Abstract
Background
Widespread sampling of vertebrates, which comprise the majority of published animal mitochondrial genomes, has led to the view that mitochondrial gene rearrangements are relatively rare, and that gene orders are typically stable across major taxonomic groups. In contrast, more limited sampling within the Phylum Mollusca has revealed an unusually high number of gene order arrangements. Here we provide evidence that the lability of the molluscan mitochondrial genome extends to the family level by describing extensive gene order changes that have occurred within the Vermetidae, a family of sessile marine gastropods that radiated from a basal caenogastropod stock during the Cenozoic Era.
Results
Major mitochondrial gene rearrangements have occurred within this family at a scale unexpected for such an evolutionarily young group and unprecedented for any caenogastropod examined to date. We determined the complete mitochondrial genomes of four species (Dendropoma maximum, D. gregarium, Eualetes tulipa, and Thylacodes squamigerus) and the partial mitochondrial genomes of two others (Vermetus erectus and Thylaeodus sp.). Each of the six vermetid gastropods assayed possessed a unique gene order. In addition to the typical mitochondrial genome complement of 37 genes, additional tRNA genes were evident in D. gregarium (trnK) and Thylacodes squamigerus (trnV, trnLUUR). Three pseudogenes and additional tRNAs found within the genome of Thylacodes squamigerus provide evidence of a past duplication event in this taxon. Likewise, high sequence similarities between isoaccepting leucine tRNAs in Thylacodes, Eualetes, and Thylaeodus suggest that tRNA remolding has been rife within this family. While vermetids exhibit gene arrangements diagnostic of this family, they also share arrangements with littorinimorph caenogastropods, with which they have been linked based on sperm morphology and primary sequence-based phylogenies.
Conclusions
We have uncovered major changes in gene order within a family of caenogastropod molluscs that are indicative of a highly dynamic mitochondrial genome. Studies of mitochondrial genomes at such low taxonomic levels should help to illuminate the dynamics of gene order change, since the telltale vestiges of gene duplication, translocation, and remolding have not yet been erased entirely. Likewise, gene order characters may improve phylogenetic hypotheses at finer taxonomic levels than once anticipated and aid in investigating the conditions under which sequence-based phylogenies lack resolution or prove misleading.
Background
Animal mitochondrial (mt) genomes typically consist of a circular molecule of DNA encoding 37 genes (2 rRNA genes, 13 protein-encoding genes, and 22 tRNA genes), the arrangement of which is often highly conserved within major taxonomic groups [1]. Consequently, when gene rearrangements occur, they may provide compelling phylogenetic markers that can corroborate or contradict hypotheses based on primary sequence data and provide resolution for deeper nodes that are often weakly supported in sequence-based phylogenies [2-6]. With recent technological and methodological advances (e.g., rolling circle amplification: [7,8]; next generation sequencing technologies: [9]), and associated decreasing costs of DNA sequencing, the amplification and sequencing of whole mt genomes has become routine. As a result, there has been a marked increase in the sequencing of whole animal mt genomes over the past decade as well as the development of computational methods to extract phylogenetic information from these genomes through inferences of past gene dynamics [10-12]. To date, 1868 complete metazoan mt genomes are available in the NCBI Genomes database http://www.ncbi.nlm.nih.gov/guide/genomes/; January 8, 2010), the majority belonging to arthropods (293) and vertebrates (1292).
Compared to other major metazoan phyla, molluscan mitochondrial genomes are poorly represented at NCBI [13], with only 78 complete mt genomes available as of January, 2010. Despite this, molluscan mt genomes are beginning to challenge the traditional view that mitochondrial gene orders are stable over long periods of evolutionary time [13-16], a view based largely on the heavily sampled and highly conserved mt genomes of vertebrates. Instead, mollusc mt genomes demonstrate substantial heterogeneity in length and "architecture" [16], reflecting differences in gene complement resulting from gene loss or duplication, as well as changes in the position and strand specificity of tRNA genes, protein-encoding genes, and rRNA genes. Changes in gene arrangement within the Mollusca have been so dramatic that representatives of four classes of molluscs (Gastropoda; Bivalvia; Cephalopoda; Scaphopoda) share remarkably few mitochondrial gene boundaries, with gene orders varying extensively even across major lineages of bivalves as well as gastropods [14]. Changes in gene arrangement have also been observed within bivalve and gastropod genera, based on changes in position of: 1) tRNAs and an rRNA gene in the oyster, Crassostrea [17], and 2) protein encoding and tRNA genes in the vermetid marine gastropod genus, Dendropoma [18]. Differences in gene order are also evident between paternally versus maternally inherited mitochondrial genomes of bivalves exhibiting doubly uniparental inheritance [19], including the unionid freshwater bivalve, Inversidens japanensis [14], and the marine venerid clam, Venerupis (Ruditapes) philippinarum (NCBI, unpublished). Similar intrageneric gene translocations have now been described in 19 of 144 genera in which two or more complete mt genomes have been sequenced [16], including representatives of the Porifera, Platyhelminthes, Nematoda, Mollusca, Arthropoda and Chordata. Thus, growing evidence suggests that mt genomes of many metazoan phyla may be considerably more plastic than originally believed, with the conserved genome architecture of vertebrates reflecting a derived stabilization of the mt genome and not an ancestral feature [16].
The discovery of mt gene order changes at lower taxonomic levels, as found within the Mollusca, is exciting for several reasons. First, gene dynamics involving translocations and inversions of genes offer the promise of new and robust characters that can be used to support phylogenetic hypotheses at the level of families, genera, and species [18]. Given the comparatively low rate of rearrangement and the astronomical number of possible gene arrangements, convergence is likely to be rare compared to four-state nucleotide sequence data [20]. Second, it is becoming increasingly apparent that the application of mitochondrial sequences and gene order data to questions of evolutionary history and phylogenetic relatedness requires a better understanding of the evolutionary dynamics of mt genomes [21]. Basic mechanisms of gene rearrangement associated with slipped-strand mispairing [22], errors in replication origins or end points [23], and intramolecular recombination [24], remain poorly understood. Likewise, tRNA remolding and tRNA recruitment events [25-28], gene rearrangement "hotspots" [29,30], the non-random loss of duplicated genes [31] and gene order homoplasy [32-34], which can act to confound phylogenetic inferences based on mtDNA sequences and gene orders, need to be explored more fully. Comparison of gene arrangements at low taxonomic levels can help to elucidate the process of gene rearrangement. For instance, the signature of specific processes such as tRNA remolding or recruitment can be most easily recognized when such events have occurred recently, since remolded or recruited tRNAs can be identified through high similarity scores and phylogenetic analyses [27,28]. Likewise, those taxonomic groupings with unusually labile genomes offer the opportunity to investigate the mechanics of gene rearrangement: telltale vestiges of gene duplication and translocation, typically erased or overwritten with time, may still be present within these genomes [35] and such intermediate stages can be critical to reconstructing the processes through which such gene rearrangements have occurred. Comparisons of mt genomes at low taxonomic levels, even within families and genera, can thus be extremely helpful in interpreting the evolutionary dynamics of these genomes and exploiting the phylogenetic signal retained within these DNA molecules [16].
