Abstract
The availability of dense molecular markers has made possible the use of genomic selection in plant and animal breeding. However, models for genomic selection pose several computational and statistical challenges and require specialized computer programs, not always available to the end user and not implemented in standard statistical software yet. The R-package BLR (Bayesian Linear Regression) implements several statistical procedures (e.g., Bayesian Ridge Regression, Bayesian LASSO) in a unifi ed framework that allows including marker genotypes and pedigree data jointly. This article describes the classes of models implemented in the BLR package and illustrates their use through examples. Some challenges faced when applying genomic-enabled selection, such as model choice, evaluation of predictive ability through cross-validation, and choice of hyper-parameters, are also addressed.
Prediction of genetic values is a central problem in quantitative genetics. Accurate predictions of genetic values of genotypes whose phenotypes are yet to be observed (e.g., newly developed lines) are needed to attain rapid genetic progress and to reduce phenotyping costs (e.g., Bernardo and Yu, 2007). Over many decades, such predictions have been obtained using phenotypic and family data, the latter usually represented by a pedigree. However, pedigree-based models do not account for Mendelian segregation, a term that in an additive model and in the absence of inbreeding explains as much as one half of the genetic variance. This sets an upper limit on the accuracy of estimates of genetic values of individuals without progeny. Dense molecular markers (MM) are now available in the genome of humans and of many plant and animal species. Unlike pedigree data, MM allow tracing back Mendelian segregation events at many points along the genome. Potentially, this information can be used to improve the accuracy of estimates of genetic values of newly developed lines.
Following the ground-breaking contribution of Meuwissen et al. (2001), genomic selection (GS) has gained ground in plant and animal breeding (e.g., Bernardo and Yu, 2007; Hayes et al., 2009; VanRaden et al., 2009; de los Campos et al., 2009; Crossa et al., 2010). In practice, implementing GS involves analyzing large amounts of phenotypic and MM data and requires specialized computer programs. The main purpose of this article is to show how the R-package (R Development Core Team, 2009) BLR (Bayesian Linear Regression, de los Campos and Pérez, 2010) can be used to implement several models for GS. A first version of the algorithms and the R-code was presented in de los Campos et al. (2009). The package was developed further and its performance was significantly improved by the first two authors of this article; the package and data set are available from the R website (http://www.r-project.org; verified 29 July 2010). We provide a brief overview of parametric models for GS and describe the type of models implemented in BLR. We show two applications that illustrate the use of the package and several features of the models implemented in it.
Parametric Models for Genomic Selection
In parametric models for GS (e.g., Meuwissen et al., 2001), phenotypic outcomes, yi (i = 1,…,n), are regressed on marker covariates xij (j = 1,…,p) using a linear model of the form , where βj is the regression of yi on the jth marker covariate and εi is a model residual. In matrix notation, the regression model is expressed as y = Xβ + ε, where y = {yi}, X = {xij}, β = {βj} and ε = {εi}. The number of molecular markers (p) is usually larger than the number of observations (n) and, because of this, estimation of marker effects via multiple regression by ordinary least squares (OLS) is not feasible. Instead, penalized estimation methods such as ridge regression (RR, Hoerl and Kennard, 1970), or the Least Absolute Shrinkage and Selection Operator (LASSO; Tibshirani, 1996) or Bayesian methods such as those of Meuwissen et al. (2001) or the Bayesian LASSO (BL) of Park and Casella (2008) (e.g., Yi and Xu, 2008; de los Campos et al., 2009) can be used to estimate marker effects.
In RR, estimates of the effects of MM are obtained as the solution to the following optimization problem:
Here, is a regularization parameter controlling the trade-offs between goodness of fit measured by the residual sum of squares, , and model complexity measured by the sum of squared marker effects (). The first order conditions of the above optimization problem are satisfied by ; equivalently, . Relative to OLS, RR adds a constant, , to the diagonal of the matrix of coefficients; this makes the solution unique and shrinks estimates of marker effects towards zero, with the extent of shrinkage increasing as increases (with the solution to the above problem is the OLS estimate of marker effects).
From a Bayesian perspective, can be viewed as the conditional posterior mode in a model with Gaussian likelihood and IID (independent and identically distributed) Gaussian marker effects, that is,
| [1] |
Above, is the a priori variance of marker effects. The prior distribution of marker effects becomes increasingly informative and concentrated around zero as decreases. The posterior mean and mode of β from [1] is equal to the ridge-regression estimate, with , that is, . Therefore, prediction of genetic values from [1] can be obtained as
| [2a] |
Alternatively, from [1] and using properties of the multivariate normal distribution, one has:
| [2b] |
Formulae [2a] and [2b] are equivalent and correspond to the Best Linear Unbiased Predictor (BLUP) under the model defined by [1]. Note that [2a] and [2b] require solving systems of p and n equations, respectively. Therefore, with p >> n, expression [2b] is computationally more convenient. However, unlike [2a], expression [2b] does not yield estimates of marker effects.
In RR-BLUP, estimates of marker effects are penalized to the same extent, and this may not be appropriate if some markers are located in regions not associated with genetic variance whereas others are linked to QTLs (Goddard and Hayes, 2007). To overcome this limitation, methods performing variable selection and shrinkage (e.g., LASSO) or Bayesian methods using marker-specific shrinkage of effects, such as methods BayesA and BayesB of Meuwissen et al. (2001) or the BL of Park and Casella (2008), have been proposed.
