Abstract
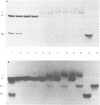
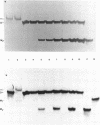
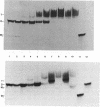
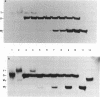
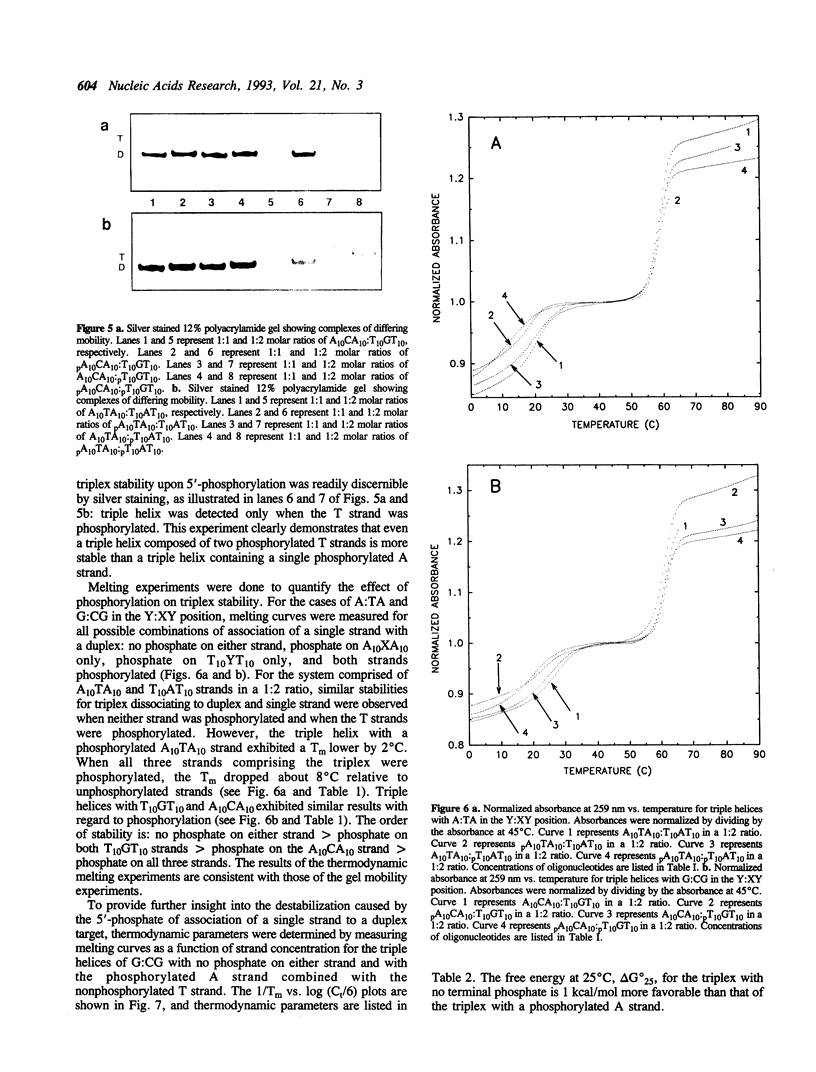
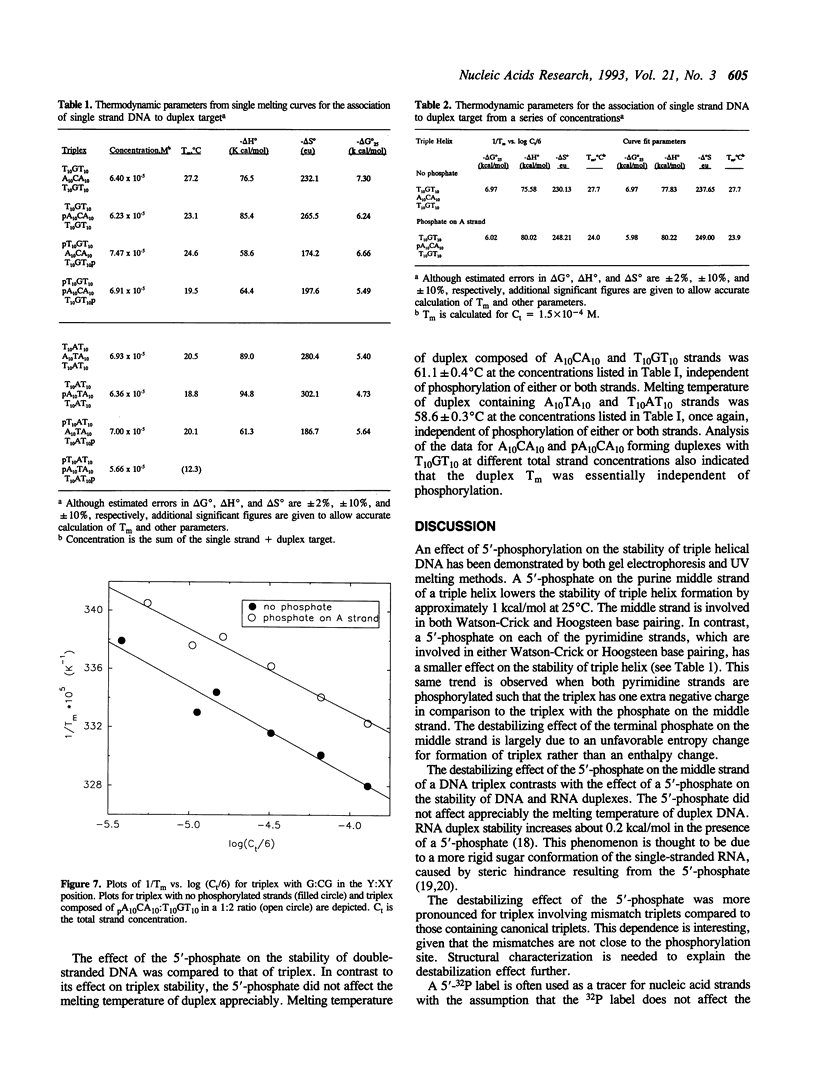
An effect of 5'-phosphorylation on the stability of triple helical DNA containing pyrimidine:purine:pyrimidine strands has been demonstrated by both gel electrophoresis and UV melting. A 5'-phosphate on the purine-rich middle strand of a triple helix lowers the stability of triple helix formation by approximately 1 kcal/mol at 25 degrees C. The middle strand is involved in both Watson-Crick and Hoogsteen base pairing. In contrast, a 5'-phosphate on the pyrimidine-rich strands, which are involved in either Watson-Crick or Hoogsteen base pairing, has a smaller effect on the stability of triple helix. The order of stability is: no phosphate on either strand > phosphate on both pyrimidine strands > phosphate on purine strand > phosphate on all three strands. Differential stability of triple helix species is postulated to stem from an increase in rigidity due to steric hindrance from the 5'-phosphate. This result indicates that labelling with 32P affect equilibrium in triplex formation.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Bond P. J. Structures for Poly(U)-poly(A)-poly(U)triple stranded polynucleotides. Nat New Biol. 1973 Jul 25;244(134):99–101. doi: 10.1038/newbio244099a0. [DOI] [PubMed] [Google Scholar]
- Beal P. A., Dervan P. B. Second structural motif for recognition of DNA by oligonucleotide-directed triple-helix formation. Science. 1991 Mar 15;251(4999):1360–1363. doi: 10.1126/science.2003222. [DOI] [PubMed] [Google Scholar]
