Abstract
The biochemical properties of mouse LSECtin, a glycan-binding receptor that is a member of the C-type lectin family found on sinusoidal endothelial cells, have been investigated. The C-type carbohydrate-recognition domain of mouse LSECtin, expressed in bacteria, has been used in solid-phase binding assays, and a tetramerized form has been used to probe a glycan array. In spite of sequence differences near the glycan-binding sites, the mouse receptor closely mimics the properties of the human receptor, showing high affinity binding to glycans bearing terminal GlcNAcβ1-2Man motifs. Site-directed mutagenesis has been used to confirm that residues near the binding site that differ between the human and the mouse proteins do not affect this binding specificity. Mouse and human LSECtin have been shown to bind Ebola virus glycoprotein with equivalent affinities, and the GlcNAcβ1-2Man disaccharide has been demonstrated to be an effective inhibitor of this interaction. These studies provide a basis for using mouse LSECtin, and knockout mice lacking this receptor, to model the biological properties of the human receptor.
Keywords: C-type lectin, glycan array, glycan-binding protein, LSECtin, viral glycoprotein
Introduction
LSECtin is one of a group of glycan-binding receptors encoded near the end of human chromosome 19 (Liu et al. 2004). Other receptors in the cluster include the dendritic and endothelial cell receptors dendritic cell-intercellular adhesion molecule 1 grabbing nonintegrin (DC-SIGN) and DC-SIGN related receptor (DC-SIGNR) (L-SIGN) and the structurally related Fc-binding receptor CD23 (Soilleux et al. 2000). All of these receptors contain C-type carbohydrate-recognition domains (CRDs). Although a related cluster is found in the syntenic region of chromosome 8 in mouse, there are major differences in the organization of several of the genes, with eight mouse DC-SIGN-related genes replacing the two human genes (Powlesland et al. 2006). These differences make it difficult to identify clear mouse-human orthologs. In contrast, the human and mouse LSECtin genes appear to be much more closely related, although the two sequences differ at key residues that have been proposed to be involved in selective binding of ligands (Powlesland et al. 2008; Tang et al. 2009).
The mRNA for LSECtin is found in a restricted pattern, with the primary sites of expression being sinusoidal endothelial cells in liver and lymph nodes (Liu et al. 2004), although expression has also been reported in suitably stimulated dendritic cells and macrophages in vitro and in the Kupffer cells (Domínguez-Soto et al. 2007; Domínguez-Soto et al. 2009). Unlike many glycan-binding receptors, LSECtin does not appear to function in clathrin-mediated endocytosis (Gramberg et al. 2008; Powlesland et al. 2008), although it can undergo antibody-induced internalization in myeloid cells (Domínguez-Soto et al. 2007). Several biological functions for LSECtin mediated by its ability to interact with specific glycans have been investigated. In vitro studies demonstrate that human LSECtin can provide a route for Ebola virus and SARS coronavirus infection of cells initiated by binding of LSECtin to glycans of the viral envelope glycoproteins (Gramberg et al. 2005; Domínguez-Soto et al. 2007; Powlesland et al. 2008). Studies in mice indicate that mouse LSECtin is able to bind T cells through glycans on the CD44 T-cell surface marker (Tang et al. 2010). This interaction leads to suppression of T-cell responses and knockout mice lacking LSECtin show increased sensitivity to liver damage mediated by T cells following acute injury, suggesting that LSECtin plays a role in protection of the liver from immune attack (Tang et al. 2009). Finally, it has been suggested that receptor expressed on myeloid cells could be involved in antigen capture and presentation (Domínguez-Soto et al. 2007).
Use of the knockout mice (Tang et al. 2009) and other mouse-specific tools to model interactions of the human receptor is based on the assumption that the human and mouse proteins function similarly. From the work with DC-SIGN and its homologues, which provides precedent for rapid evolution of function in this region of the genome, as well as the presence of some potentially important differences in the sequences of human and mouse LSECtin, there is need for empirical evidence comparing the biochemical properties of the two receptors in order to provide a foundation for extrapolating results from mice to humans.