The BLR package implements Bayesian regression with marker-specific or marker homogenous shrinkage of estimates effects. The package allows inclusion of covariates other than markers and regressions on a pedigree as well. In BLR, phenotypes are expressed as follows:
| [3] |
where y, the response, is a (n × 1) vector (missing values are allowed); μ is an intercept; , , , and are incidence matrices for the vectors of effects βF, u, βR, and βL, whose dimensions are pF, pU, pR, and pL, respectively. These vectors of effects differ with respect to the prior distributions assigned, as discussed later on. Finally, ε is a vector of model residuals assumed to be distributed as , where is an (unknown) variance parameter and the wi’s are (known) weights that allow for heterogeneous-residual variances. From these assumptions, the conditional distribution of the data, given the location effects, the residual variance and the weights, is:
| [4] |
Any of the elements on the right-hand side of [3], except μ and ε, can be excluded in BLR; by default, the program runs an intercept model, i.e., yi = μ + εi. If weights are not provided, all weights are set equal to one.
The Bayesian model is completed by assigning a prior to all model unknowns, . The intercept, μ, and the vector of “fixed” effects, βF, are assigned flat priors, that is, p(μ, βF) ∝ constant. This treatment yields posterior means of these unknowns that are similar to those obtained with OLS, provided that μ and βF are the only effects included in the model.
The vector u is modeled using the standard assumptions of the infinitesimal additive model (e.g., Fisher, 1918; Wright, 1921; Henderson, 1975), that is, , where A is a positive-definite matrix (usually a numerator relationship matrix computed from a pedigree) and is an unknown variance, whose prior is a scaled inverse-χ2 density with degrees of freedom dfu and scale Su, that is, ; the hyper-parameters are user provided. In the parameterization used in BLR, . The multivariate normal prior assigned to u induces shrinkage of estimates of effects uj toward zero and borrowing of information between levels of the random effect, .
The vector of regression coefficients βR is assigned a Gaussian prior with variance common to all effects, that is, . This prior induces estimates that are the Bayesian counterpart of those obtained with Ridge Regression; we refer to this as “Bayesian Ridge Regression” (BRR). The variance parameter, , is treated as unknown and is assigned a scaled inverse-χ2 prior density, that is, with degrees of freedom, dfβR, and scale, SβR, provided by the user.
The vector of regression coefficients βL is treated as in the Bayesian LASSO of Park and Casella (2008); the conditional prior distribution of marker effects, , is Gaussian with marker-specific prior variances, that is, . This prior induces marker-specific shrinkage of estimates of effects, whose extent depends on . The variance parameters, , are assigned exponential IID priors, . Finally, in the BLR package, the prior distribution of the regularization parameter λ, p(λ), can be:
a mass-point at some value (i.e., fixed λ),
p(λ2 )~ Gamma(r,δ), as suggested by Park and Casella (2008), or
With the above assumptions, the marginal prior of regression coefficients βLj, , is Double-Exponential (DE). Figure 1 displays the Gaussian and Double-Exponential density functions of random variables with zero mean and unit variance. Relative to the Gaussian, the DE distribution places a higher density at zero and thicker tails, inducing stronger shrinkage of estimates for markers with relatively small effect and less shrinkage of estimates for markers with sizable effect.
Figure 1.
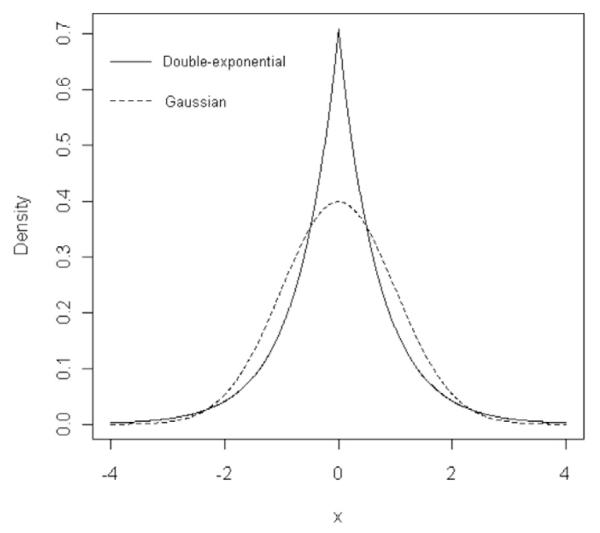
Gaussian and Double-Exponential densities, both for a random variable with zero mean and unit variance.
Finally, the residual variance is assigned a scaled inverse-χ2 prior density with degrees of freedom, dfε, and scale parameter, Sε, provided by the user that is:
Collecting the aforementioned assumptions, the prior distribution in BLR is:
The prior distribution is indexed by several hyper-parameters; in the Appendix, we provide guidelines for choosing these parameters on the basis of prior expectations about the proportion of phenotypic variance that can be attributed to each of the components on the righthand side of [3].