Here we present further evidence of highly dynamic molluscan mt genomes by revealing extensive gene order changes within members of one caenogastropod family: the Vermetidae. Vermetids are a group of sessile, irregularly coiled, suspension-feeding gastropods found in warm temperate to tropical oceans around the world that radiated from a basal caenogastropod stock in the early Cenozoic Era. They are currently classified as members of the Hypsogastropoda [36,37], a large and diverse group with a fossil record extending back to the Permo-Triassic boundary that includes all extant caenogastropods, except for the Architaenioglossa, Cerithioidea and Campaniloidea. While relationships within the Hypsogastropoda are not well resolved, vermetids are typically positioned within the infraorder Littorinimorpha. More specifically, molecular analyses suggest that vermetids are members of a largely asiphonate clade of gastropods including the Littorinidae, Eatoniellidae, Rissoidae, Anabathridae, Hipponicidae, Pterotracheidae, Epitoniidae, Cerithiopsidae, Eulimidae, and Naticidae [37]. This association is also supported by morphological similarities in euspermatozoa shared by many members of this clade [38].
Gene order rearrangements have been recognized previously in this family [18] based on small (<3.5 kb) portions of the mt genome sequenced from several species within the genus Dendropoma. In this paper, we expand upon these earlier results by providing complete mt genomes for two Dendropoma species as well as for representatives of two other vermetid genera, Thylacodes and Eualetes. We also reveal additional gene rearrangements within this family through the partial genomes of the vermetid genera Thylaeodus and Vermetus. The extent of gene rearrangement within the family offers great potential for improving our phylogenetic hypothesis for the enigmatic Vermetidae as well as for understanding more fully the mechanics of gene order change within metazoan mt genomes.
Methods
Amplification and sequencing
Selection of taxa was based on: 1) the discovery of novel gene orders in these species following PCR amplifications spanning gene boundaries [18] and 2) those genomes that successfully amplified using long and accurate PCR (LAPCR). The collecting locality, tissue source, and Field Museum of Natural History (FMNH) voucher information for each specimen, are presented in Table 1, along with GenBank accession numbers, primer sequences, and lengths of amplification products. Other gene arrangements have been identified in additional vermetids based on partial genome sequences (<3 kb), but these results are not presented here (Rawlings et al., in prep).
Table 1.
Sampling localities, tissue sources, and PCR primer sequences for the six vermetid species examined in this study
| Species | FMNH voucher # GenBank accession # |
Locality | Tissue | Primer Set A (5'-3') rrnL-F/cox1-R |
Fragment Size (bp) | Primer Set B (5'-3') rrnL-R/cox1-F |
Fragment Size (bp) |
|---|---|---|---|---|---|---|---|
| Dendropoma maximum (Sowerby1825) | FMNH 318221 HM174253 |
Gulf of Aqaba, Jordan | Foot tissue; buccal mass | CGAATTGAAAGGGGGGCTTGTGACCTCGATGTTG TTTCGATCCGTTAAAAGCATAGTGATAGCTCC |
6877 | ACGCTACCTTCGCACGGTCAAAGTACCGCGGC TTGTTATGCCAATAATGATTGGTGGTTTCGG |
8809 |
| Dendropoma gregarium Hadfield & Kay 1972 | FMNH 318222 HM174252 |
HI, USA | Foot tissue; buccal mass | CAAATCGAAAAAAGGGTTTGCGACCTCGATGTTG TTACGGTCAGTTAAGAGTATGGTAATAGCACC |
6490 | ATGCTACCTTTGCACGGTCAGGGTACCGCGGC TGGTAATACCCATGATAATTGGAGGTTTTGG |
9254 |
| Eualetes tulipa (Chenu1843) | FMNH 318223 HM174254 |
Peanut Island, Palm Beach Co., FL, USA | Buccal mass | CATATCGAAAGAATAGTTTGCGACCTCGATGTTG TTTCGGTCCGTCAACAATATTGTAATTGCCCC |
6880 | TTCAACGAGAGCGACGGGCGATATGTACAC (rrnS-R) TGGTAATGCCTATAATGATTGGGGGGTTCGG |
7472 |
| Thylacodes squamigerus1 (Carpenter, 1857) | FMNH 318997 HM174255 |
Corona del Mar, CA, USA | Foot tissue | CCCATCGAAAGAAGAGTTTGTGACCTCGATGTTG TTTCGGTCCGTCAACAGCATAGTAATAGCTCC |
6648 | ATGCTACCTTTGCACGGTCAGAGTACCGCGGC TGGTTATACCAATAATAATTGGTGGCTTCGG |
9034 |
|
Thylaeodus sp. |
FMNH 318224 HM174256 |
Kewalo Marine Laboratory, HI, USA | Head, foot & mantle margin | CATATTGAAAAAAAAGTTTGTGACCTCGATGTTG TTTCGATCAGTCAATAACATAGTAATTGCGCC |
4703 | ATGCTACCTTTGCACGGTCAGAGTACCGCGGC TAGTAATACCTATAATAATTGGTGGATTTGG |
N/A |
| Vermetus erectus Dall, 1888 | FMNH 318225 HM174257 |
Pourtales Terrace, Florida Keys; FL, USA | Foot tissue; buccal mass | CTAATCGAAGAAAAGGCTTGTGACCTCGATGTTG TTTCGGTCAGTCAGTAGTATAGTAATAGCACC |
3107 | ATGCTACCTTAGCACGGTTAAAATACCGCGGC TTGTCATGCCTATAATAATTGGCGGATTTGG |
N/A |
1Thylacodes has been demonstrated to have priority over Serpulorbis; hence Serpulorbis squamigerus is now correctly referred to as Thylacodes squamigerus (see [62]).
