Abstract
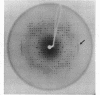
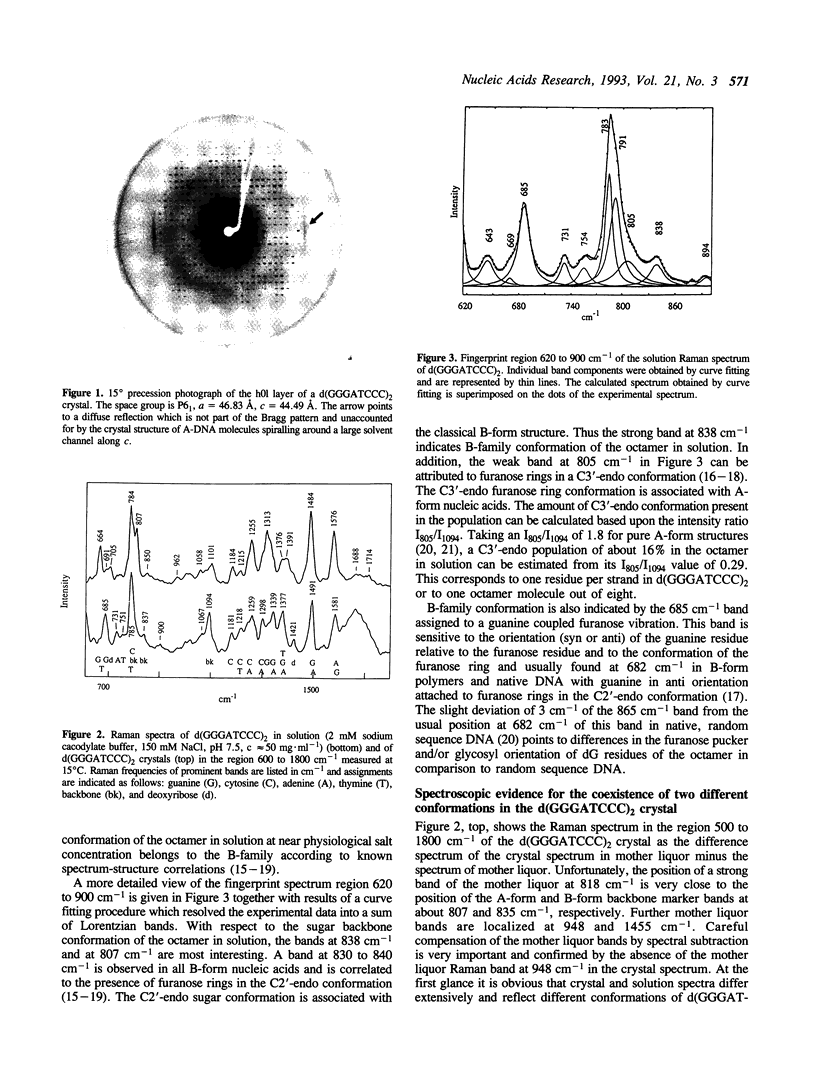
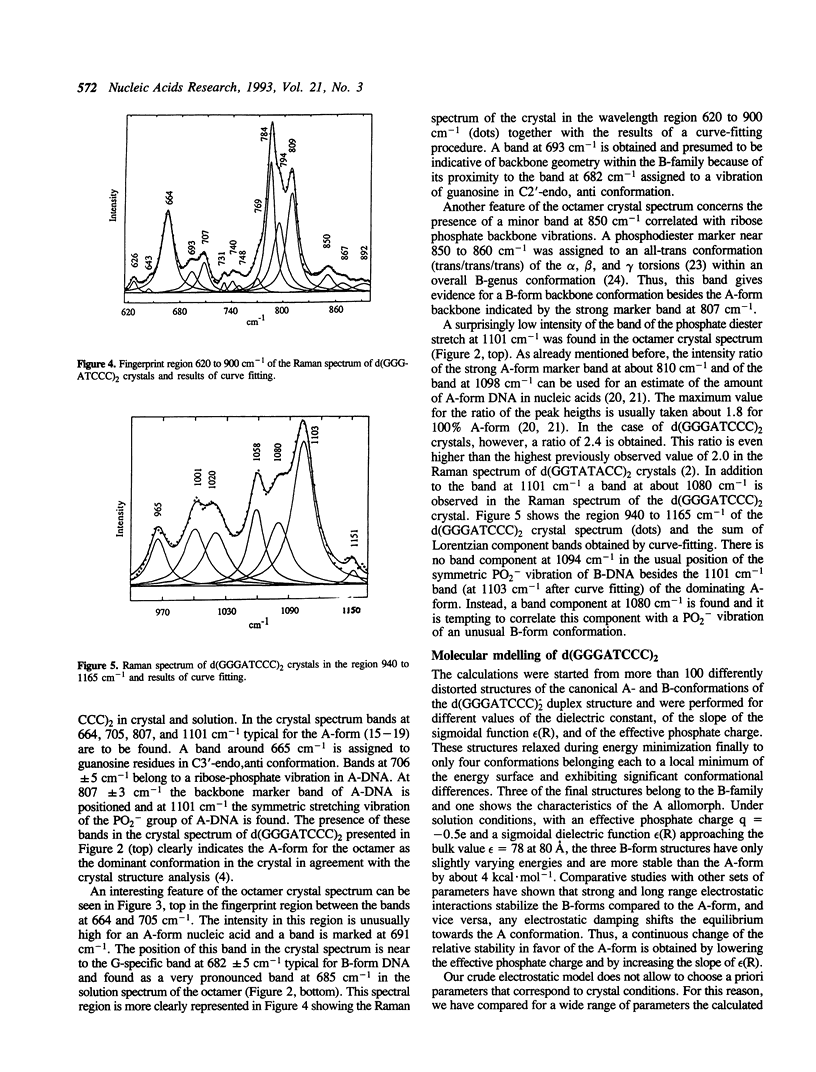
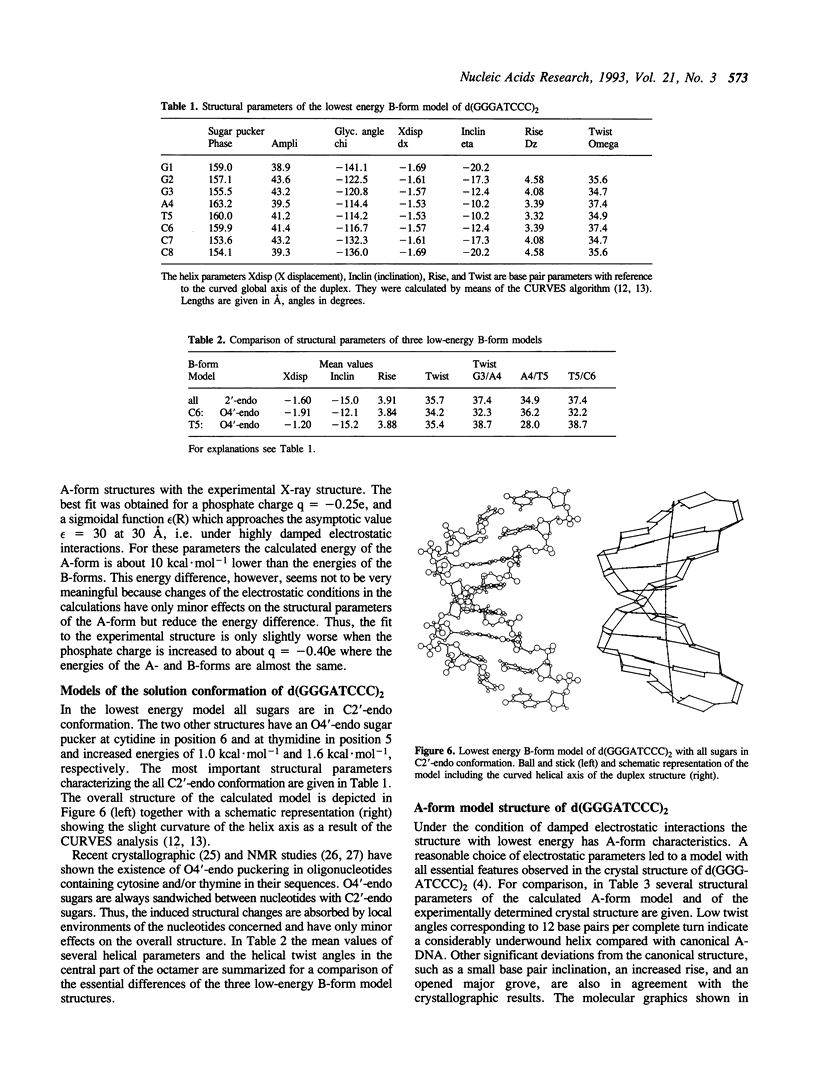
In the crystal, d(GGGATCCC)2 forms an A-DNA double helix as known from a single crystal X-ray diffraction study. Accordingly, in the Raman spectra of crystals the A-family marker bands at 664, 705, 807 and 1101 cm-1 and the spectral characteristics in the region 1200 to 1500 cm-1 clearly demonstrate the A-form as the dominant conformation. Bands at 691, 850, and 1080 cm-1, however, indicate that a minor fraction of the octamer molecules in the crystal is in an unusual, still not unequivocally identified conformation possibly belonging to the B-family. In solution, the octamer is in B-like conformation as shown by the presence of B-DNA Raman marker bands at 685, 837, 1094 and 1421 cm-1. Molecular modelling techniques lead to three structures with slightly different B-form geometries as the lowest energies models when a sigmoidal dielectric function with the bulk dielectric constant epsilon = 78 and the value q = -0.5e for the effective phosphate charges was used in the calculations. An A-form structure bearing a strong resemblance to the experimentally determined crystal structure becomes the lowest energy model structure when the electrostatic parameters are changed to epsilon = 30 and q = -0.25e, respectively.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benevides J. M., Wang A. H., Rich A., Kyogoku Y., van der Marel G. A., van Boom J. H., Thomas G. J., Jr Raman spectra of single crystals of r(GCG)d(CGC) and d(CCCCGGGG) as models for A DNA, their structure transitions in aqueous solution, and comparison with double-helical poly(dG).poly(dC). Biochemistry. 1986 Jan 14;25(1):41–50. doi: 10.1021/bi00349a007. [DOI] [PubMed] [Google Scholar]