In the studies described in this manuscript, biochemical characterization of mouse LSECtin is presented and close similarity in the sugar-binding properties of the two receptors, and their ability to interact with Ebola virus glycoprotein, is documented. In addition, it is demonstrated that the simple disaccharide GlcNAcβ1-2Man acts as an effective inhibitor of binding to the viral glycoprotein at micromolar concentrations.
Results
Expression and characterization of the CRD from mouse LSECtin
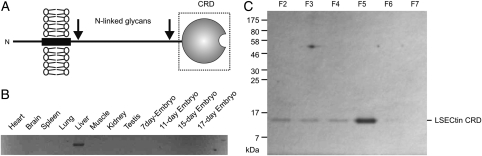
LSECtin is a type 2 transmembrane protein in which the C-type CRD resides at the C-terminal end of the protein (Figure 1A). An initial survey of the distribution of LSECtin expression was undertaken using the polymerase chain reaction to amplify the LSECtin-coding region from a panel of cDNAs with a pair of primers that lie just outside the protein-coding region at each end (Figure 1B). Expression was detected exclusively in the liver, as was previously observed for human LSECtin in a survey of a similar set of human tissues (Liu et al. 2004). This distribution is consistent with the idea that mouse LSECtin performs a function similar to human LSECtin in sinusoidal endothelial cells, although it was not possible to document expression in lymph node as well because of the difficulty of obtaining a suitable cDNA library.
Fig. 1.
Expression of the CRD from mouse LSECtin. (A) Summary of LSECtin domain organization. The portion expressed and used in binding studies is shown in the dotted box. (B) Analysis of tissue expression of LSECtin using the polymerase chain reaction. A library of cDNAs was used as a template for amplification from a pair of primers flanking the 5′ and 3′ ends of the coding region of the mouse LSECtin mRNA. The resulting 1050 bp DNA fragment was resolved by agarose gel electrophoresis. (C) SDS–polyacrylamide gel electrophoresis of the CRD expressed in E. coli and purified on a mannose-Sepharose affinity column. The protein was eluted from the column with EDTA-containing elution buffer in 2 mL fractions. Aliquots (20 µL) of elution fractions F2 through F7 were run on the gel.
The CRD from mouse LSECtin was initially expressed in a bacterial system previously used for production of the human protein (Powlesland et al. 2008), in which the portion of the cDNA encoding the CRD is inserted immediately after the bacterial ompA signal sequence. The signal sequence allows folding of the CRD in the periplasm, so the protein could be extracted and purified directly by affinity chromatography on immobilized sugars (data not shown). However, for the mouse CRD, more efficient expression was obtained by production of the CRD as inclusion bodies that were dissolved in guanidine hydrochloride and renatured by dilution and dialysis (Figure 1C). Similar yields were obtained by purification on either fucose- or mannose-Sepharose. However, due to the relatively weak affinity of individual CRDs for the monosaccharide ligands, a 10-mL column was required to achieve effective purification. The CRD was used directly in the solid-phase binding assays.
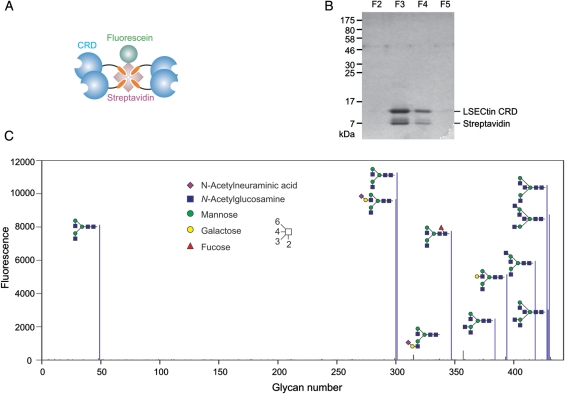
For characterization of the binding specificity of mouse LSECtin by glycan array analysis, a tetrameric form of the CRD was created by attaching a C-terminal biotinylation sequence to the CRD and co-expressing the CRD and biotin ligase (Figure 2A). The resulting protein was purified by affinity chromatography as for the untagged protein. Incubation of an excess of biotin-tagged CRD with fluorescently labeled streptavidin resulted in formation of tetramers that could be purified away from uncomplexed CRDs by re-chromatography on a fucose-Sepharose. The tetrameric protein is efficiently retained on a 1-mL affinity column, whereas uncomplexed CRD washes through. The bound CRD–streptavidin complex was then eluted from the affinity column with ethylenediaminetetraacetic acid (EDTA) (Figure 2B).
