Abstract
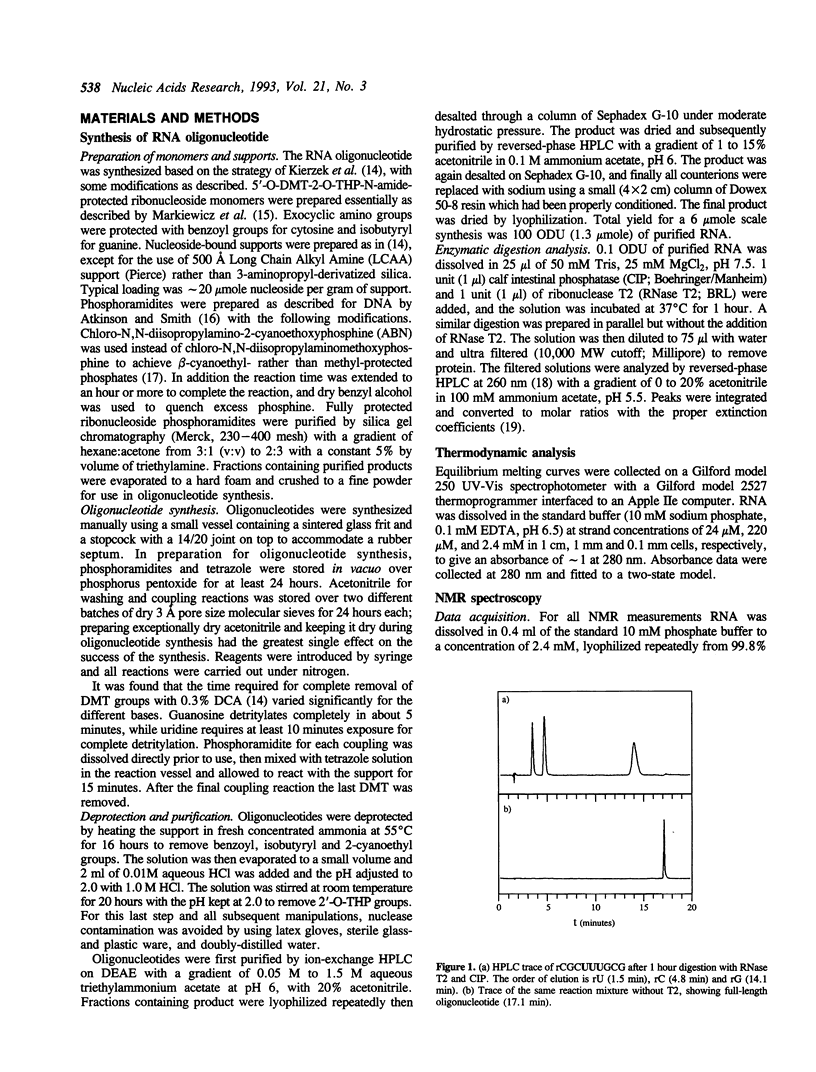
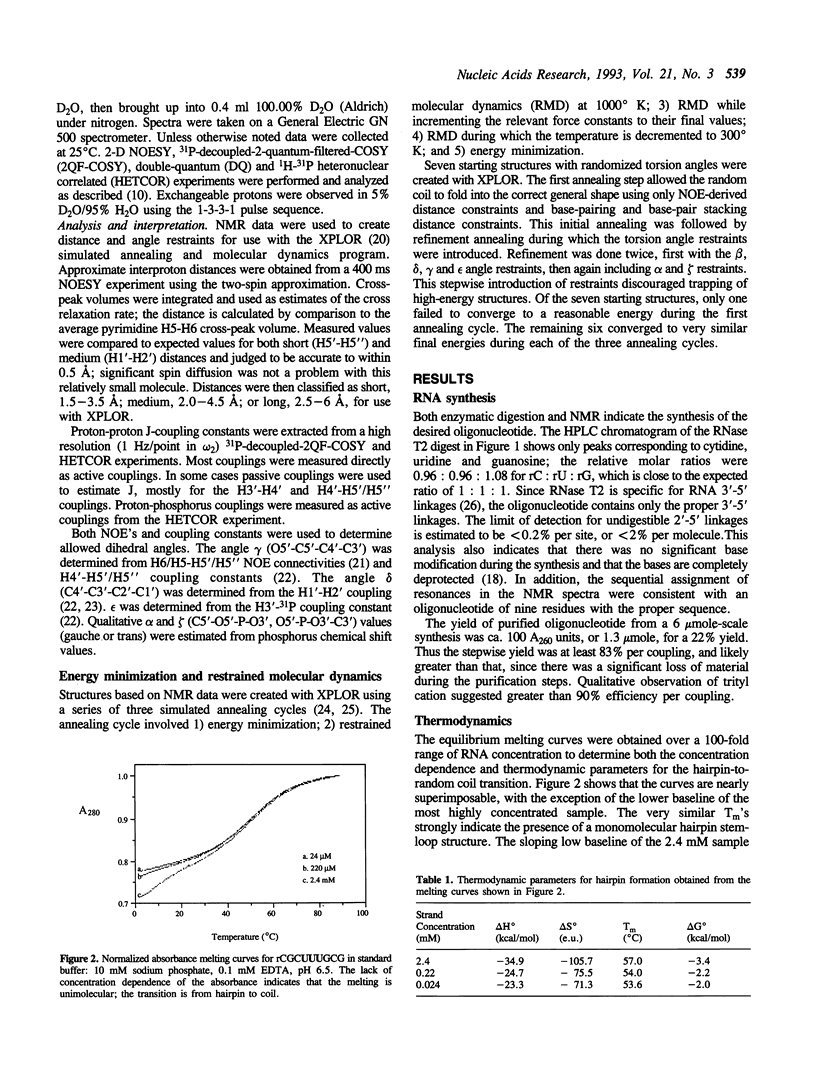
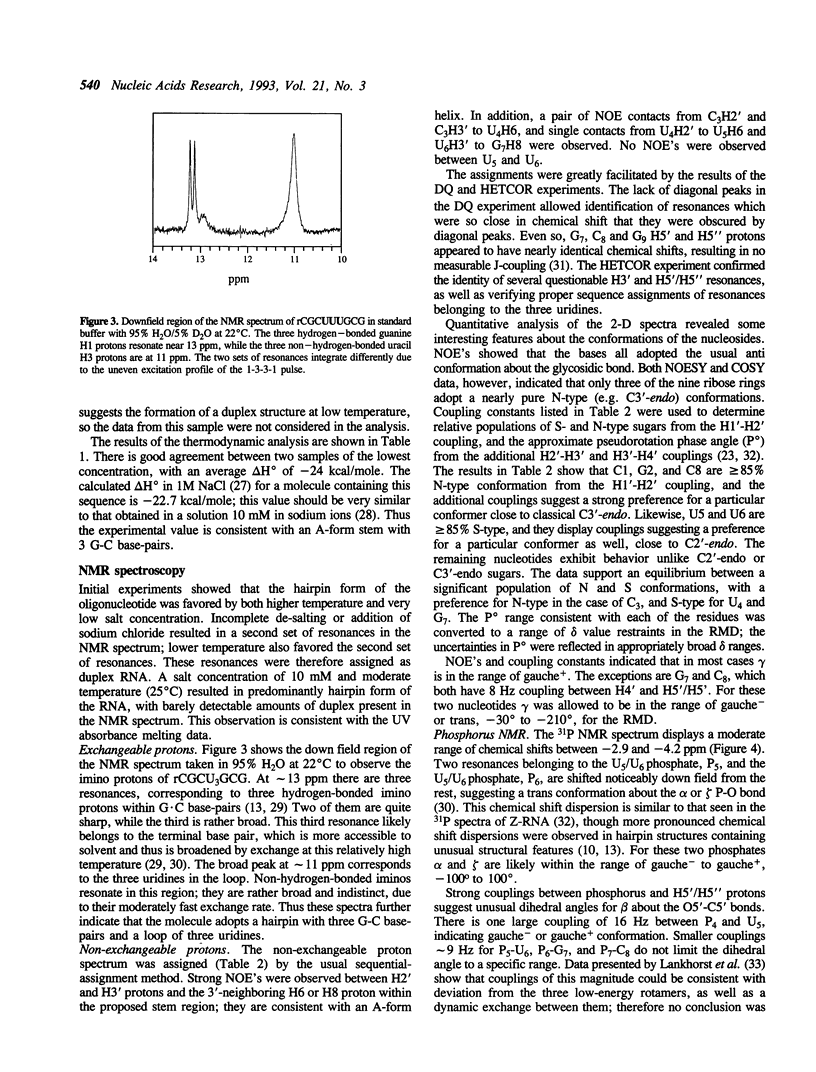
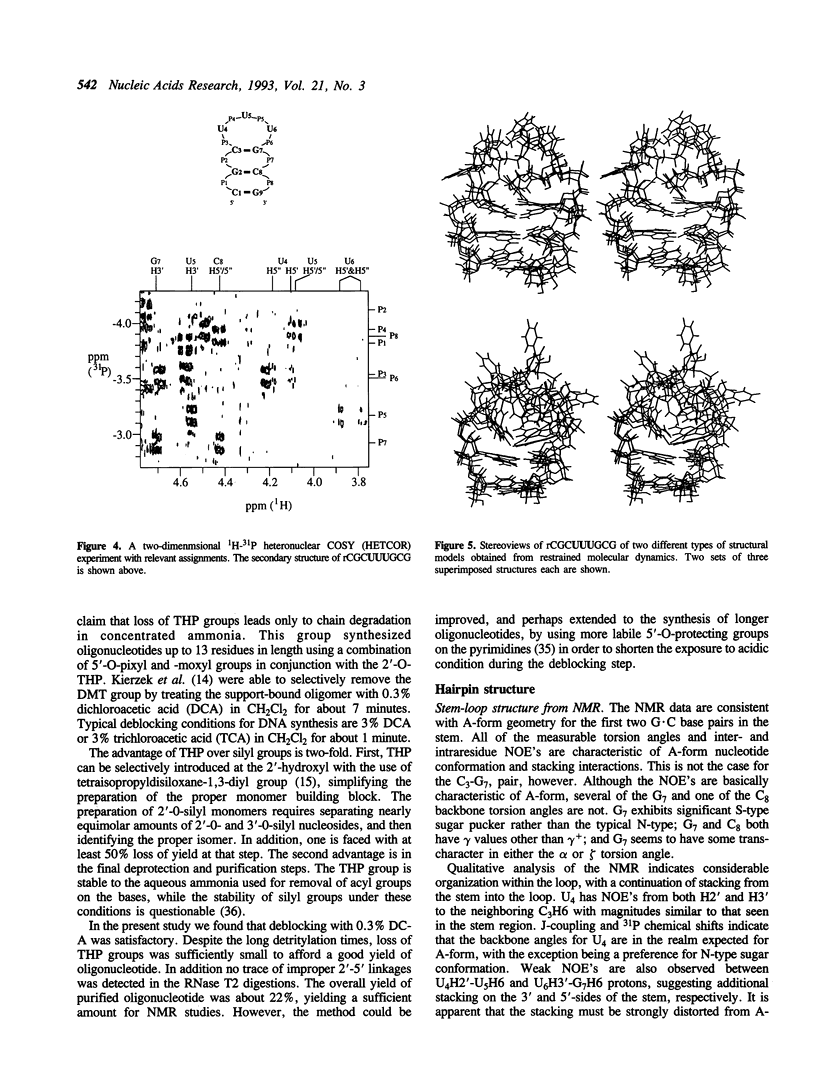
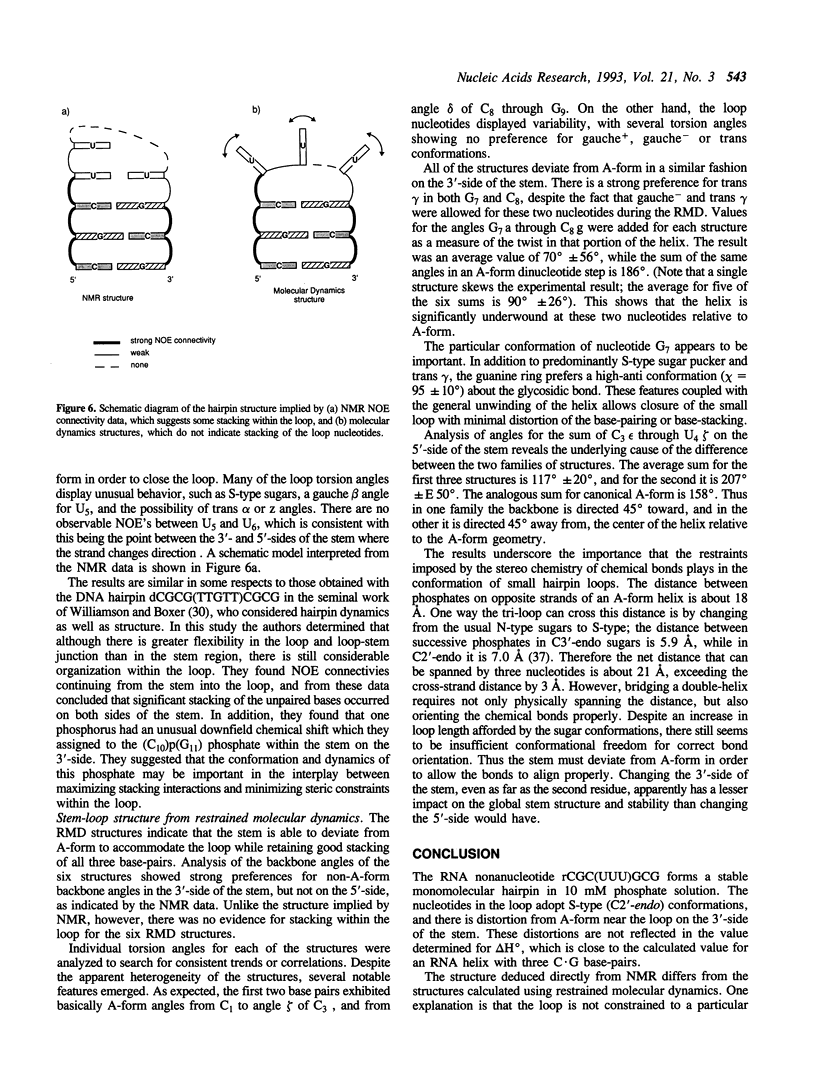
The hairpin stem-loop form of the RNA oligonucleotide rCGC(UUU)GCG has been studied by NMR spectroscopy. In 10 mM phosphate buffer this RNA molecule forms a unimolecular hairpin with a stem of three base pairs and a loop of three uridines, as judged by both NMR and UV absorbance melting behavior. Distance and torsion angle restraints were determined using homonuclear proton-proton and heteronuclear proton-phosphorus 2-D NMR. These values were used in restrained molecular dynamics to determine the structure of the hairpin. The stem has characteristics of A-form geometry, although distortion from A-form occurs in the 3'-side of the stem, presumably to aid in accommodating the small loop. The loop nucleotides adopt C2'-endo conformations. NOE's strongly suggest stacking of the uracils with the stem, especially the first uracil on the 5'-side of the loop. The reversal of the chain direction in the loop seems to occur between U5 and U6. Loop structures produced by molecular dynamics simulations had a wide range of conformations and did not show stacking of the uracils. A flexible loop with significant dynamics is consistent with all the data.