- Borer P. N., Dengler B., Tinoco I., Jr, Uhlenbeck O. C. Stability of ribonucleic acid double-stranded helices. J Mol Biol. 1974 Jul 15;86(4):843–853. doi: 10.1016/0022-2836(74)90357-x. [DOI] [PubMed] [Google Scholar]
- Broitman S. L., Im D. D., Fresco J. R. Formation of the triple-stranded polynucleotide helix, poly(A.A.U). Proc Natl Acad Sci U S A. 1987 Aug;84(15):5120–5124. doi: 10.1073/pnas.84.15.5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlin M. J. Comparative properties of DNA, RNA, and hybrid homopolymer pairs. Fed Proc. 1965 Nov-Dec;24(6):1446–1457. [PubMed] [Google Scholar]
- Cooney M., Czernuszewicz G., Postel E. H., Flint S. J., Hogan M. E. Site-specific oligonucleotide binding represses transcription of the human c-myc gene in vitro. Science. 1988 Jul 22;241(4864):456–459. doi: 10.1126/science.3293213. [DOI] [PubMed] [Google Scholar]
- Durland R. H., Kessler D. J., Gunnell S., Duvic M., Pettitt B. M., Hogan M. E. Binding of triple helix forming oligonucleotides to sites in gene promoters. Biochemistry. 1991 Sep 24;30(38):9246–9255. doi: 10.1021/bi00102a017. [DOI] [PubMed] [Google Scholar]
- Freier S. M., Alkema D., Sinclair A., Neilson T., Turner D. H. Contributions of dangling end stacking and terminal base-pair formation to the stabilities of XGGCCp, XCCGGp, XGGCCYp, and XCCGGYp helixes. Biochemistry. 1985 Aug 13;24(17):4533–4539. doi: 10.1021/bi00338a008. [DOI] [PubMed] [Google Scholar]
- Griffin L. C., Dervan P. B. Recognition of thymine adenine.base pairs by guanine in a pyrimidine triple helix motif. Science. 1989 Sep 1;245(4921):967–971. doi: 10.1126/science.2549639. [DOI] [PubMed] [Google Scholar]
- Mergny J. L., Sun J. S., Rougée M., Montenay-Garestier T., Barcelo F., Chomilier J., Hélène C. Sequence specificity in triple-helix formation: experimental and theoretical studies of the effect of mismatches on triplex stability. Biochemistry. 1991 Oct 8;30(40):9791–9798. doi: 10.1021/bi00104a031. [DOI] [PubMed] [Google Scholar]