DNA was extracted from ethanol-preserved tissues using a phenol chloroform extraction protocol as described in [39]. Initially, an 1800 bp region of the mitochondrial genome spanning the rrnS to nad1 region was amplified as part of a phylogenetic analysis of Vermetidae [18]. This sequence was subsequently used to design outwardly facing primers for LAPCR (Table 1: rrnL-F; rrnL-R). Because attempts to amplify the genome in one piece were not successful, we amplified a 650 bp fragment of cox1 using modifications of Folmer's widely used cox1 primers [40]. This cox1 sequence was then used as a template for designing a second pair of primers (cox1-F; cox1-R). Successful amplifications were associated with the primer combinations: rrnL-F/cox1-R ("A" fragment) and rrnL-R/cox1-F ("B" fragment). LAPCR reactions were undertaken using GeneAmp XL PCR kits (Applied Biosystems; N8080193). Typically, 25 μL reactions were set up in two parts separated by a wax bead following the manufacturer's recommendations, using a [Mg(OAc)2] of 1.2 mM and an annealing temperature specific to the primer combination. Typical conditions consisted of a 94°C denaturation period lasting 60 s, followed by: 16 cycles at 94°C for 25 s, 60°C for 60 s, and 68°C for 10 min; 18 cycles at 94°C for 25 s, 60°C for 60 s, and 68°C for 12 min; and a final extension period at 72°C for 10 min. Amplifications were run on a Stratagene Gradient Robocycler. PCR products were cleaned by separating high molecular weight products from primers and sequencing reagents using Millipore Ultrafree filter columns [7]. Samples were added to 200 μL of sterile water and then spun in a picofuge for 15 min or until the filter membranes were dry. PCR products were eluted from the membrane in 20 μL of sterile distilled water, and 5 μL of this product was run out on a 0.8% agarose gel to confirm the presence of a band of the appropriate size. Typically, products from several replicate PCR reactions were pooled prior to quantitation. Once 3 ng of PCR product had been obtained, samples were dried down in a vacuum centrifuge and sent to the Joint Genome Institute, Walnut Creek, CA, where they were sequenced using standard shotgun sequencing protocols [7,8].
Genome annotation and analysis
Genome annotation
The approximate locations of the rRNA and protein-encoding genes were determined by aligning each unannotated genome with genes from other caenogastropod mt genomes. The precise boundaries of rRNA genes could not be determined due to the lack of sequence similarity at the 5' and 3' ends; therefore, the location of each rRNA gene was assumed to extend from the boundary of the upstream flanking gene to the boundary of the downstream flanking gene, as in [41]. A standard initiation codon was located at the beginning of each protein-encoding gene (either ATG, ATA, or GTG), and the derived amino acid sequence was aligned with homologous protein sequences to ensure that this was a suitable initiation codon (based on length). When possible, the first proper stop codon (TAG or TAA) downstream of the initiation codon was chosen to terminate translation; however, to reduce overlap with downstream genes, abbreviated stop codons (T or TA) were selected for some genes. In these instances, polyadenylation of the mRNA was assumed to restore a full TAA stop codon [7]. Regardless, some genes appeared to overlap based on the conservation of their open reading frame sequences and the lack of a potential abbreviated stop codon. Gene locations and secondary structures of tRNAs were identified using tRNAscanSE [42] and ARWEN [43]. On the rare occasion that these programs did not find all of the expected tRNAs, the remaining tRNAs were found by eye and folded manually. We produced tRNA drawings manually using Canvas (ACD Systems). Using cox1 as the conventional starting point for the four genomes, linear maps of the circular genomes were created to facilitate comparison of gene orders amongst vermetids, across available caenogastropods, and between caenogastropods and other select molluscs, including the abalone, Haliotis rubra (Class Gastropoda,Superorder Vetigastropoda), the octopus, Octopus vulgaris (Class Cephalopoda), and the chiton, Katharina tunicata (Class Polyplacophora).
Nucleotide composition
The nucleotide composition of each complete and partial genome was described by calculating the overall base composition, %AT content, AT-skew, and GC-skew for the strand encoding cox1, hereafter referred to as the "+" strand. Base composition and %AT content were determined using MacVector (MacVector, Inc.), and strand skews (AT skew = (A-T)/(A+T); GC skew = (G-C)/G+C)) were calculated using the formulae of [44]. For complete genomes, the %AT content, AT-skew, and GC-skew were also calculated for rRNA genes, protein-encoding genes (for all bases, third codon positions, and third positions of four-fold degenerate (4FD) codons, as in [44]), tRNA genes (separately for those coded for on the "+" and "-" strand), and intergenic (unassigned) regions. Values were compared across categories within each genome, and within categories across genomes. In addition, we explored the nucleotide composition at third positions of 4FD codons in relation to the position of each protein-encoding gene within the genome [45]. Gene positions were determined by calculating the distance (number of nucleotides) from the midpoint of each gene to a reference point chosen here as the start of nad1.
A base composition plot (%A+C and %G+T along the length of the genome) was created for each complete genome using a sliding window of 100 nucleotides. Typically, the leading strand is G+T rich associated with its protracted single-stranded state during replication and transcription; deviations from this pattern can signify switches in the assignments of these strands [41]. Plots were aligned with linear representations of the matching genome to signify base composition trends for protein and tRNA genes encoded on the "+" strand, tRNA genes encoded on the "-" strand, and rRNA genes.
Genetic code
To ensure that all codons were being utilized, codon usage frequencies were analyzed using MacVector (MacVector, Inc.); unused codons can potentially signal a change in the genetic code [46]. Codons whose amino acid identity has changed in the mt genetic code of other metazoans (e.g. flatworms, echinoderms, and hemichordates; [47]) were investigated using a procedure similar to [48]: the derived amino acids of five codons (TGA, ATA, AGA, AGG, AAA) were examined in the alignment of three highly conserved proteins (cox1, cox2, and cox3) from the mt genomes of four vermetids, D. gregarium, D. maximum, E. tulipa and T. squamigerus, and several other caenogastropods. Within the vermetids, if the derived amino acid from any of these five codons occurred in a conserved position (present at that location in >50% of caenogastropod sequences examined) this was scored as a positive result. Amino acids that occurred in a non-conserved position (<50%) were scored as negative. The proportion of appearances in conserved vs. non-conserved locations was calculated for each codon across the vermetids examined. High percentages in conserved locations likely indicate the retention of amino acid identity by the specific codon [48]; low percentages can be suggestive of a change in amino acid identity, although other explanations are also possible.