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blommers M. J., van de Ven F. J., van der Marel G. A., van Boom J. H., Hilbers C. W. The three-dimensional structure of a DNA hairpin in solution two-dimensional NMR studies and structural analysis of d(ATCCTATTTATAGGAT). Eur J Biochem. 1991 Oct 1;201(1):33–51. doi: 10.1111/j.1432-1033.1991.tb16253.x. [DOI] [PubMed] [Google Scholar]
- Chastain M., Tinoco I., Jr Structural elements in RNA. Prog Nucleic Acid Res Mol Biol. 1991;41:131–177. doi: 10.1016/S0079-6603(08)60008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore G. M., Gronenborn A. M., Piper E. A., McLaughlin L. W., Graeser E., van Boom J. H. The solution structure of a RNA pentadecamer comprising the anticodon loop and stem of yeast tRNAPhe. A 500 MHz 1H-n.m.r. study. Biochem J. 1984 Aug 1;221(3):737–751. doi: 10.1042/bj2210737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis P. W., Adamiak R. W., Tinoco I., Jr Z-RNA: the solution NMR structure of r(CGCGCG). Biopolymers. 1990 Jan;29(1):109–122. doi: 10.1002/bip.360290116. [DOI] [PubMed] [Google Scholar]
- Eadie J. S., McBride L. J., Efcavitch J. W., Hoff L. B., Cathcart R. High-performance liquid chromatographic analysis of oligodeoxyribonucleotide base composition. Anal Biochem. 1987 Sep;165(2):442–447. doi: 10.1016/0003-2697(87)90294-6. [DOI] [PubMed] [Google Scholar]