- Morgan A. R., Wells R. D. Specificity of the three-stranded complex formation between double-stranded DNA and single-stranded RNA containing repeating nucleotide sequences. J Mol Biol. 1968 Oct 14;37(1):63–80. doi: 10.1016/0022-2836(68)90073-9. [DOI] [PubMed] [Google Scholar]
- Moser H. E., Dervan P. B. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987 Oct 30;238(4827):645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- Petersheim M., Turner D. H. Base-stacking and base-pairing contributions to helix stability: thermodynamics of double-helix formation with CCGG, CCGGp, CCGGAp, ACCGGp, CCGGUp, and ACCGGUp. Biochemistry. 1983 Jan 18;22(2):256–263. doi: 10.1021/bi00271a004. [DOI] [PubMed] [Google Scholar]
- Postel E. H., Flint S. J., Kessler D. J., Hogan M. E. Evidence that a triplex-forming oligodeoxyribonucleotide binds to the c-myc promoter in HeLa cells, thereby reducing c-myc mRNA levels. Proc Natl Acad Sci U S A. 1991 Sep 15;88(18):8227–8231. doi: 10.1073/pnas.88.18.8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley M., Maling B. Physical and chemical characterization of two- and three-stranded adenine-thymine and adenine-uracil homopolymer complexes. J Mol Biol. 1966 Sep;20(2):359–389. doi: 10.1016/0022-2836(66)90069-6. [DOI] [PubMed] [Google Scholar]
- Yoon K., Hobbs C. A., Koch J., Sardaro M., Kutny R., Weis A. L. Elucidation of the sequence-specific third-strand recognition of four Watson-Crick base pairs in a pyrimidine triple-helix motif: T.AT, C.GC, T.CG, and G.TA. Proc Natl Acad Sci U S A. 1992 May 1;89(9):3840–3844. doi: 10.1073/pnas.89.9.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]