The posterior distribution of model unknowns is proportional to the product of the likelihood and the prior distribution, that is:
| [5] |
This posterior distribution does not have a closed form; however, a Gibbs sampler can be used to draw samples from it. The Gibbs sampler is as in de los Campos et al. (2009) but extended to accommodate “fixed” effects and BRR.
Using Bayesian Linear Regression
This section gives two examples that illustrate the use of the BLR package and describe features of the models. It is assumed that the reader is familiar with the R-language/environment. In Example 1, we study the impact of different shrinkage methods (BRR vs. BL) using simulated data. Example 2 illustrates how the package can be used to implement cross-validation (CV) using Bayesian methods. Cross-validation can be used for model comparison (e.g., to compare predictive ability of pedigree-based models versus marker-based models) or for selecting model parameters (e.g., λ in the BL). Both examples make use of a wheat data set made available with the package, whose main features are described next.
Wheat Data Set
These data contain a historical set of 599 wheat lines from CIMMYT’s Global Wheat Program that were genotyped for 1447 Diversity Array Technology markers (DArT, Triticarte Pty. Ltd, Canberra, Australia; http://www.triticarte.com.au; verified 26 July 2010) and evaluated for grain yield (GY) in four macroenvironments. The dataset becomes available in the R environment by running the following R-code:
library(BLR) data(wheat)
Function library() loads the package, and data() loads datasets included in the package into the environment. The above code loads the following objects [type objects() in the R console to list them] into the environment: - Y, a matrix (599 × 4) containing the 2-yr average grain yield of each of these lines in each of the four environments (phenotypes were standardized to a unit variance within each environment); - A (599 × 599) is a numerator relationship matrix computed from a pedigree that traced back many generations. This relationship matrix was derived using the Browse application of the International Crop Information System (ICIS), as described in http://cropwiki.irri.org/icis/index.php/TDM_GMS_Browse; verified 26 July 2010 (McLaren, 2005); - X (599 × 1279) is a matrix with DArT genotypes; data are from pure lines and genotypes were coded as 0/1 denoting the absence/presence of the DArT. Markers with a minor allele frequency lower than 0.05 were removed, and missing genotypes were imputed with samples from the marginal distribution of marker genotypes, that is, xij ~ Bernoulli ( ), where is the estimated allele frequency computed from the nonmissing genotypes. The number of DArT MMs after edition was 1279. - sets (599 × 1) is a vector that assigns observations to 10 disjoint sets; the assignment was generated at random. This is used later to conduct a 10-fold CV.
Example 1: The Nature of Different Shrinkage Methods
As stated, BRR or BL use different priors for marker effects; this induces different types of shrinkage of estimates of such effects. The simulation presented in this section aims at illustrating these differences. Data were generated using marker genotypes from the wheat dataset (X) with marker effects and model residuals simulated as described below.
Data Simulation
Data were simulated under an additive model of the form, , i = 1,…,599, where μ = 100 is an effect common to all individuals; {xij} are marker genotypes from a collection of wheat lines described previously;{βj} are marker effects; and εi ~ N(εi|0,1) are IID standard normal residuals. We assumed that most markers (1267) had a relatively small effect and that only a few markers (12) had a sizable effect. Specifically, marker effects were sampled from the following mixture model:
where k = 1279−1. The R-code used to implement this simulation was:
set.seed(12345)
data(wheat)
p<-ncol(X)
nQTL<-12
n<-nrow(X)
b0<-rnorm(p,sd=1/sqrt(p))
isQTL<-seq(from=3,to=p,length=nQTL)
b0[isQTL]<-rep(c(−1,1),times=nQTL/2)*
runif(min=.5,max=.8,n=12)
yHat0<-100+X%*%b0
e0<-rnorm(n,sd=1)
y<-yHat0+e0
Function set.seed() initializes the random number generator; X is the matrix with information on molecular markers. Functions rnorm() and runif() generate random draws from the uniform and normal distributions, respectively. Figure 2 shows realized marker effects obtained with the above R-code. In this example, the sample variance of phenotypes (y) was 1.78, and the ratio of the sample variance of genetic values relative to phenotypic variance was 0.43.
Figure 2.
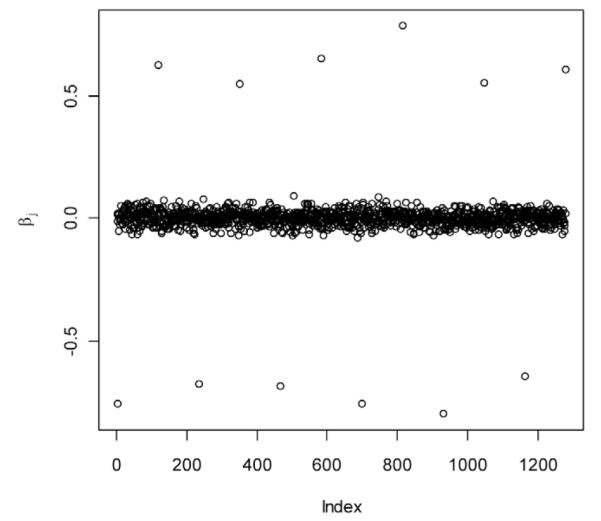
Realized marker effects.