- Feng S., Holland E. C. HIV-1 tat trans-activation requires the loop sequence within tar. Nature. 1988 Jul 14;334(6178):165–167. doi: 10.1038/334165a0. [DOI] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groebe D. R., Uhlenbeck O. C. Characterization of RNA hairpin loop stability. Nucleic Acids Res. 1988 Dec 23;16(24):11725–11735. doi: 10.1093/nar/16.24.11725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot C. A., Hilbers C. W., van der Marel G. A., van Boom J. H., Singh U. C., Pattabiraman N., Kollman P. A. On loop folding in nucleic acid hairpin-type structures. J Biomol Struct Dyn. 1986 Apr;3(5):843–857. doi: 10.1080/07391102.1986.10508468. [DOI] [PubMed] [Google Scholar]
- Haasnoot C. A., Westerink H. P., van der Marel G. A., van Boom J. H. Discrimination between A-type and B-type conformations of double helical nucleic acid fragments in solution by means of two-dimensional nuclear Overhauser experiments. J Biomol Struct Dyn. 1984 Oct;2(2):345–360. doi: 10.1080/07391102.1984.10507572. [DOI] [PubMed] [Google Scholar]
- Haasnoot C. A., de Bruin S. H., Berendsen R. G., Janssen H. G., Binnendijk T. J., Hilbers C. W., van der Marel G. A., van Boom J. H. Structure, kinetics and thermodynamics of DNA hairpin fragments in solution. J Biomol Struct Dyn. 1983 Oct;1(1):115–129. doi: 10.1080/07391102.1983.10507429. [DOI] [PubMed] [Google Scholar]
