] [Google Scholar]
- Funato N., Chapman S. L., McKee M. D., Funato H., Moriss J. A., Shelton J. M., Richardson J. A., Yanagisawa H. (2009). Hand2 controls osteoblast differentiation in the branchial arch by inhibiting DNA binding of Runx2. Development 136, 615-625 [DOI] [PubMed] [Google Scholar]
- Gage P. J., Suh H., Camper S. A. (1999). Dosage requirement of Pitx2 for development of multiple organs. Development 126, 4643-4651 [DOI] [PubMed] [Google Scholar]
- Goldring M. B., Tsuchimochi K., Ijiri K. (2006). The control of chondrogenesis. J. Cell. Biochem. 97, 33-44 [DOI] [PubMed] [Google Scholar]
- Hall B. K. (1982). Mandibular morphogenesis and craniofacial malformations. J. Craniofac. Genet. Dev. Biol. 2, 309-322 [PubMed] [Google Scholar]
- Hendershot T. J., Liu H., Sarkar A. A., Giovannucci D. R., Clouthier D. E., Abe M., Howard M. J. (2007). Expression of Hand2 is sufficient for neurogenesis and cell type-specific gene expression in the enteric nervous system. Dev. Dyn. 236, 93-105 [DOI] [PubMed] [Google Scholar]
- Hendershot T. J., Liu H., Clouthier D. E., Shepherd I. T., Coppola E., Studer M., Firulli A. B., Pittman D. L., Howard M. J. (2008). Conditional deletion of Hand2 reveals critical functions in neurogenesis and cell type-specific gene expression for development of neural crest-derived noradrenergic sympathetic ganglion neurons. Dev. Biol. 319, 179-191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinoi E., Bialek P., Chen Y.-T., Rached M.-T., Groner Y., Behringer R. R., Ornitz D. M., Karsenty G. (2006). Runx2 inhibits chondrocyte proliferation and hypertrophy through its expression in the perichondrium. Genes Dev. 20, 2937-2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler K. L., Hendershot T. J., Troy S. E., Vincentz J. W., Firulli A. B., Howard M. J. (2010). Targeted deletion of Hand2 in cardiac neural crest-derived cells influences cardiac gene expression and outflow tract development. Dev. Biol. 341, 291-304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holleville N., Mateos S., Bontoux M., Bollerot K., Monsoro-Burq A.-H. (2007). Dlx5 drives Runx2 expression and osteogenic differentiation in developing cranial suture mesenchyme. Dev. Biol. 304, 860-874 [DOI] [PubMed] [Google Scholar]
- Hosokawa R., Oka K., Yamaza T., Iwata J., Urata M., Xu X., Bringas P., Jr, Nonaka K., Chai Y. (2010). TGF-β mediated FGF10 signaling in cranial neural crest cells controls development of myogenic progenitor cells through tissue-tissue interactions during tongue morphogenesis. Dev. Biol. 341, 186-195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J., Mao J., Tenzen T., Kottmann A. H., McMahon A. P. (2004). Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia. Genes Dev. 18, 937-951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J., Li X., McEvilly R. J., Rosenfeld M. G., Lufkin T., Rubenstein J. L. R. (2008). Dlx genes pattern mammalian jaw primordium by regulating both lower jaw-specific and upper jaw-specific genetic programs. Development 135, 2905-2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R. T., Gao Y. H., Inada M., et al. (1997). Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 15, 367-371 [DOI] [PubMed] [Google Scholar]
- Kontges G., Lumsden A. (1996). Rhombencephalic neural crest segmentation is preserved throughout craniofacial ontogeny. Development 122, 3229-3242 [DOI] [PubMed] [Google Scholar]
- Le Douarin N. M. (1982). The Neural Crest. New York: Cambridge University Press; [Google Scholar]
- Lin C. R., Kioussi C., O'Connor S., Briata P., Szeto D., Liu F., Izpisua Belmonte J. C., Rosenfeld M. G. (1999). Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature 401, 279-282 [DOI] [PubMed] [Google Scholar]
- Liu W., Selever J., Lu M.-F., Martin J. F. (2003). Genetic dissection of Pitx2 in craniofacial development uncovers new functions in branchial arch morphogenesis, late aspects of tooth morphogenesis and cell migration. Development 130, 6375-6385 [DOI] [PubMed] [Google Scholar]
- Maemura K., Kurihara H., Kurihara Y., Oda H., Ishikawa T., Copeland N. G., Gilbert D. J., Jenkins N. A., Yazaki Y. (1996). Sequence analysis, chromosomal location, and developmental expression of the mouse preproendothelin-1 gene. Genomics 31, 177-184 [DOI] [PubMed] [Google Scholar]
- McFadden D. G., McAnally J., Richardson J. A., Charité J., Olson E. N. (2002). Misexpression of dHAND induces ectopic digits in the developing limb bud in the absence of direct DNA binding. Development 129, 3077-3088 [DOI] [PubMed] [Google Scholar]
- McLellan A. S., Kealey T., Langlands K. (2006). An E box in the exon 1 promoter regulates insulin-like growth factor-I expression in differentiating muscle cells. Am. J. Physiol. Cell Physiol. 291, 300-307 [DOI] [PubMed] [Google Scholar]
- Miller C. T., Schilling T. F., Lee K.-H., Parker J., Kimmel C. B. (2000). sucker encodes a zebrafish Endothelin-1 required for ventral pharyngeal arch development. Development 127, 3815-3838 [DOI] [PubMed] [Google Scholar]
- Miller C. T., Yelon D., Stainier D. Y., Kimmel C. B. (2003). Two endothelin 1 effectors, hand2 and bapx1, pattern ventral pharyngeal cartilage and the jaw joint. Development 130, 1353-1365 [DOI] [PubMed] [Google Scholar]