Unassigned regions
As metazoan mt genomes are typically compact with minimal non-coding DNA [49], unassigned stretches of nucleotides often contain control elements for transcription or replication or remnants of duplicated protein, rRNA or tRNA genes. We examined putative non-coding regions (> 20 bp) for gene remnants, repeat sequences, inverted repeats, palindromes, and secondary structure features, all of which can be associated with signaling elements of the mt genome. Protein, rRNA and tRNA remnants were identified by aligning unassigned regions with annotated genes from caenogastropod mitochondrial genomes. MEME [50] and M-Fold [51] were used to identify potential sequence motifs and structural features, respectively.
tRNA remolding/recruitment
To search for close matches between tRNA genes indicative of gene remolding/recruitment, each tRNA gene sequence was aligned to tRNA genes from other available caenogastropod genomes (including new vermetid tRNA sequences). Similarity scores were based on initial tRNA alignments undertaken in MacVector that were subsequently adjusted by eye according to secondary structure features (stems vs. loops). As in [28], the third base of the anticodon triplet was excluded in the calculation of % similarity between two tRNA genes, but gaps were counted as mismatches.
Results and Discussion
Caenogastropod mitochondrial genomes
To date, comparisons across published caenogastropod mt genomes have suggested a model of gene order conservation unusual for the Gastropoda [15,48,52,53]. Among the 16 complete caenogastropod genomes available at NCBI as of January 8, 2010, gene order rearrangements within this clade appear minor, involving only changes in position of trnLUUR, trnLCUN, trnV and trnS2, and an inversion of trnT ([53]). Likewise, only two rearrangements, one inversion and one transposition, separate the reconstructed ancestral gastropod gene order from that of most caenogastropods [15,53]. While this conservation in gene order within the Caenogastropoda may be real, it could also reflect a strong sampling bias for members of the Neogastropoda (12 complete genomes) - a largely Cenozoic radiation of predatory snails - with only four complete genomes from two genera (Cymatium and Oncomelania), examined from other caenogastropod groups. In contrast, wider sampling of mt gene orders within the Heterobranchia, a sister clade to the Caenogastropoda, has demonstrated highly dynamic mt genomes [15]. Of 13 genomes sampled across a disparate array of taxa, including 5 opisthobranchs (including an unpublished Elysia genome), 7 pulmonates (including a second Biomphalaria genome not included in [15]), and 1 basal heterobranch [15], numerous changes in gene order have been observed, with few mt gene boundaries shared between heterobranchs and the hypothetical ancestral gastropod mt genome inferred by [15].
Here, based on a detailed sampling of mt genomes within one family of caenogastropods outside the Neogastropoda, we provide a new and very different picture of gene order dynamics within the Caenogastropoda. Our results increase the number of caenogastropod mt genomes sequenced to 20, substantially increase the sampling of mt genomes outside the Neogastropoda, and present the first direct evidence of major gene order rearrangements within the Littorinimorpha based on complete genome sequences. Full mt genomes were successfully sequenced for the vermetids, Dendropoma gregarium, D. maximum, Eualetes tulipa, and Thylacodes squamigerus (Figure 1; Tables 2, 3, 4 and 5). Amplifications of the "B" fragment (cox1-F/rrnL-R) were not successful for Thylaeodus sp. and V. erectus and consequently only partial mt genomes are described for these two taxa (Figure 1; Tables 6 and 7). Nevertheless, extensive gene rearrangements were evident within these species compared to other vermetids and caenogastropods. Characterization of the vermetid mt genomes presented below is based only on the four complete genomes, except where noted otherwise.
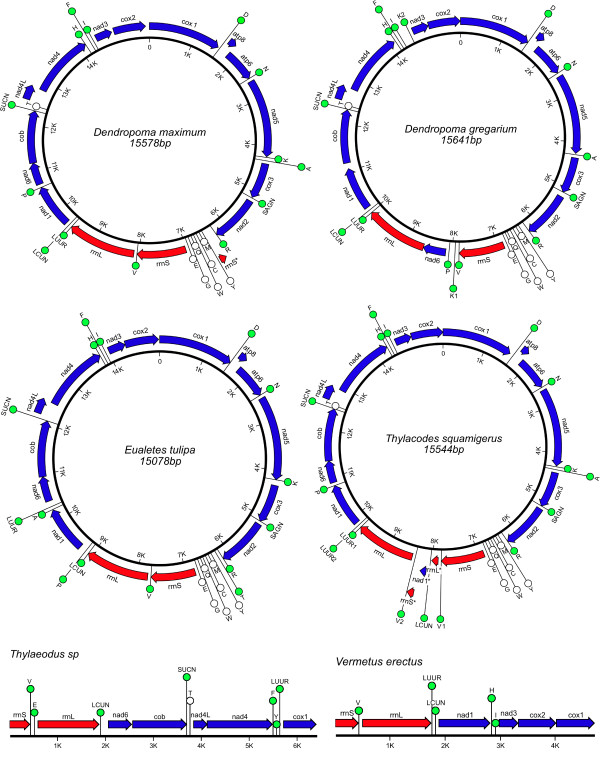
Figure 1.
Gene maps of four complete and two partial vermetid mt genomes. Blue arrows represent protein-encoding genes; red arrows represent rRNA genes; and circles represent tRNA genes. Arrows indicate the direction of transcription (clockwise = "+" strand). Genes are labeled according to their standard abbreviations. For tRNA genes, green circles represent those genes located on the "+" strand and white circles those located on the "-" strand. Regions of unassigned nucleotides (presumed non-coding sequences) are not identified on these maps. As is the standard convention for metazoan mt genomes, cox1 has been designated the start point for the "+" strand. The asterisks (*) in D. maximum and T. squamigerus denote stretches of sequences (presumed pseudogenes) with high similarity to portions of complete genes. For exact locations of each gene, as well as unassigned regions, see Tables 2-7.