- Happ C. S., Happ E., Nilges M., Gronenborn A. M., Clore G. M. Refinement of the solution structure of the ribonucleotide 5'r(GCAUGC)2: combined use of nuclear magnetic resonance and restrained molecular dynamics. Biochemistry. 1988 Mar 8;27(5):1735–1743. doi: 10.1021/bi00405a053. [DOI] [PubMed] [Google Scholar]
- Heus H. A., Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991 Jul 12;253(5016):191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- Kierzek R., Caruthers M. H., Longfellow C. E., Swinton D., Turner D. H., Freier S. M. Polymer-supported RNA synthesis and its application to test the nearest-neighbor model for duplex stability. Biochemistry. 1986 Dec 2;25(24):7840–7846. doi: 10.1021/bi00372a009. [DOI] [PubMed] [Google Scholar]
- Kraulis J., Clore G. M., Nilges M., Jones T. A., Pettersson G., Knowles J., Gronenborn A. M. Determination of the three-dimensional solution structure of the C-terminal domain of cellobiohydrolase I from Trichoderma reesei. A study using nuclear magnetic resonance and hybrid distance geometry-dynamical simulated annealing. Biochemistry. 1989 Sep 5;28(18):7241–7257. doi: 10.1021/bi00444a016. [DOI] [PubMed] [Google Scholar]
- Lankhorst P. P., Haasnoot C. A., Erkelens C., Altona C. Carbon-13 NMR in conformational analysis of nucleic acid fragments. 2. A reparametrization of the Karplus equation for vicinal NMR coupling constants in CCOP and HCOP fragments. J Biomol Struct Dyn. 1984 Jun;1(6):1387–1405. doi: 10.1080/07391102.1984.10507527. [DOI] [PubMed] [Google Scholar]