- Mitsiadis T. A., Drouin J. (2008). Deletion of the Pitx1 genomic locus affects mandibular tooth morphogenesis and expression of the Barx1 and Tbx1 genes. Dev. Biol. 313, 887-896 [DOI] [PubMed] [Google Scholar]
- Miyama K., Yamada G., Yamamoto T. S., Takagi C., Miyado K., Sakai M. (1999). A BMP-inducible gene, Dlx5, regulates osteoblast differentiation and mesoderm induction. Dev. Biol. 208, 123-133 [DOI] [PubMed] [Google Scholar]
- Morikawa Y., D'Autreaux F., Gershon M. D., Cserjesi P. (2007). Hand2 determines the noradrenergic phenotype in the mouse sympathetic nervous system. Dev. Biol. 307, 114-126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noden D. M., Francis-West P. (2006). Differentiation and morphogenesis of craniofacial muscles. Dev. Dyn. 235, 1194-1218 [DOI] [PubMed] [Google Scholar]
- Otto F., Thornell A. P., Crompton T., Denzel A., Gilmour K. C., Rosewell I. R., Stamp G. W., Beddington R. S., Mundlos S., Olsen B. R., et al. (1997). Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89, 765-771 [DOI] [PubMed] [Google Scholar]
- Ozeki H., Kurihara Y., Tonami K., Watatani S., Kurihara H. (2004). Endothelin-1 regulates the dorsoventral branchial arch patterning in mice. Mech. Dev. 121, 387-395 [DOI] [PubMed] [Google Scholar]
- Rinon A., Lazar S., Marshall H., Buchmann-Meller S., Neufeld A., Elhanany-Tamir H., Taketo M. M., Sommer L., Krumlauf R., Tzahor E. (2007). Cranial neural crest cells regulate head muscle patterning and differentiation during vertebrate embryogenesis. Development 134, 3065-3075 [DOI] [PubMed] [Google Scholar]
- Rivera-Perez J. A., Mallo M., Gendron-Maguire M., Gridley T., Behringer R. R. (1995). goosecoid is not an essential component of the mouse gastrula organizer but is required for craniofacial and rib development. Development 121, 3005-3012 [DOI] [PubMed] [Google Scholar]
- Ruest L. B., Clouthier D. E. (2009). Elucidating timing and function of endothelin-A receptor signaling during craniofacial development using neural crest cell-specific gene deletion and receptor antagonism. Dev. Biol. 328, 94-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruest L.-B., Dager M., Yanagisawa H., Charité J., Hammer R. E., Olson E. N., Yanagisawa M., Clouthier D. E. (2003a). dHAND-Cre transgenic mice reveal specific potential functions of dHAND during craniofacial development. Dev. Biol. 257, 263-277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruest L.-B., Hammer R. E., Yanagisawa M., Clouthier D. E. (2003b). Dlx5/6-enhancer directed expression of Cre recombinase in the pharyngeal arches and brain. Genesis 37, 188-194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruest L. B., Xiang X., Lim K. C., Levi G., Clouthier D. E. (2004). Endothelin-A receptor-dependent and -independent signaling pathways in establishing mandibular identity. Development 131, 4413-4423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. (1999). Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21, 70-71 [DOI] [PubMed] [Google Scholar]
- Talbot J. C., Johnson S. L., Kimmel C. B. (2010). hand2 and Dlx genes specify dorsal, intermediate and ventral domains within zebrafish pharyngeal arches. Development 137, 2507-2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirumangalathu S., Harlow D. E., Driskell A. L., Krimm R. F., Barlow L. A. (2009). Fate mapping of mammalian embryonic taste bud progenitors. Development 136, 1519-1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T., Kurihara H., Yamagishi H., Kurihara Y., Yazaki Y., Olson E. N., Srivastava D. (1998). A signaling cascade involving endothelin-1, dHAND and Msx1 regulates development of neural-crest-derived branchial arch mesenchyme. Development 125, 3005-3014 [DOI] [PubMed] [Google Scholar]
- Xiong W., He F., Morikawa Y., Yu X., Zhang Z., Lan Y., Jiang R., Cserjesi P., Chen Y. (2009). Hand2 is required in the epithelium for palatogenesis in mice. Dev. Biol. 330, 131-141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada G., Mansouri A., Torres M., Stuart E. T., Blum M., Schultz M., De Robertis E. M., Gruss P. (1995). Targeted mutation of the murine goosecoid gene results in craniofacial defects and neonatal death. Development 121, 2917-2922 [DOI] [PubMed] [Google Scholar]
- Yamagishi H., Olson E. N., Srivastava D. (2000). The basic helix-loop-helix transcription factor, dHAND, is required for vascular development. J. Clin. Invest. 105, 261-270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa H., Yanagisawa M., Kapur R. P., Richardson J. A., Williams S. C., Clouthier D. E., de Wit D., Emoto N., Hammer R. E. (1998). Dual genetic pathways of endothelin-mediated intercellular signaling revealed by targeted disruption of endothelin converting enzyme-1 gene. Development 125, 825-836 [DOI] [PubMed] [Google Scholar]
- Yanagisawa H., Clouthier D. E., Richardson J. A., Charité J., Olson E. N. (2003). Targeted deletion of a branchial arch-specific enhancer reveals a role of dHAND in craniofacial development. Development 130, 1069-1078 [DOI] [PubMed] [Google Scholar]
- Zerucha T., Stuhmer T., Hatch G., Park B. K., Long Q., Yu G., Gambarotta A., Schultz J. R., Rubenstein J. L. R., Ekker M. (2000). A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J. Neurosci. 20, 709-721 [DOI] [PMC free article] [PubMed] [Google Scholar]