Table 2.
Detailed description of genes and unassigned regions (UR) within the complete mt genome (15578 bp) of Dendropoma maximum
| Gene | Starta | End | Length | Amino acids | Start codon | End codonb | UR (%AT)c |
|---|---|---|---|---|---|---|---|
| cox1 | 1 | 1536 | 1536 | 512 | ATG | TAG | 38 (76.3) |
| trnD | 1575 | 1642 | 68 | - | - | - | 0 |
| atp8 | 1643 | 1798 | 156 | 52 | ATG | TAG | -7 |
| atp6 | 1792 | 2479 | 688 | 229 | ATG | T-- | 0 |
| trnN | 2480 | 2550 | 71 | - | - | - | 0 |
| nad5 | 2551 | 4251 | 1701 | 567 | ATG | TAA | 14 (78.6) |
| trnK | 4266 | 4340 | 75 | - | - | - | -2 |
| trnA | 4339 | 4404 | 66 | - | - | - | 1 |
| cox3 | 4406 | 5185 | 780 | 260 | ATG | TAA | 15 (86.7) |
| trnSAGN | 5201 | 5274 | 74 | - | - | - | 1 |
| nad2 | 5276 | 6295 | 1020 | 340 | ATG | TAG | 0 |
| trnR | 6296 | 6364 | 69 | - | - | - | 11 (54.5)/167 (69.3)d |
| rrnS* (pseudo) | 6376 | 6496 | 121 | - | - | - | 35 (74.3) |
| trnY* | 6532 | 6598 | 67 | - | - | - | 0 |
| trnM* | 6599 | 6668 | 70 | - | - | - | 2 |
| trnC* | 6671 | 6739 | 69 | - | - | - | 0 |
| trnW* | 6740 | 6807 | 68 | - | - | - | -4 |
| trnQ* | 6804 | 6874 | 71 | - | - | - | 0 |
| trnG* | 6875 | 6943 | 69 | - | - | - | 0 |
| trnE* | 6944 | 7016 | 73 | - | - | - | 0 |
| rrnS | 7017 | 8049 | 1033 | - | - | - | 0 |
| trnV | 8050 | 8116 | 67 | - | - | - | 0 |
| rrnL | 8117 | 9563 | 1447 | - | - | - | 0 |
| trnLUUR | 9564 | 9633 | 70 | - | - | - | -4 |
| trnLCUN | 9630 | 9698 | 69 | - | - | - | 1 |
| nad1 | 9700 | 10632 | 933 | 311 | ATG | TAA | 2 |
| trnP | 10635 | 10707 | 73 | - | - | - | 1 |
| nad6 | 10709 | 11189 | 481 | 160 | ATG | T-- | 0 |
| cob | 11190 | 12329 | 1140 | 380 | ATG | TAG | 2 |
| trnSUCN | 12332 | 12399 | 68 | - | - | - | 33 (54.5) |
| trnT* | 12433 | 12508 | 76 | - | - | - | 8 |
| nad4L | 12517 | 12810 | 294 | 98 | ATG | TAG | -7 |
| nad4 | 12804 | 14161 | 1358 | 452 | GTG | TA- | 0 |
| trnH | 14162 | 14231 | 70 | - | - | - | 46 (47.8) |
| trnF | 14278 | 14346 | 69 | - | - | - | 0 |
| trnI | 14347 | 14418 | 72 | - | - | - | 2 |
| nad3 | 14421 | 14774 | 354 | 118 | ATG | TAG | 46 (65.2) |
| cox2 | 14821 | 15498 | 678 | 226 | ATG | TAG | 80 (63.8) |
a Genes are arranged relative to cox1. Those encoded on the "-" strand are indicated with an asterisk (*).
b T, and TA, refer to instances where incomplete stop codons are inferred.
c Unassigned regions are identified by positive values. Negative values indicate overlap between adjacent genes. Values in brackets refer to %AT of the associated UR (only those UR >10 bp in length were analyzed).
d For the unassigned region between trnR and trnY spanning the pseudogene rrnS.
Table 3.
Detailed description of genes and unassigned regions (UR) within the complete mt genome (15641 bp) of Dendropoma gregarium
| Gene | Starta | End | Length | Amino acids | Start codon | End codonb | UR (%AT)c |
|---|---|---|---|---|---|---|---|
| cox1 | 1 | 1533 | 1533 | 511 | ATG | TAG | 3 |
| trnD | 1537 | 1611 | 75 | - | - | - | 2 |
| atp8 | 1614 | 1769 | 156 | 52 | ATG | TAA | -7 |
| atp6 | 1763 | 2452 | 690 | 230 | ATG | TAA | 7 |
| trnN | 2460 | 2535 | 76 | - | - | - | 2 |
| nad5 | 2538 | 4259 | 1722 | 574 | ATA | TAA | 27 (74.1) |
| trnA | 4287 | 4354 | 68 | - | - | - | 1 |
| cox3 | 4356 | 5134 | 779 | 260 | ATG | TA- | 0 |
| trnSAGN | 5135 | 5201 | 67 | - | - | - | 38 (60.5) |
| nad2 | 5240 | 6252 | 1013 | 338 | ATG | TA- | 0 |
| trnR | 6253 | 6321 | 69 | - | - | - | 55 (72.7) |
| trnY* | 6377 | 6443 | 67 | - | - | - | 1 |
| trnM* | 6445 | 6514 | 70 | - | - | - | 10 (30.0) |
| trnC* | 6525 | 6597 | 73 | - | - | - | 1 |
| trnW* | 6599 | 6667 | 69 | - | - | - | 11 (50.0) |
| trnQ* | 6679 | 6735 | 57 | - | - | - | -3 |
| trnG* | 6733 | 6798 | 66 | - | - | - | 2 |
| trnE* | 6801 | 6872 | 72 | - | - | - | 0 |
| rrnS | 6873 | 7851 | 979 | - | - | - | 0 |
| trnV | 7852 | 7922 | 71 | - | - | - | 16 (50.0) |
| trnK1 | 7939 | 8013 | 75 | - | - | - | 55 (65.5) |
| trnP | 8069 | 8139 | 71 | - | - | - | 1 |
| nad6 | 8141 | 8629 | 489 | 163 | ATG | TAG | 0 |
| rrnL | 8630 | 9978 | 1349 | - | - | - | 0 |
| trnLUUR | 9979 | 10049 | 71 | - | - | - | -2 |
| trnLCUN | 10048 | 10116 | 69 | - | - | - | 2 |
| nad1 | 10119 | 11049 | 931 | 310 | ATG | T-- | 125 (52.8) |
| cob | 11175 | 12323 | 1149 | 383 | ATG | TAA | 13 (84.6) |
| trnSUCN | 12337 | 12407 | 71 | - | - | - | 4 |
| tRNA T* | 12412 | 12479 | 68 | - | - | - | 7 |
| nad4L | 12487 | 12780 | 294 | 98 | ATG | TAA | -7 |
| nad4 | 12774 | 14129 | 1356 | 452 | ATG | TAG | 30 (66.7) |
| trnH | 14160 | 14227 | 68 | - | - | - | 0 |
| trnF | 14228 | 14296 | 69 | - | - | - | 6 |
| trnI | 14303 | 14375 | 73 | - | - | - | 137 (58.4) |
| trnK2 | 14513 | 14586 | 74 | - | - | - | 2 |
| nad3 | 14589 | 14939 | 351 | 117 | ATG | TAG | 7 |
| cox2 | 14947 | 15636 | 690 | 230 | ATG | TAG | 5 |
a Genes are arranged relative to cox1. Those encoded on the "-" strand are indicated with an asterisk (*).