Choice of Hyper-Parameters
The Appendix provides guidelines on how to choose values of hyper-parameters. It is assumed that the user has prior beliefs about the proportion of phenotypic variance that can be attributed to each of the components of the regression. In the simulation, the variance of phenotypes was about 1.78, and the variance of model residuals was 1. In practice, one does not know the true proportion of phenotypic variance that can be assigned to the genetic signal and model residuals, unless a precise estimate of heritability is available. Suppose our prior belief is that 50% of the phenotypic variance can be attributed to the genetic signal. Using dfε = 3 and = 0.9 in formula [1A] of the appendix, we set Sε = 4.5. Values of hyper-parameters of the prior distribution of and λ2 can be chosen using formulae [3A] and [4A] in the Appendix. Using these formulae requires computing the sum of squares (over markers) of the average genotype, that is, , where . In the wheat dataset, this quantity is approximately 504; using this and dfβR = 3, VR = 0.9 in formula [3A], we set . Finally, 504 using in formula [4A] of the Appendix, we find . Choosing λ2 ~ G(λ2|r = 2×10−5,δ = 0.52) gives a prior density for λ that has high density and is relatively flat around (Fig. 3).
Figure 3.
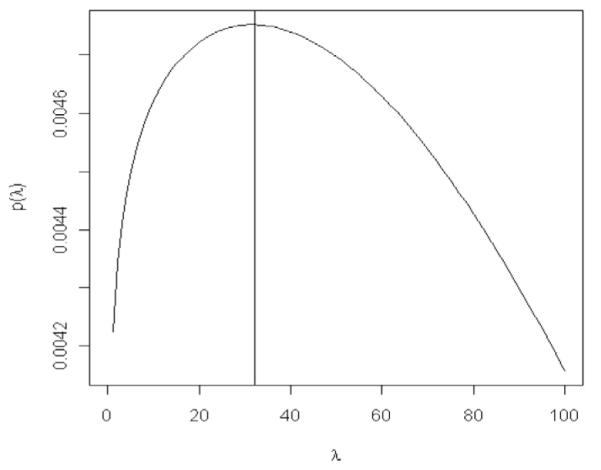
Prior density of λ when λ2 is assigned a gamma prior with rate equal to 2 × 10−5 and shape equal to 0.52. When λ2 ~ G(r,δ), ; this is the density displayed above.
Fitting the Model
Using the aforementioned values of hyper-parameters, BL and BRR were fitted using the following R-code:
prior=list(
varE=list(S=4.5,df=3),
varBR=list(S=.009,df=3),
lambda=list(type="random",
value=30,shape=.52,rate=2e–5))
nIter<-60000
burnIn<-10000
fmR<-BLR(y=y,XR=X,nIter=nIter,burnIn=burnIn,
thin=10,saveAt="R _ ",prior=prior)
dput(fmR,file="fmR.out")
fmL<-BLR(y=y,XL=X,nIter=nIter,
burnIn=burnIn,thin=10,
saveAt="L _ ",prior=prior)
dput(fmL,file="fmL.out")
In the above code, y is the response vector, X is the matrix of genotypes, and nIter and burnIn define the number of iterations and burn-in period, respectively, used in the Gibbs sampler. The prior is provided as a list; type “help(BLR)” in the R console for more details. The BLR function returns a list with posterior means, posterior standard deviations, and Deviance Information Criterion (DIC, Spiegelhalter et al., 2002). Function dput() saves this list to the hard drive. The fitted model can then be retrieved using function dget(). In addition to returning posterior means and posterior standard deviations, as the Gibbs sampler runs, samples of the intercept, of the fixed effects, of the variance parameters and of λ are saved to the hard-drive using the thinning specified by the user (which is set to 10 by default).
Results of Example 1
We ran the processes in an Intel Xeon 5530 2.4 GHz Quad Core Processor (R was executed in a single thread) with 6 GB of RAM memory. With this dataset (599 subjects, 1279 markers) the process took about 1% of RAM memory. BRR took about 5.5 s for every 1000 iterations of the sampler; BL takes about twice as much time.
Figure 4 shows the estimated posterior density of the residual variance for each of the models; the prior density (up to a constant) is included as well. The posterior distributions moved away from the prior and were sharp. The estimated posterior means (standard deviation) of were 1.00 (0.0883) and 0.930 (0.0814) for BRR and BL, respectively. These values are close to the true value of the parameter (one) and suggest that BL over-fitted the data slightly. The posterior standard deviation (SD) was 8% smaller in BL, and mixing of the residual variance was better in BRR. BL gives a slightly “less informative” posterior distribution and worse mixing for two reasons. First, because of use of marker-specific variances, the number of unknowns in BL is much larger than in BRR. Second, the BL has an extra level in which the regularization parameter (λ) indexing the prior distribution of the marker-specific variances is inferred from the data. In BRR, the counterparts of λ are the hyper-parameters indexing the prior assigned to ( dfβR and SβR ) which are specified by the user.
Figure 4.