- Benevides J. M., Weiss M. A., Thomas G. J., Jr DNA recognition by the helix-turn-helix motif: investigation by laser Raman spectroscopy of the phage lambda repressor and its interaction with operator sites OL1 and OR3. Biochemistry. 1991 Jun 18;30(24):5955–5963. doi: 10.1021/bi00238a020. [DOI] [PubMed] [Google Scholar]
- Doucet J., Benoit J. P., Cruse W. B., Prange T., Kennard O. Coexistence of A- and B-form DNA in a single crystal lattice. Nature. 1989 Jan 12;337(6203):190–192. doi: 10.1038/337190a0. [DOI] [PubMed] [Google Scholar]
- Haran T. E., Shakked Z., Wang A. H., Rich A. The crystal structure of d(CCCCGGGG): a new A-form variant with an extended backbone conformation. J Biomol Struct Dyn. 1987 Oct;5(2):199–217. doi: 10.1080/07391102.1987.10506390. [DOI] [PubMed] [Google Scholar]
- Heinemann U., Lauble H., Frank R., Blöcker H. Crystal structure analysis of an A-DNA fragment at 1.8 A resolution: d(GCCCGGGC). Nucleic Acids Res. 1987 Nov 25;15(22):9531–9550. doi: 10.1093/nar/15.22.9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauble H., Frank R., Blöcker H., Heinemann U. Three-dimensional structure of d(GGGATCCC) in the crystalline state. Nucleic Acids Res. 1988 Aug 25;16(16):7799–7816. doi: 10.1093/nar/16.16.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavery R., Parker I., Kendrick J. A general approach to the optimization of the conformation of ring molecules with an application to valinomycin. J Biomol Struct Dyn. 1986 Dec;4(3):443–462. doi: 10.1080/07391102.1986.10506361. [DOI] [PubMed] [Google Scholar]