- Puglisi J. D., Tinoco I., Jr Absorbance melting curves of RNA. Methods Enzymol. 1989;180:304–325. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- Puglisi J. D., Wyatt J. R., Tinoco I., Jr Solution conformation of an RNA hairpin loop. Biochemistry. 1990 May 1;29(17):4215–4226. doi: 10.1021/bi00469a026. [DOI] [PubMed] [Google Scholar]
- Romaniuk P. J., Lowary P., Wu H. N., Stormo G., Uhlenbeck O. C. RNA binding site of R17 coat protein. Biochemistry. 1987 Mar 24;26(6):1563–1568. doi: 10.1021/bi00380a011. [DOI] [PubMed] [Google Scholar]
- Scaringe S. A., Francklyn C., Usman N. Chemical synthesis of biologically active oligoribonucleotides using beta-cyanoethyl protected ribonucleoside phosphoramidites. Nucleic Acids Res. 1990 Sep 25;18(18):5433–5441. doi: 10.1093/nar/18.18.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha N. D., Biernat J., McManus J., Köster H. Polymer support oligonucleotide synthesis XVIII: use of beta-cyanoethyl-N,N-dialkylamino-/N-morpholino phosphoramidite of deoxynucleosides for the synthesis of DNA fragments simplifying deprotection and isolation of the final product. Nucleic Acids Res. 1984 Jun 11;12(11):4539–4557. doi: 10.1093/nar/12.11.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani G., Cheong C., Tinoco I., Jr Structure of an unusually stable RNA hairpin. Biochemistry. 1991 Apr 2;30(13):3280–3289. doi: 10.1021/bi00227a016. [DOI] [PubMed] [Google Scholar]