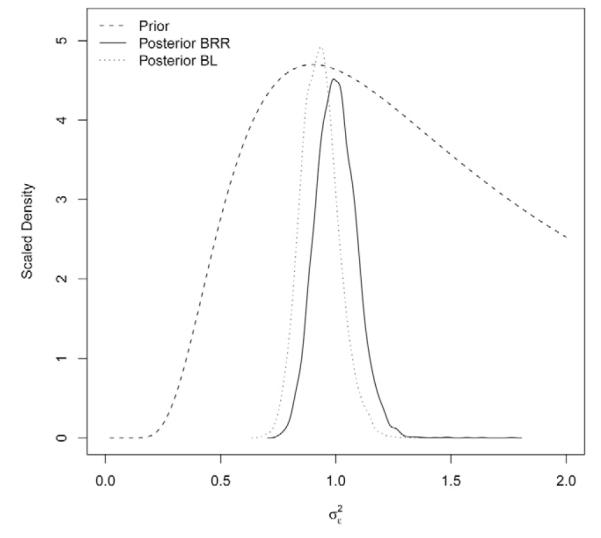
Prior and estimated posterior density of the residual variance, by model (BRR = Bayesian Ridge Regression; BL = Bayesian LASSO).
The posterior mean of λ in BL was 20.1, and a 95th highest posterior density confidence region was bounded by [15.7, 26.2]. These results also indicate that the posterior distribution of λ moved away from the prior (Fig. 3), indicating that Bayesian learning takes place.
Measures of goodness of fit and model complexity [pD = estimated effective number of parameters, Spiegelhalter et al. (2002), and DIC] are included in the fitted object. This can be assessed with the following code:
fmR$fit ## Bayesian Ridge Regression fmL$fit ## Bayesian LASSO
Table 1 provides estimates of the log-likelihood evaluated at the posterior mean of model unknowns, , the posterior mean of the log-likelihood, , the estimated effective number of parameters, pD, and DIC for BRR and BL. The Bayesian LASSO fitted the data better, and had a higher estimated number of effective parameters; DIC, which balances goodness of fit and complexity, favored the BL.
Table 1.
Measures of goodness of fit, model complexity, and Deviance Information Criterion (DIC) by model.
| Bayesian Ridge Regression | Bayesian LASSO | |
|---|---|---|
| † | −776.1 | −748.7 |
| −847.9 | −825.8 | |
| pD | 143.6 | 154.3 |
| DIC | 1839.5 | 1806.1 |
= log-likelihood evaluated at the estimated posterior mean of model unknowns; = estimated posterior mean of the log-likelihood; pD = estimated effective number of parameters.
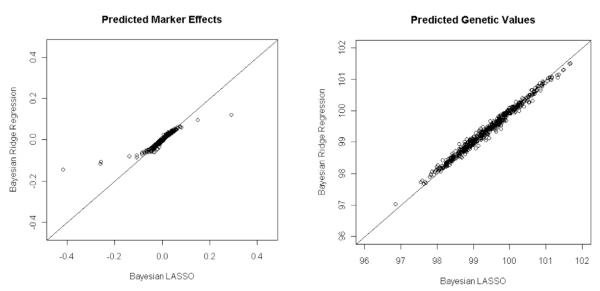
The mean-squared error of estimates of marker effects, , where (.) can be either L or R, were 0.0046 and 0.0039 for BRR and BL, respectively. The mean-squared error of estimates of genetic values, , were 0.3712 and 0.3116 for BRR and BL, respectively. Therefore, BL outperformed BRR in this simulation. The difference between methods is more pronounced at the level of marker effects (19% difference in MSE between methods for genetic values). Figure 5 shows scatter plots of the estimates of marker effects (left) and of genetic values (right) obtained with BL (horizontal axis) and BRR (vertical axis). The BRR shrinks estimates of markers with sizable effects to a larger extent than BL (see left panel in Fig. 5). On the other hand, the two models yielded similar predictions of genetic values (see right panel in Fig. 5). This occurs because with p >> n, one can arrive at similar predictions of genetic values either with a model where genetic values are highly dependent on a few markers with sizable effect (something that occurs in LASSO and, to a lesser extent, in BL) or with a model where a large number of markers make a small contribution to genetic values (something that occurs in RR-BLUP and BRR). Which model yields better prediction of genetic values will depend on the underlying architecture of the trait and on the available marker data.
Figure 5.
Estimates of marker effects (left panel) and of genetic values (right panel) obtained with the Bayesian Ridge Regression (BRR, vertical axis) versus those obtained with the Bayesian LASSO (BL, horizontal axis).
Example 2: Assessing Predictive Ability by Cross-Validation
Predicting genetic values of lines with yet-to-be observed phenotypes is a central problem in plant breeding programs. Such predictions can be used, for example, to decide which of the newly developed lines will be included in field trials or which of these lines will be parents for the next breeding cycle. Either of the models described above, BL or BRR, can be used to obtain these predictions. The rate of genetic progress will depend on how accurate such predictions are, i.e., on the ability of the model to predict future outcomes. Cross-validation (CV) methods can be used to assess predictive ability and for for tuning-up values of certain parameters. In this section, we illustrate how the regularization parameter of the BL, λ, can be chosen using CV methods, and compare the performance of this approach with that obtained with the fully Bayesian approach that consists of assigning a prior to λ. The comparison is made using the wheat dataset previously described.