b T, and TA, refer to instances where incomplete stop codons are inferred.
c Unassigned regions are identified by positive values. Negative values indicate overlap between adjacent genes. Values in brackets refer to %AT of the associated UR (only those UR >10 bp in length were analyzed).
Table 4.
Detailed description of genes and unassigned regions (UR) within the complete mt genome (15078 bp) of Eualetes tulipa
| Gene | Starta | End | Length | Amino acids | Start codon | End codonb | UR (%AT)c |
|---|---|---|---|---|---|---|---|
| cox1 | 1 | 1531 | 1531 | 510 | ATG | T-- | 0 |
| trnD | 1532 | 1596 | 65 | - | - | - | 0 |
| atp8 | 1597 | 1752 | 156 | 49 | ATG | TAG | -7 |
| atp6 | 1746 | 2430 | 685 | 228 | ATG | T-- | 0 |
| trnN | 2431 | 2497 | 67 | - | - | - | 0 |
| nad5 | 2498 | 4205 | 1708 | 569 | ATG | T-- | 0 |
| trnK | 4206 | 4270 | 65 | - | - | - | 51 (62.1) |
| cox3 | 4322 | 5104 | 783 | 261 | ATG | TAA | 0 |
| trnSAGN | 5105 | 5168 | 64 | - | - | - | 0 |
| nad2 | 5169 | 6176 | 1008 | 336 | ATG | TAA | 0 |
| trnR | 6177 | 6242 | 66 | - | - | - | 4 |
| trnT | 6247 | 6312 | 66 | - | - | - | 33 (66.7) |
| trnY* | 6346 | 6411 | 66 | - | - | - | 2 |
| trnM* | 6414 | 6480 | 67 | - | - | - | 0 |
| trnC* | 6481 | 6546 | 66 | - | - | - | 0 |
| trnW* | 6547 | 6609 | 63 | - | - | - | -5 |
| trnQ* | 6605 | 6671 | 67 | - | - | - | 0 |
| trnG* | 6672 | 6735 | 64 | - | - | - | 6 |
| trnE* | 6742 | 6807 | 66 | - | - | - | 0 |
| rrnS | 6808 | 7716 | 909 | - | - | - | 0 |
| trnV | 7717 | 7783 | 67 | - | - | - | 0 |
| rrnL | 7784 | 9087 | 1304 | - | - | - | 0 |
| trnLCUN | 9088 | 9152 | 65 | - | - | - | 9 |
| trnP | 9162 | 9225 | 64 | - | - | - | 73 (71.2) |
| nad1 | 9299 | 10231 | 933 | 311 | ATA | TAG | 10 (50.0) |
| trnA | 10242 | 10304 | 63 | - | - | - | 2 |
| trnLUUR | 10307 | 10371 | 65 | - | - | - | 56 (58.9) |
| nad6 | 10428 | 10934 | 507 | 169 | ATG | TAG | 2 |
| cob | 10937 | 12076 | 1140 | 380 | ATG | TAG | 0 |
| trnSUCN | 12077 | 12143 | 67 | - | - | - | 18 (66.7) |
| nad4L | 12162 | 12452 | 291 | 97 | ATG | TAG | -7 |
| nad4 | 12446 | 13810 | 1365 | 455 | ATG | TAG | -1 |
| trnH | 13810 | 13875 | 66 | - | - | - | -1 |
| trnF | 13875 | 13940 | 66 | - | - | - | 0 |
| trnI | 13941 | 14012 | 72 | - | - | - | 0 |
| nad3 | 14013 | 14366 | 354 | 118 | ATG | TAG | 3 |
| cox2 | 14370 | 15058 | 689 | 229 | ATG | TA- | 20 (65.0) |
a Genes are arranged relative to cox1. Those encoded on the "-" strand are indicated with an asterisk (*).
b T, and TA, refer to instances where incomplete stop codons are inferred.
c Unassigned regions are identified by positive values. Negative values indicate overlap between adjacent genes. Values in brackets refer to %AT of the associated UR (only those UR >10 bp in length were analyzed).
Table 5.