Fig. 2.
Glycan array analysis of the binding specificity of LSECtin. (A) Schematic diagram of a tetrameric form of the CRD from mouse LSECtin created by binding of the biotin-tagged CRD to streptavidin. (B) Re-purification of the CRD–streptavidin complex on a fucose-Sepharose column. (C) Results of probing version 4.0 of the glycan array of the Consortium for Functional Glycomics. The tetrameric complex was used at 0.5 µg mL−1. Oligosaccharides that give positive signals are depicted in symbol representation.
Probing of version 4.0 of the glycan array created by the Consortium for Functional Glycomics (Blixt et al. 2004) revealed strong signals for a very selected subset of glycans (Figure 2C). The common feature of all of these glycan is the presence of terminal GlcNAcβ1-2Man disaccharides. Every oligosaccharide that gives a positive signal on the array contains one or more such terminal structures. Only two glycans that contain terminal GlcNAcβ1-2Man yield poor signals on the array, and in each case, these are bi-antennary oligosaccharides in which one branch is extended, which may lead to steric hindrance of binding of the remaining exposed GlcNAcβ1-2Man group.
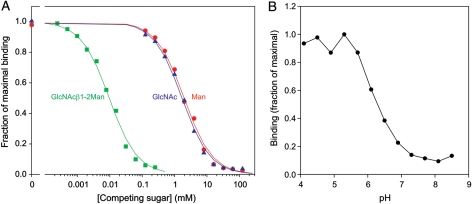
The specificity of mouse LSECtin for the GlcNAcβ1-2Man disaccharide was further demonstrated in competition binding assays (Figure 3A and Table I). In these experiments, the CRD was immobilized on polystyrene wells and probed with 125I-labelled mannose-bovine serum albumin (BSA), a neoglycoprotein ligand with a high density of terminal mannose residues. Based on the measured inhibition constants, the affinity for the disaccharide was ∼1000-fold higher than for any of the monosaccharides tested.
Fig. 3.
Characterization of ligand binding to mouse LSECtin by solid-phase binding assays. CRD immobilized on polystyrene wells was probed with 125I-mannose-BSA. (A) Relative affinities for monosaccharide and disaccharide ligands were determined by competition with the radiolabeled reporter ligand. Results are summarized in Table I. (B) pH dependence of ligand binding was measured at a fixed concentration of the reporter ligand.
Table I.
Affinities of monosaccharide and disaccharide ligands for mouse LSECtin determined by solid-phase binding assays
| Competing sugar | KI (mM) |
|---|---|
| Mannose | 2.2 ± 0.3 |
| GlcNAc | 2.2 ± 0.5 |
| Fucose | 3.1 ± 0.3 |
| Glucose | 5.5 ± 0.6 |
| α-Methyl mannoside | 2.0 ± 0.7 |
| α-Methyl galactoside | >500 |
| GlcNAcβ1-2Man | 0.0102 ± 0.0008 |
CRD immobilized on polystyrene wells was probed with 125I-mannose-BSA in the presence of competing sugars.
Comparison of ligand-binding properties of human and mouse LSECtin
The initial characterization of mouse LSECtin binding to glycan ligands suggested that its ligand-binding specificity is closely analogous to that of the human protein (Powlesland et al. 2008). Testing of the pH sensitivity of ligand binding indicated that, as with human LSECtin, the binding activity of mouse LSECtin is enhanced at low pH (Figure 3B). The combined results thus suggest close similarity in the mechanism of ligand binding to human and mouse LSECtin.
The observed high degree of similarity between the human and mouse proteins was not necessarily predicted based on sequence comparisons. Previous modeling studies led to the hypothesis that an important contribution to high affinity binding of terminal GlcNAcβ1-2Man residues might be interaction of the N-acetyl group with the side chain of residue Trp259 (Powlesland et al. 2008). Surprisingly, this residue was found to be substituted by arginine in the mouse protein (Figure 4A). The similarity of the ligand-binding activities of the human and mouse proteins does not support the hypothesis that interactions of the aromatic side chain are important. The role of this residue was tested directly by creating mutant versions of the human CRD in which Trp259 is substituted with arginine, as found in the mouse protein, or with alanine. Comparison of binding of the wild-type and mutant proteins to the exposed terminal GlcNAcβ1-2Man disaccharides of Ebola virus glycoprotein confirmed that the absence of the tryptophan side chain does not significantly affect the interaction (Figure 4B).