Model
Here, the linear model is y = 1μ + XLβL + u + ε. We chose values of hyper-parameters using formulae presented in the Appendix. As in Example 1, it was assumed a priori that 50% of the phenotypic variance (which equals to one because phenotypes were standardized to a unit variance) could be attributed to genetic values. With this, and using dfε = 3 in formula [1A] in the Appendix, we obtained Sε = 2.5. Further, we assumed a priori that one half of the variance of genetic values can be accounted for by the regression on markers, XLβL, and that the regression on the pedigree, u, accounted for the other half. With this, and using dfu = 3 and in [2A], we obtained Su = 0.63. Finally, using in formula [4A] in the Appendix, we obtained . Choosing λ2 ~ G(λ2|r = 1×10−5,δ = 0.52) gives a prior for λ that has a maximum and is relatively flat in the neighborhood of 45.
The R-code in Fig. 6 illustrates how this CV was performed for the first trait. The vector sets assign lines to folds of the CV. The code involves two loops: the outer loop runs over folds of the CV; the inner loop fits models over a grid of values of λ. For every fold in the outer loop, the phenotypes of approximately 60 lines are declared as missing values; the fitted model yields a prediction of the performance of these lines.
Figure 6.
R-code used to fit models of example 2.
Results of Example 2
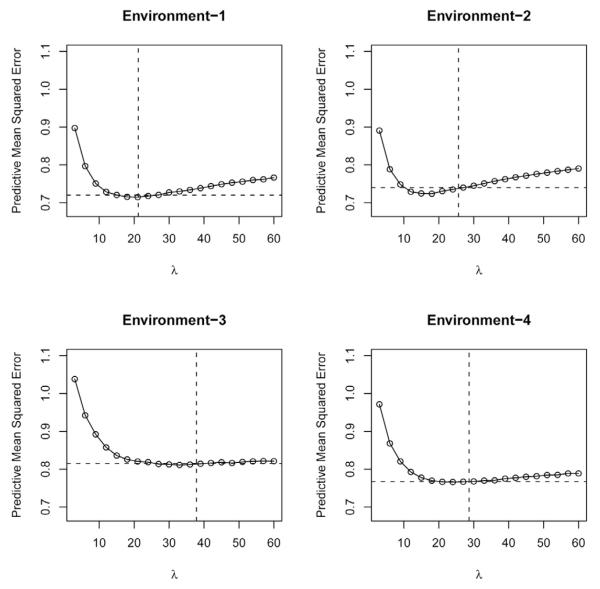
Figure 7 gives the estimated mean-squared error of predictive residuals (PMSE, vertical axis) versus values of the regularization parameter (λ), by environment. The vertical and horizontal dashed lines give the average (across 10 folds of the CV) estimated posterior mean of λ and the estimated PMSE obtained when a prior was assigned to λ (i.e., the fully-Bayesian LASSO). In all environments except E3, the curve relating PMSE and λ was U-shaped, with an optimum λ (minimum PMSE) near 20. However, the absolute value of the slope of the curve was higher for low values of λ, indicating that over-fitting, something that occurs with small values of λ, is more problematic. This may occur because the conditional expectation function in these models included two random components: the regression on markers and the regression on the pedigree. The presence of u in the linear model cannot prevent the over-fitting occurring when λ is “too small”. However, as values of λ increase (i.e., placing a higher penalty on the regression on markers), the contribution of the regression on the pedigree to the conditional expectation increases as well, preventing lack of fit. Environment 3 constitutes an extreme example of this; here the curve relating PMSE and λ looks L-shaped.
Figure 7.
Predictive mean squared error (PMS, vertical axis) versus values of the regularization parameter of the Bayesian LASSO (horizontal axis), by environment. The vertical and horizontal dashed lines give the average (across 10 folds of the cross-validation) estimated posterior mean of λ and the estimated PMSE obtained when a prior was assigned to λ.
The posterior modes of λ were always considerably smaller than the prior mode (45), indicating that Bayesian learning took place. In all environments, the fully Bayesian treatment yielded posterior means of λ that were in the neighborhood of the optimal values found when models were run over a grid of values of this unknown. Also, predictive ability of models with random λ was as good as the best obtained when CV was used for choosing λ. These results suggest that, at least for these traits and this population, the fully Bayesian treatment, which consists of inferring λ from the data, yields good results.
Conclusions
The BLR package allows fitting high-dimensional linear regression models including dense molecular markers, pedigree information, and covariates other than markers. The interface allows the user to choose models (e.g., BL versus BRR) and prior hyper-parameters easily. The algorithms implemented are relatively efficient and models with a modest number of markers (e.g., ~1000) can be fitted in a standard PC easily. The routines implemented in the package have also been used successfully in problems with larger numbers of molecular markers. For example, Weigel et al. (2009) used an earlier version of the package to fit models using data from the 50k Bovine Illumina Bead Chip, and our experience indicates that the software can be used with an even larger number of markers. Computational time is expected to increase linearly with the number of markers; the user also needs to be aware that marker information is loaded in the memory. Therefore, as the number of marker increases, so do the memory requirements.