Detailed description of genes and unassigned regions (UR) within the complete mt genome (15544 bp) of Thylacodes squamigerus
| Gene | Starta | End | Length | Amino acids | Start codon | End codonb | UR (%AT)c |
|---|---|---|---|---|---|---|---|
| cox1 | 1 | 1537 | 1537 | 512 | ATG | T-- | 78 (57.5) |
| trnD | 1616 | 1682 | 67 | - | - | - | 1 |
| atp8 | 1684 | 1839 | 156 | 52 | ATG | TAG | -7 |
| atp6 | 1833 | 2531 | 699 | 233 | ATG | TAA | 12 (83.3) |
| trnN | 2544 | 2610 | 67 | - | - | - | 1 |
| nad5 | 2612 | 4306 | 1695 | 565 | ATG | TAG | 17 (47.1) |
| trnK | 4324 | 4389 | 66 | - | - | - | -3 |
| trnA | 4387 | 4453 | 67 | - | - | - | 0 |
| cox3 | 4454 | 5236 | 783 | 261 | ATG | TAA | -2 |
| trnSAGN | 5235 | 5299 | 65 | 0 | |||
| nad2 | 5300 | 6298 | 999 | 333 | ATG | TAG | -1 |
| trnR | 6298 | 6365 | 68 | - | - | - | 35 (74.3) |
| trnY* | 6401 | 6467 | 67 | - | - | - | -2 |
| trnM* | 6466 | 6531 | 66 | - | - | - | 6 |
| trnC* | 6538 | 6599 | 62 | - | - | - | 2 |
| trnW* | 6602 | 6669 | 68 | - | - | - | -5 |
| trnQ* | 6665 | 6729 | 65 | - | - | - | 8 |
| trnG* | 6738 | 6802 | 65 | - | - | - | 3 |
| trnE* | 6806 | 6873 | 68 | - | - | - | 0 |
| rrnS | 6874 | 7809 | 936 | - | - | - | 0 |
| trnV1 | 7810 | 7877 | 68 | - | - | - | 0/163 (69.9)d |
| rrnL (pseudo) | 7878 | 8017 | 140 | - | - | - | 23 (82.6) |
| trnLCUN | 8041 | 8105 | 65 | - | - | - | 0/270 (58.9)e |
| nad1 (pseudo) | 8106 | 8225 | 120 | - | - | - | 36 (61.1) |
| rrnS (pseudo) | 8262 | 8375 | 114 | - | - | - | 0 |
| trnV2 | 8376 | 8443 | 68 | - | - | - | 0 |
| rrnL | 8444 | 9733 | 1290 | - | - | - | 0 |
| trnLUUR1 | 9734 | 9801 | 68 | - | - | - | -4 |
| trnLUUR2 | 9798 | 9862 | 65 | - | - | - | 0 |
| nad1 | 9863 | 10798 | 936 | 312 | ATG | TAG | 1 |
| trnP | 10800 | 10867 | 68 | - | - | - | 2 |
| nad6 | 10870 | 11350 | 481 | 160 | ATG | T-- | 0 |
| cob | 11351 | 12490 | 1140 | 380 | ATG | TAG | 0 |
| trnSUCN | 12491 | 12561 | 71 | - | - | - | -2 |
| trnT* | 12560 | 12623 | 64 | - | - | - | 5 |
| nad4L | 12629 | 12919 | 291 | 97 | ATG | TAG | -7 |
| nad4 | 12913 | 14277 | 1365 | 455 | ATG | TAA | 5 |
| trnH | 14283 | 14344 | 62 | - | - | - | 2 |
| trnF | 14347 | 14416 | 70 | - | - | - | 6 |
| trnI | 14423 | 14492 | 70 | - | - | - | 2 |
| nad3 | 14495 | 14848 | 354 | 118 | ATG | TAG | 6 |
| cox2 | 14855 | 15544 | 690 | 230 | ATG | TAA | 0 |
a Genes are arranged relative to cox1. Those encoded on the "-" strand are indicated with an asterisk (*).
b T, and TA, refer to instances where incomplete stop codons are inferred.
c Unassigned regions are identified by positive values. Negative values indicate overlap between adjacent genes. Values in brackets refer to %AT of the associated UR (only those UR >10 bp in length were analyzed).
d For the unassigned region between trnV1 and trnLCUN spanning the pseudogene rrnL.
e For the unassigned region between trnLCUN and trnV2 spanning the pseudogenes nad1 and rrnS.
Table 6.
Detailed description of genes and unassigned regions (UR) within the partial mt genome of Thylaeodus sp.
| Gene | Starta | End | Length | Amino acids | Start codon | End codonb | UR (%AT)c |
|---|---|---|---|---|---|---|---|
| rrnS (partial) | 1 | 431 | 431 | - | 5 | ||
| trnV | 437 | 504 | 68 | - | 13 (53.8) | ||
| trnE | 518 | 591 | 74 | - | 0 | ||
| rrnL | 592 | 1882 | 1291 | - | 16 (56.3) | ||
| trnLCUN | 1899 | 1964 | 66 | - | 99 (70.7) | ||
| nad6 | 2064 | 2562 | 499 | 166 | ATG | T-- | 0 |
| cob | 2563 | 3703 | 1141 | 380 | ATG | T-- | -6 |
| trnSUCN | 3698 | 3766 | 69 | - | 1 | ||
| trnT* | 3768 | 3834 | 67 | - | 11 (100.0) | ||
| nad4L | 3846 | 4131 | 286 | 95 | ATG | T-- | -2 |
| nad4 | 4130 | 5491 | 1362 | 454 | ATG | TAA | 6 |
| trnF | 5498 | 5565 | 68 | - | - | - | 9 |
| trnY | 5575 | 5639 | 65 | - | - | - | -1 |
| trnLUUR | 5639 | 5708 | 70 | - | - | - | 11 (63.6) |
| cox1 (partial) | 5720 | 6415 | 696 | 232 | ATG | N/A | 0 |
a Genes are not arranged relative to cox1 because only a partial genome is available. Those genes encoded on the "-" strand are indicated with an asterisk (*).
b T, and TA, refer to instances where incomplete stop codons were inferred.
c Unassigned regions are identified by positive values. Negative values indicate overlap between adjacent genes. Values in brackets refer to %AT of the associated UR (only those UR >10 bp in length were analyzed).
Table 7.