Fig. 4.
Effect of mutating binding-site residues on activity of the CRD from human LSECtin. (A) Sequence alignment of the region of the portions of human and mouse LSECtin believed to form the ligand-binding site. The tryptophan residue at position 259 in the human protein has been suggested to contribute to high affinity binding of the GlcNAcβ1–2Man epitope. (B) The effects of changing Trp259 to arginine, as in the mouse CRD, or to alanine on binding of Ebola glycoprotein-Fc fusion protein to the immobilized CRD.
Human and mouse LSECtin binding to Ebola virus glycoprotein and characterization of simple sugars as inhibitors
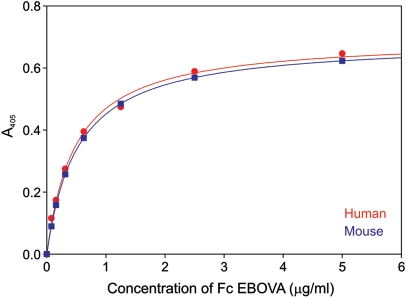
The similarity in the binding specificities of human and mouse LSECtin suggested that, like the human protein, mouse LSECtin might be a target for binding to the surface glycoprotein of Ebola virus. The Ebola glycoprotein has been shown to bear an unusually large number of under-processed glycans with terminal GlcNAcβ1-2Man that are not further modified by addition of galactose (Powlesland et al. 2008). A direct binding assay confirmed that human and mouse LSECtin interact with the Ebola glycoprotein-Fc fusion protein with very similar affinities of 0.52 µg mL−1 for the mouse protein and 0.48 µg mL−1 for the human protein (Figure 5). This result provides important evidence that the mouse protein is a potentially useful model for human Ebola virus interaction with human sinusoidal endothelial cells.
Fig. 5.
Comparison of Ebola virus glycoprotein binding to human and mouse LSECtin. Saturation binding curves were obtained in parallel assays in which immobilized CRD was probed with Ebola glycoprotein-Fc fusion glycoprotein.
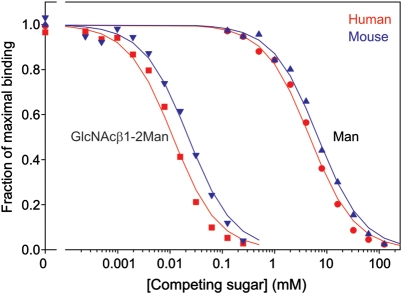
Based on the results in Figure 3, showing that the simple GlcNAcβ1-2Man disaccharide competes at micromolar concentrations for binding of neoglycoprotein ligands to mouse LSECtin, as well as published work showing similar affinities of the human protein (Powlesland et al. 2008), the disaccharide was tested for its ability to compete for binding of both receptors to the Ebola virus glycoprotein. The results demonstrate that the KI of the disaccharide is in the low micromolar range for both human and mouse receptors (Figure 6 and Table II). This result provides an important lead toward the development of high-affinity inhibitors of Ebola virus binding to sinusoidal endothelial cells as well as confirming that studies in the mouse will be relevant to humans as well.
Fig. 6.
Characterization of a low-molecular-weight inhibitor of Ebola virus glycoprotein binding to LSECtin by solid-phase binding competition assay. Monosaccharide and disaccharide ligands were used to compete with 125I-Ebola virus glycoprotein-Fc fusion for binding to the immobilized CRDs from human and mouse LSECtin. Results are summarized in Table II.
Table II.
Inhibition of Ebola virus glycoprotein binding to LSECtin by simple sugars
| Competing sugar | LSECtin | KI (mM) |
|---|---|---|
| Mannose | Human | 5.8 ± 1.5 |
| Mouse | 6.2 ± 0.7 | |
| GlcNAcβ1-2Man | Human | 0.00110 ± 0.0005 |
| Mouse | 0.00166 ± 0.008 |
Competition with 125I-labelled Ebola virus glycoprotein-Fc fusion for binding to immobilized CRDs was measured.