In models for genomic selection, with p >> n, marker effects cannot be estimated uniquely from the likelihood. A unique solution can be obtained by using penalized estimation methods or, in a Bayesian framework, by assigning informative priors to marker effects. Because of lack of identification at the level of the likelihood, the choice of prior is expected to play a role. As illustrated in Example 1, different priors yield different estimates of marker effects. Although models per se cannot solve the intrinsic identification problem, one can use model comparison criteria, such as DIC, or the principle of parsimony, or CV methods, to choose among prior distributions of marker effects. Even if the choice of prior affects estimates of marker effects, it is still possible to obtain similar estimates of genetic values using different priors (see Example 1, or the simulation study presented in de los Campos et al., 2009). This occurs because, with p >> n, one can arrive at similar predictions either with a model where genetic values are highly dependent on some markers or with a model where all markers make a small contribution to genetic values.
Simulation studies (e.g., Habier et al., 2007) have suggested that models using marker-specific shrinkage of estimates of effects such as the BL or models Bayes A or Bayes B of Meuwissen et al. (2001) may capture linkage disequilibrium between markers and QTLs better than a BRR. Therefore, Habier et al. (2007) conclude, when estimates of effects are used to perform several rounds of selection and without retraining, those models should perform better than a BRR. However, as pointed out by Jannink et al. (2010), the superiority of these models over a BRR has not always been confirmed by real-data analysis.
Finally, estimates of marker effects obtained with BL or BRR could be used to assess the relative contribution of each region to genetic variability. However, one needs to be aware that the estimated marker effect reflects not only linkage between markers and genes affecting the trait but also the density of markers in the region. If a region containing a QTL has a high density of markers, the effect of the QTL is expected to be “distributed” across linked markers; conversely, if in the same region markers are sparse, estimated marker effects are expected to be larger in absolute value.
Acknowledgments
The authors thank the numerous cooperators in national agricultural research institutes who carried out the Elite Spring Wheat Yield Trials (ESWYT) and provided the phenotypic data analyzed in this article. We also thank the International Nursery and Seed Distribution Units in CIMMYT-Mexico for preparing and distributing the seed and computerizing the data. Financial support by the Wisconsin Agriculture Experiment Station and grant DMS-NSF DMS-044371 to Gustavo de los Campos and Daniel Gianola and of NIH grant R01GM077490 to Gustavo de los Campos is acknowledged.
Abbreviations
- BL
Bayesian LASSO
- BLR
Bayesian Linear Regression
- BLUP
Best Linear Unbiased Prediction
- BRR
Bayesian Ridge Regression
- CV
cross-validation
- DE
Double-Exponential
- DIC
Deviance Information Criterion
- GY
grain yield
- GS
Genomic selection
- IID
independent and identically distributed
- LASSO
Least Absolute Shrinkage and Selection Operator
- MM
molecular markers
- OLS
ordinary least squares
- PMSE
mean-squared error of predictive residuals
- RR
ridge regression
APPENDIX
The prior distribution of the BLR model is indexed by several hyper-parameters. We provide guidelines for choosing their values on the basis of beliefs as to what proportion of the variance of phenotypes can be attributed to each of the random components of the regression, that is, ui, , and εi. Other authors (e.g., Meuwissen et al., 2001) have discussed how to choose hyper-parameters in the context of models for genomic selection. However, the derivation used by those authors assumes that genotypes are random and marker effects are fixed quantities, while in fact the opposite is true in the Bayesian models that have been proposed (e.g., Gianola et al., 2009). Here, we derive formulae that are consistent with the standard treatment of marker and marker effects in Bayesian models for GS, where marker genotypes are observed quantities and marker effects are random unknowns. Unlike the formulae in Meuwissen et al. (2001), the ones presented here do not require making any assumption about the extent of linkage disequilibrium between markers.
Residual Variance
The prior distribution of the residual variance is indexed by two parameters, {Sε, dfε}. One can choose these parameters so that the prior mode of , matches our prior beliefs about the variance of model residuals. In practice, dfε can be chosen to be a small value, usually greater than two, to guarantee a finite prior expectation, e.g., dfε = 3, then the prior scale can be:
| [1A] |
where Vε is chosen to reflect our expectation of the variance of model residuals. Formulas similar to that presented in this appendix can be derived using formulas for the prior expectation.
Variance of the Infinitesimal Effect
From the prior distribution, the variance of ui is , where aii is the ith diagonal element of A, which in the absence of inbreeding equals one. If we let be the average diagonal value of A and Vu be our prior expectation about , then we can choose dfu = 3 and set the prior scale to be:
| [2A] |
Prior Variance of Marker Effects
The contributions of XR and XL to phenotypes is . Here, (.) stands for R or L, depending on whether BRR or BL is being used. In regressions for GS, marker genotypes are fixed and marker effects are random. Furthermore, at the level of the marginal distribution, marker effects are IID; therefore, . For the “average” genotype, this formula becomes where V. represent user’s beliefs about the variance that can be assigned to the regression on X., and denotes the average value of the jth column of X.
In BRR, the prior distribution of marker effects is Gaussian and . As before, dfβR can be chosen to have a relatively small value, e.g., dfβR = 3 then the prior scale can be set to be
| [3A] |
where VR is set to reflect our expectation of the variance of phenotypes that can be attributed to the regression on XR.