- Lavery R., Sklenar H. Defining the structure of irregular nucleic acids: conventions and principles. J Biomol Struct Dyn. 1989 Feb;6(4):655–667. doi: 10.1080/07391102.1989.10507728. [DOI] [PubMed] [Google Scholar]
- Lavery R., Sklenar H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J Biomol Struct Dyn. 1988 Aug;6(1):63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- Lavery R., Sklenar H., Zakrzewska K., Pullman B. The flexibility of the nucleic acids: (II). The calculation of internal energy and applications to mononucleotide repeat DNA. J Biomol Struct Dyn. 1986 Apr;3(5):989–1014. doi: 10.1080/07391102.1986.10508478. [DOI] [PubMed] [Google Scholar]
- Liquiers J., Taillandier E., Peticolas W. L., Thomas G. A. The infrared and Raman spectra of the duplex of d(GGTATACC) in the crystal show bands due to both the A-form and the B-form of DNA. J Biomol Struct Dyn. 1990 Oct;8(2):295–302. doi: 10.1080/07391102.1990.10507806. [DOI] [PubMed] [Google Scholar]
- Martin J. C., Wartell R. M. Changes in raman vibrational bands of calf thymus DNA during the B-to-A transition. Biopolymers. 1982 Mar;21(3):499–512. doi: 10.1002/bip.360210303. [DOI] [PubMed] [Google Scholar]
- Metzler W. J., Wang C., Kitchen D. B., Levy R. M., Pardi A. Determining local conformational variations in DNA. Nuclear magnetic resonance structures of the DNA duplexes d(CGCCTAATCG) and d(CGTCACGCGC) generated using back-calculation of the nuclear Overhauser effect spectra, a distance geometry algorithm and constrained molecular dynamics. J Mol Biol. 1990 Aug 5;214(3):711–736. doi: 10.1016/0022-2836(90)90288-W. [DOI] [PubMed] [Google Scholar]