Discussion
The close similarity in the biochemical properties of human and mouse LSECtin is in stark contrast to the widely divergent properties of the mouse orthologs of DC-SIGN. The difference in the evolutionary history of these two protein families, in spite of their proximity in mammalian genomes, indicates that these proteins are subject to different evolutionary pressures. One interpretation of the conserved binding properties of LSECtin is that this receptor may bind a conserved endogenous glycan ligand, whereas DC-SIGN is evolving to interact with different pathogen glycans. This interpretation is consistent with the fact that DC-SIGN functions as an endocytic receptor able to a direct pathogen uptake and presentation to the immune system (Engering et al. 2002; Guo et al. 2004), whereas LSECtin does not have constitutive endocytic activity (Gramberg et al. 2008; Powlesland et al. 2008). In this scenario, the binding of endogenous ligands, such as CD44 on T cells, would be the primary function of LSECtin, which has been hijacked by certain pathogens such as Ebola virus.
Few other glycan-binding receptors show such conserved properties between species as is documented here for LSECtin. DC-SIGN is one of many receptors that show extreme divergence between human and mouse (Powlesland et al. 2006). The CD33 family of siglecs represent another example of independent radiation of a group of glycan-binding receptors (Crocker et al. 2007), and some receptors, such as the Kupffer cell receptor and prolectin, are present in rodents but not humans or vice versa (Fadden et al. 2003; Graham et al. 2009). Aside from the evolutionary and functional implications of the conservation of LSECtin function, the close similarities in their ligand-binding properties, as well as the similar restricted distribution of tissues in which they are expressed, provide a firm foundation for use of mouse LSECtin as a model for the human receptor, which is not always possible in the case of more divergent receptors. The development of low-molecular-weight inhibitors that work equally well on the human and mouse proteins, as well as the description of mice in which LSECtin has been knocked out (Tang et al. 2009), thus provide opportunities for exploring the physiological functions of the human receptor using mouse models.
Materials and methods
Materials
The GlcNAcβ1-2Man disaccharide was purchased from Dextra Laboratories (Reading, UK). Soluble Ebola glycoprotein-Fc fusion protein was produced by transient transfection of 293T cells as described previously (Powlesland et al. 2008). Purified protein was iodinated by the chloramine T method (Greenwood et al. 1963).
Analysis of LSECtin expression
The polymerase chain reaction was carried out using a mouse cDNA panel and Advantage 2 DNA polymerase from ClonTech-Takara Bio Europe (Saint-Germain-en-Laye, France) under the standard reaction conditions described by the manufacturer. Forward and reverse oligonucleotide primers, obtained from Invitrogen (Paisley, UK), were 5-ccagggctggacgccaccaccacc-3′ and 5′-ctggggtcactaaagcatgcactggtcagg-3′. Following an initial denaturation at 95°C for 1 min, 40 cycles of 95°C for 30 s and 68°C for 1 min were executed, and the products were resolved on a 2% agarose gel containing ethidium bromide.
Expression system for CRD from mouse LSECtin
A cDNA for mouse LSECtin was isolated by polymerase chain reaction amplification from a mouse liver cDNA library from BD Biosciences (Oxford, UK). Initially, an expression system using the ompA signal sequence was constructed by direct analogy with the method previously used for the human protein (Powlesland et al. 2008). For more efficient expression, synthetic oligonucleotides were combined with restriction fragments from the cDNA to insert the region encoding the CRD into vector T5T (Eisenberg et al. 1990), with codons for an initiator methionine followed by an alanine codon and then the cDNA sequence beginning at residue 161 of mouse LSECtin, which encodes the sequence SerCysGlu at the N-terminal end of the CRD. For expression of biotin-tagged protein, the C-terminal end of the cDNA was similarly modified by insertion of synthetic oligonucleotides so that the expression vector codes for a protein in which the C-terminal cysteine residue of LSECtin is followed by the sequence GlyLeuAsnAspIlePheGluAlaGlnLysIleGluTrpHisGlu (Schatz 1993). All sequences were confirmed by sequencing on an Applied Biosystems 310 genetic analyzer.