In the BL, the marginal prior density of marker effects is Double-Exponential, and the prior variance of marker effects is . Plugging this in
we obtain . Solving for λ, we get:
| [4A] |
where is a noise-to-signal variance ratio. With [4A] we can choose a target value for the regularization parameter. Then we can choose hyper-parameters so that the prior has a mode and is relatively flat, in the neighborhood of . Note that when λ2 ~ G(r,δ), p(λ|r,δ) = G(λ2|r,δ)2λ.
The above formulas constitute guidelines for choosing values of hyper-parameters. In practice, if Bayesian learning takes place, one would expect the posterior distribution to move away from the prior. Furthermore, with small n, it is always useful to check the sensitivity of inferences with respect to the choice of hyper-parameters.
Footnotes
Permission for printing and for reprinting the material contained herein has been obtained by the publisher.
References
- Bernardo R, Yu J. Prospects for genome-wide selection for quantitative traits in maize. Crop Sci. 2007;47:1082–1090. [Google Scholar]
- Crossa J, de los Campos G, Pérez P, Gianola D, Dreisigacker S, Burgueño J, Araus JL, Makumb D, Yan J, Singh R, Arief V, Banziger M, Braun H-J. Prediction of genetic values of quantitative traits in plant breeding using pedigree and molecular markers. Genetics. 2010 doi: 10.1534/genetics.110.118521. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Campos G, Naya H, Gianola D, Crossa J, Legarra A, et al. Predicting quantitative traits with regression models for dense molecular markers and pedigrees. Genetics. 2009;182:375–385. doi: 10.1534/genetics.109.101501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Campos G, Pérez P. [verified 22 July 2010];BLR: Bayesian linear regression. R package version 1.1. 2010 http://www.r-project.org/
- Fisher RA. The correlation between relatives on the supposition of Mendelian inheritance. Trans. R. Soc. Edinb. 1918;52:399–433. [Google Scholar]
- Gianola D, de los Campos G, Hill WG, Manfredi E, Fernando RL. Additive genetic variability and the Bayesian alphabet. Genetics. 2009;183:347–363. doi: 10.1534/genetics.109.103952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard ME, Hayes BJ. Genomic selection. J. Anim. Breed. Genet. 2007;124:323–330. doi: 10.1111/j.1439-0388.2007.00702.x. [DOI] [PubMed] [Google Scholar]
- Habier D, Fernando RL, Dekkers JCM. The impact of genetic relationship information on genome-assisted breeding values. Genetics. 2007;177:2389–2397. doi: 10.1534/genetics.107.081190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes BJ, Bowman PJ, Chamberlain AJ, Goddard ME. Invited review: Genomic selection in dairy cattle: Progress and challenges. J. Dairy Sci. 2009;92:433–443. doi: 10.3168/jds.2008-1646. [DOI] [PubMed] [Google Scholar]
- Henderson CR. Best linear unbiased estimation and prediction under a selection model. Biometrics. 1975;31:423–447. [PubMed] [Google Scholar]
- Jannink JL, Lorenz AJ, Hiroyoshi I. Genomic selection in plant breeding: From theory to practice. Brief. Funct. Genomics. 2010;9:166–177. doi: 10.1093/bfgp/elq001. [DOI] [PubMed] [Google Scholar]
- McLaren CG. The International Rice Information System. A platform for meta-analysis of rice crop data. Plant Physiol. 2005;139:637–642. doi: 10.1104/pp.105.063438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen THE, Hayes BJ, Goddard ME. Prediction of total genetic values using genome-wide dense marker maps. Genetics. 2001;157:1819–1829. doi: 10.1093/genetics/157.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park T, Casella G. The Bayesian LASSO. J. Am. Stat. Assoc. 2008;103:681–686. [Google Scholar]
- R Development Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: [verified 22 July 2010]. 2009. ISBN 3–900051–07–0, URL http://www.r-project.org/ [Google Scholar]
- Spiegelhalter DJ, Best NG, Carlin BP, van der Linde A. Bayesian measures of model complexity and fit (with discussion) J. R. Stat. Soc. Ser. B Stat. Methodol. 2002;64:583–639. [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the LASSO. J. Royal Stat. Soc. B. 1996;58:267–288. [Google Scholar]
- VanRaden PM, Van Tassell CP, Wiggans GR, Sonstegard TS, Schnabel RD, et al. Invited review: Reliability of genomic predictions for North American Holstein bulls. J. Dairy Sci. 2009;92:16–24. doi: 10.3168/jds.2008-1514. [DOI] [PubMed] [Google Scholar]
- Weigel K, de los Campos G, Vazquez AI, González-Recio O, Naya H, Wu XL, Long N, Rosa GJM, Gianola D. Predictive ability of direct genomic values for lifetime net merit of Holstein sires using selected subsets of single nucleotide polymorphism markers. J. Dairy Sci. 2009;92:5248–5257. doi: 10.3168/jds.2009-2092. [DOI] [PubMed] [Google Scholar]
- Wright S. Systems of mating. I. The biometric relations between parents and offspring. Genetics. 1921;6:111–123. doi: 10.1093/genetics/6.2.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi N, Xu S. Bayesian LASSO for quantitative trait loci mapping. Genetics. 2008;179:1045–1055. doi: 10.1534/genetics.107.085589. [DOI] [PMC free article] [PubMed] [Google Scholar]