- Wang Y. Y., Lyttle M. H., Borer P. N. Enzymatic and NMR analysis of oligoribonucleotides synthesized with 2'-tert-butyldimethylsilyl protected cyanoethylphosphoramidite monomers. Nucleic Acids Res. 1990 Jun 11;18(11):3347–3352. doi: 10.1093/nar/18.11.3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson J. R., Boxer S. G. Multinuclear NMR studies of DNA hairpins. 1. Structure and dynamics of d(CGCGTTGTTCGCG). Biochemistry. 1989 Apr 4;28(7):2819–2831. doi: 10.1021/bi00433a012. [DOI] [PubMed] [Google Scholar]
- Williamson J. R., Boxer S. G. Synthesis of a thymidine phosphoramidite labelled with 13C at C6: relaxation studies of the loop region in a 13C labelled DNA hairpin. Nucleic Acids Res. 1988 Feb 25;16(4):1529–1540. doi: 10.1093/nar/16.4.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witherell G. W., Uhlenbeck O. C. Specific RNA binding by Q beta coat protein. Biochemistry. 1989 Jan 10;28(1):71–76. doi: 10.1021/bi00427a011. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Winker S., Gutell R. R. Architecture of ribosomal RNA: constraints on the sequence of "tetra-loops". Proc Natl Acad Sci U S A. 1990 Nov;87(21):8467–8471. doi: 10.1073/pnas.87.21.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolters J. The nature of preferred hairpin structures in 16S-like rRNA variable regions. Nucleic Acids Res. 1992 Apr 25;20(8):1843–1850. doi: 10.1093/nar/20.8.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]