The CRD of LSECtin was expressed in Escherichia coli strain BL21(DE3) grown in the Luria–Bertani broth containing 50 µg mL−1 ampicillin. For addition of biotin, the cells were also transformed with plasmid birA, which encodes the gene for biotin ligase (Chapman-Smith and Cronan 1999), and chloramphenicol at 20 µg mL−1 was included in the medium. For protein production, a 200 mL starter culture was diluted to 6 L and grown at 37°C to OD550 of 0.7, at which point additions were made to achieve final concentrations of 100 µg mL−1 isopropyl-β-d-thiogalactoside and 12.5 µg mL−1 biotin. After further growth for 2.5 h at 37°C, bacteria were harvested by centrifugation for 15 min at 3000 × g, washed in 400 mL of 10 mM Tris–Cl, pH 7.8, and collected by centrifugation for 10 min at 6000 × g. The final pellet was resuspended in 200 mL of 10 mM Tris–Cl, pH 7.8, and sonicated at full power in a Branson Model 250 sonicator for 6 × 30 s, with cooling on ice between sonication steps. The insoluble inclusion bodies were collected by centrifugation for 15 min at 10,000 × g and dissolved in 100 mL of 6 M guanidine hydrochloride containing 100 mM Tris–Cl, pH 7.0, by brief sonication. Following addition of 10 µL of 2-mercaptoethanol, the mixture was stirred for 30 min at 4°C and centrifuged for 30 min at 10,000 × g. The supernatant was dialyzed against three changes of 2 L of loading buffer (150 mM NaCl, 25 mM CaCl2, 25 mM Tris–Cl, pH 7.8), centrifuged for 5 min at 8000 × g and 30 min at 1,00,000 × g, the supernatant was filtered through glass wool and applied to a 10 mL column of mannose- or fucose-Sepharose (Fornstedt and Porath 1975). The column was washed with 16 mL of loading buffer and eluted in 2 mL fractions with elution buffer (150 mM NaCl, 2.5 mM EDTA, 25 mM Tris–Cl, pH 7.8).
Site-directed mutagenesis of human LSECtin
Mutations were introduced into the expression plasmid for the CRD from human LSECtin by the overlap extension method with the polymerase chain reaction (Ho et al. 1989), using flanking forward primer gtcgactctagataacgaggcgc and flanking reverse primer ggtcagcagttgtgccttttctcac and mutagenic primers gggagagcccaatgacgctagggggcgcgagaactgtg and cacagttctcgcgccccctagcgtcattgggctctccc for the W259R mutation and gggagagcccaatgacgctgcggggcgcgagaactgtg and cacagttctcgcgccccgcagcgtcattgggctctccc for the W259A mutation. The wild-type and mutant human proteins were expressed in the folded form in the periplasm of E. coli and were purified as described previously.
Screening of glycan array
Approximately 250 µg of biotin-tagged CRD in 6 mL of eluting buffer was adjusted to 25 mM CaCl2 and incubated overnight at 4°C with 150 µg of Alexa 488-labeled streptavidin (Invitrogen). The complex was re-applied to a 1-mL column of mannose-Sepharose, which was rinsed with 5 mL of loading buffer and eluted with 0.5-mL aliquots of eluting buffer. Version 4.0 of the glycan array of the Consortium for Functional Glycomics was screened in buffer containing 150 mM NaCl, 2 mM CaCl2, 2 mM MgCl2, 20 mM Tris–Cl pH 7.4, 0.05% Tween 20 and 1% BSA following their standard protocol.
Binding assays
For competition binding assays and pH-dependence assays, polystyrene wells coated with CRD from LSECtin at a concentration of 50 µg mL−1 were probed with 125I-labeled mannose-BSA or Ebola glycoprotein-Fc fusion protein (Mitchell et al. 2001). For direct binding assays, biotin-tagged CRD was applied to streptavidin-coated wells that were incubated with unlabeled Ebola glycoprotein-Fc fusion protein at various dilutions followed alkaline phosphatase-protein A conjugate and p-nitrophenylphosphate substrate (Powlesland et al. 2008). Data were fitted to binding equations using the nonlinear least-squares fitting function of SigmaPlot (Systat Software, Hounslow, UK) to determine the half-maximal concentration for binding (KD) or for inhibition of binding (KI).