- Privé G. G., Yanagi K., Dickerson R. E. Structure of the B-DNA decamer C-C-A-A-C-G-T-T-G-G and comparison with isomorphous decamers C-C-A-A-G-A-T-T-G-G and C-C-A-G-G-C-C-T-G-G. J Mol Biol. 1991 Jan 5;217(1):177–199. doi: 10.1016/0022-2836(91)90619-h. [DOI] [PubMed] [Google Scholar]
- Schmitz U., Zon G., James T. L. Deoxyribose conformation in [d(GTATATAC)]2: evaluation of sugar pucker by simulation of double-quantum-filtered COSY cross-peaks. Biochemistry. 1990 Mar 6;29(9):2357–2368. doi: 10.1021/bi00461a021. [DOI] [PubMed] [Google Scholar]
- Shakked Z., Rabinovich D., Kennard O., Cruse W. B., Salisbury S. A., Viswamitra M. A. Sequence-dependent conformation of an A-DNA double helix. The crystal structure of the octamer d(G-G-T-A-T-A-C-C). J Mol Biol. 1983 May 15;166(2):183–201. doi: 10.1016/s0022-2836(83)80005-9. [DOI] [PubMed] [Google Scholar]
- Sklenar H., Lavery R., Pullman B. The flexibility of the nucleic acids: (I). "SIR", a novel approach to the variation of polymer geometry in constrained systems. J Biomol Struct Dyn. 1986 Apr;3(5):967–987. doi: 10.1080/07391102.1986.10508477. [DOI] [PubMed] [Google Scholar]
- Thomas G. A., Kubasek W. L., Peticolas W. L., Greene P., Grable J., Rosenberg J. M. Environmentally induced conformational changes in B-type DNA: comparison of the conformation of the oligonucleotide d(TCGCGAATTCGCG) in solution and in its crystalline complex with the restriction nuclease EcoRI. Biochemistry. 1989 Mar 7;28(5):2001–2009. doi: 10.1021/bi00431a007. [DOI] [PubMed] [Google Scholar]
- Wartell R. M., Harrell J. T. Characteristics and variations of B-type DNA conformations in solution: a quantitative analysis of Raman band intensities of eight DNAs. Biochemistry. 1986 May 6;25(9):2664–2671. doi: 10.1021/bi00357a056. [DOI] [PubMed] [Google Scholar]