Other analytical procedures
Sodium dodecylsulfate (SDS)-polyacrylamide gels were performed by the method of Laemmli (1970). Protein concentrations were determined by the method of Bradford (1976).
Funding
The Wellcome Trust (075565 to M.E.T. and K.D), National Institute of General Medical Sciences (GM62116 to the Consortium for Functional Glycomics) and the Center for Infection Biology (to I.S.).
Conflict of interest
None declared.
Abbreviations
CRD, carbohydrate-recognition domain; BSA, bovine serum albumin; EDTA, ethylenediaminetetraacetic acid; SDS, sodium dodecylsulfate.
Acknowledgements
We thank David Smith and Jamie Heimburg-Molinaro of Core H of the Consortium for Functional Glycomics, Emory University, for performing the glycan array analysis.
References
- Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, et al. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci USA. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. doi:10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. doi:10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chapman-Smith A, Cronan JE., Jr. The enzymatic biotinylation of proteins: A post-translational modification of exceptional specificity. Trends Biochem Sci. 1999;24:359–363. doi: 10.1016/s0968-0004(99)01438-3. doi:10.1016/S0968-0004(99)01438-3. [DOI] [PubMed] [Google Scholar]
- Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7:255–266. doi: 10.1038/nri2056. doi:10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- Domínguez-Soto A, Aragoneses-Fenoll L, Gómez-Aguado F, Corcuera MT, Clária J, García-Monzón C, Bustos M, Corbí AL. The pathogen receptor liver and lymph node sinusoidal endothelial cell C-type lectin is expressed in human Kupffer cells and regulated by PU.1. Hepatology. 2009;49:287–296. doi: 10.1002/hep.22678. doi:10.1002/hep.22678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez-Soto Á, Aragoneses-Fenoll L, Martín-Gayo E, Martinez-Prats L, Colmenares M, Naranjo-Gómez M, Borrás FE, Muñoz P, Zubiaur M, Toribio ML, et al. The DC-SIGN-related lectin LSECtin mediates antigen capture and pathogen binding by human myeloid cells. Blood. 2007;109:5337–5345. doi: 10.1182/blood-2006-09-048058. doi:10.1182/blood-2006-09-048058. [DOI] [PubMed] [Google Scholar]
- Eisenberg SP, Evans RJ, Arend WP, Verderber E, Brewer MT, Hannum CH, Thompson RC. Primary structure and functional expression from complementary DNA of a human interleukin-1 receptor antagonist. Nature. 1990;343:341–346. doi: 10.1038/343341a0. doi:10.1038/343341a0. [DOI] [PubMed] [Google Scholar]
- Engering A, Geijtenbeek TBH, van Vliet SJ, Wijers M, van Liempt E, Demaurex N, Lanzacecchia A, Fransen J, Figdor CG, Piguet V, et al. The dendritic cell-specific adhesion receptor DC-SIGN internalizes antigen for presentation to T cells. J Immunol. 2002;168:2118–2126. doi: 10.4049/jimmunol.168.5.2118. [DOI] [PubMed] [Google Scholar]
- Fadden AJ, Holt OJ, Drickamer K. Molecular characterisation of the rat Kupffer cell glycoprotein receptor. Glycobiology. 2003;13:529–537. doi: 10.1093/glycob/cwg068. doi:10.1093/glycob/cwg068. [DOI] [PubMed] [Google Scholar]
- Fornstedt N, Porath J. Characterization studies on a new lectin found in seed of Vicia ervilia. FEBS Lett. 1975;57:187–191. doi: 10.1016/0014-5793(75)80713-7. doi:10.1016/0014-5793(75)80713-7. [DOI] [PubMed] [Google Scholar]
- Graham SA, Jégouzo SAF, Yan S, Powlesland AS, Brady JP, Taylor ME, Drickamer K. Prolectin: A glycan-binding receptor on dividing B cells in germinal centers. J Biol Chem. 2009;284:18537–18544. doi: 10.1074/jbc.M109.012807. doi:10.1074/jbc.M109.012807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramberg T, Hofmann H, Möller P, Lalor PF, Marzi A, Geier M, Krumbiegel M, Winkler T, Kirchhoff F, Adams DH, et al. LSECtin interacts with filovirus glycoproteins and the spike protein of SARS coronavirus. Virology. 2005;340:224–236. doi: 10.1016/j.virol.2005.06.026. doi:10.1016/j.virol.2005.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramberg T, Soilleux E, Fisch T, Lalor PF, Hofmann H, Wheeldon S, Cotterill A, Wegele A, Winkler T, Adams DH, et al. Interactions of LSECtin and DC-SIGN/DC-SIGNR with viral ligands: Differential pH dependence, internalization and virion binding. Virology. 2008;373:189–201. doi: 10.1016/j.virol.2007.11.001. doi:10.1016/j.virol.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood FC, Hunter WM, Glover JS. The preparation of 131I-labelled human growth hormone of high specific radioactivity. Biochem J. 1963;89:114–123. doi: 10.1042/bj0890114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Feinberg H, Conroy E, Mitchell DA, Alvarez R, Blixt O, Taylor ME, Weis WI, Drickamer K. Structural basis for distinct ligand-binding and targeting properties of the receptors DC-SIGN and DC-SIGNR. Nat Struct Mol Biol. 2004;11:591–598. doi: 10.1038/nsmb784. doi:10.1038/nsmb784. [DOI] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:151–159. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. doi:10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Liu W, Tang L, Zhang G, Wei H, Cui Y, Guo L, Gou Z, Chen X, Jiang D, Zhu Y, et al. Characterization of a novel C-type lectin gene, LSECtin: Demonstration of carbohydrate binding and expression in sinusoidal endothelial cells of liver and lymph node. J Biol Chem. 2004;279:18748–18758. doi: 10.1074/jbc.M311227200. doi:10.1074/jbc.M311227200. [DOI] [PubMed] [Google Scholar]
- Mitchell DA, Fadden AJ, Drickamer K. A novel mechanism of carbohydrate recognition by the C-type lectins DC-SIGN and DC-SIGNR: Subunit organisation and binding to multivalent ligands. J Biol Chem. 2001;276:28939–28945. doi: 10.1074/jbc.M104565200. doi:10.1074/jbc.M104565200. [DOI] [PubMed] [Google Scholar]
- Powlesland AS, Fisch T, Taylor ME, Smith DF, Tissot B, Dell A, Pöhlmann S, Drickamer K. A novel mechanism for LSECtin binding to Ebola virus surface glycoprotein through truncated glycans. J Biol Chem. 2008;283:593–602. doi: 10.1074/jbc.M706292200. doi:10.1074/jbc.M706292200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powlesland AS, Ward EM, Sadhu SK, Guo Y, Taylor ME, Drickamer K. Novel mouse homologs of human DC-SIGN: Widely divergent biochemical properties of the complete set of mouse DC-SIGN-related proteins. J Biol Chem. 2006;281:20440–20449. doi: 10.1074/jbc.M601925200. doi:10.1074/jbc.M601925200. [DOI] [PubMed] [Google Scholar]
- Schatz PJ. Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: A 13 residue consensus peptide specifies biotinylation in Escherichia coli. Biotechnology. 1993;11:1138–1143. doi: 10.1038/nbt1093-1138. [DOI] [PubMed] [Google Scholar]
- Soilleux EJ, Barten R, Trowsdale J. Cutting edge: DC-SIGN; a related gene DC-SIGNR; and CD23 form a cluster on 19p13. J Immunol. 2000;165:2937–2942. doi: 10.4049/jimmunol.165.6.2937. [DOI] [PubMed] [Google Scholar]
- Tang L, Yang J, Liu W, Tang X, Chen J, Zhao D, Wang M, Xu F, Lu Y, Liu B, et al. Liver sinusoidal endothelial cell lectin, LSECtin, negatively regulates hepatic T-cell immune response. Gastroenterology. 2009;137:1498–1508. doi: 10.1053/j.gastro.2009.07.051. doi:10.1053/j.gastro.2009.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Yang J, Tang X, Ying W, Qian X, He F. The DC-SIGN family member LSECtin is a novel ligand of CD44 on activated T cells. Eur J Immunol. 2010;40:1185–1191. doi: 10.1002/eji.200939936. doi:10.1002/eji.200939936. [DOI] [PubMed] [Google Scholar]