Detailed description of genes and unassigned regions (UR) within the partial mt genome of Vermetus erectus
| Gene | Starta | End | Length | Amino acids | Start codon | End codonb | UR (%AT)c |
|---|---|---|---|---|---|---|---|
| rrnS | 1 | 429 | 429 | - | - | - | 0 |
| trnV | 430 | 499 | 70 | - | - | - | 0 |
| rrnL | 500 | 1756 | 1257 | - | - | - | 0 |
| trnLUUR | 1757 | 1819 | 63 | - | - | - | 3 |
| trnLCUN | 1823 | 1887 | 65 | - | - | - | 1 |
| nad1 | 1889 | 2824 | 936 | 312 | ATG | TAG | 18 (66.7) |
| trnH | 2843 | 2904 | 62 | - | - | - | 10 (60.0) |
| trnI | 2915 | 2980 | 66 | - | - | - | 0 |
| nad3 | 2981 | 3331 | 351 | 117 | ATG | TAG | 3 |
| cox2 | 3335 | 4022 | 688 | 229 | ATG | T-- | 0 |
| cox1 (partial) | 4023 | 4718 | 696 | 232 | ATG | N/A | 0 |
a Genes are not arranged relative to cox1 because only a partial genome is available. Those genes encoded on the "-" strand are indicated with an asterisk (*).
b T, and TA, refer to instances where incomplete stop codons were inferred.
c Unassigned regions are identified by positive values. Negative values indicate overlap between adjacent genes. Values in brackets refer to %AT of the associated UR (only those UR >10 bp in length were analyzed).
Genome organization
The mitochondrial genomes of D. gregarium, D. maximum, E. tulipa, and T. squamigerus varied in size from 15078 bp to 15641 bp, similar in length to other caenogastropods (range: 15182 - 16648 bp, n = 16) but considerably larger than most heterobranchs (range: 13670 - 14745 bp, n = 13). Each genome contained the 37 genes typical of most animal mitochondrial genomes (Figure 1; Tables 2, 3, 4 and 5), with all 13 protein-encoding genes and 2 rRNA genes located on the "+" strand along with either 14 or 15 tRNA genes depending on the species. In each of the four genomes, the tRNA genes, trnY, trnM, trnC, trnW, trnQ, trnG, and trnE, were located on the "-" strand forming a cassette of 7 adjacent tRNAs. The only difference across genomes in the strand specific coding of a gene was for trnT, which was located on the "+" strand of E. tulipa, but on the "-" strand of D. gregarium, D. maximum, and T. squamigerus. The tendency for protein and rRNA genes to be coded for on the same strand has been found in all caenogastropod taxa examined so far, but this is atypical for molluscs (excluding bivalves) and other metazoan mt genomes described to date (137 of 1428 genomes as of Dec 17, 2008; [54]). The predominance of this single-strand-dependence for protein and rRNA genes among sponges and cnidarians has led to the proposition that this is the plesiomorphic metazoan condition [54].
The tRNA genes identified within each genome and their secondary structures are presented as additional files (see Additional file 1, Figure S1; Additional file 2, Figure S2; Additional file 3, Figure S3; Additional file 4, Figure S4; Additional file 5, Figure S5; Additional file 6, Figure S6). Extra tRNA genes were found in two vermetid genomes. D. gregarium had a second trnK located between genes trnI and nad3 (see Figure 1). This second trnK can be folded in to a typical tRNA cloverleaf structure (see Additional file 2, Figure S2), and thus may be functional. The genome of T. squamigerus also contained additional trnV and trnLUUR genes (see Figure 1). These genes were associated with two large stretches of unassigned sequence positioned between the two copies of trnV (see below).
Overlapping adjacent genes were common in vermetid mt genomes. Two pairs of protein-encoding genes overlapped in all four species (atp8 and atp6; nad4L and nad4); overlap of nad4L and nad4 was also evident in the partial genome of Thylaeodus. These gene pairs commonly overlap in animal mt genomes. This phenomenon was more variable between tRNA genes (see Tables 2, 3, 4, 5, 6 and 7 for specific examples). In addition, in three instances tRNA genes overlapped with protein-encoding genes (trnH and nad4 in E. tulipa; trnSAGN with cox3 and trnR with nad2 in T. squamigerus). Similar comparisons could not be made for the two rRNA genes since their boundaries were only imprecisely defined by boundaries with neighbouring genes.
Gene initiation and termination
Nearly all protein-encoding genes (58/61) were initiated by the canonical ATG start codon, although ATA (twice) and GTG (once) also acted as start codons (Tables 2, 3, 4, 5, 6 and 7). These start codons are not unusual in molluscan mt genomes, but their usage, as found elsewhere, appears to be much less frequent than ATG [7] (but see [15]). TAG was the most common termination codon (28/59), but TAA (15/59) and abbreviated stop codons (a single T or TA terminating the open reading frame; 16/59) were used slightly more frequently when considered together. The sequences for the cox1 genes of Thylaeodus and Vermetus were incomplete so the termination codons for these genes are currently unknown.
Unassigned regions
The number of nucleotides unassignable to any gene ranged from 289 (1.9% of the genome) in Eualetes tulipa to 625 (4.0% of the genome) in Thylacodes squamigerus (Table 8). No unassigned stretch of sequence was greater than 270 bp in length, although intergenic regions ranging in size from 10 - 99 bp were common (Table 8). There was little consistency in size and position of unassigned regions across genomes, with the position of the largest region found in different areas for each species. Short (≤ 15 bp) AT-rich (>80%) intergenic regions were evident within the genomes of Dendropoma maximum, D. gregarium, and T. squamigerus, but not in E. tulipa (Tables 2, 3, 4 and 5), and again, the position of these sequences varied across taxa. Stretches of unassigned sequence including inverted repetitive elements are known to reside between trnF and cox3 in the mt genomes of several caenogastropods, likely representing the control region for replication and transcription in these genomes [52,53]. In our comparisons across the four complete vermetid genomes, we were unable to identify any similar regions associated with repetitive sequence motifs or palindromes. As is common in mt genomes, however, secondary structure elements were found by MFOLD within many of these intergenic regions, suggestive of a role in post-transcriptional modification of the polycistronic transcript.
Table 8.
A comparison of unassigned regions (UR) within the complete mt genomes of four vermetid gastropods
| UR length distributionc | Longest URc | |||||||
|---|---|---|---|---|---|---|---|---|
| Species | % UR (bp)a | #b | 10-19 bp | 20-39 bp | 40-99 bp | > 99 bp | bp | Location |
| Dendropoma maximum d | 2.95 (459) | 8 | 2 | 2 | 3 | 1 | 167 | trnR - trnY |
| Dendropoma gregarium | 3.65 (571) | 11 | 4 | 3 | 2 | 2 | 137 | trnI - trnK2 |
| Eualetes tulipa | 1.92 (289) | 7 | 2 | 2 | 3 | 0 | 73 | trnP - nad1 |
| Thylacodes squamigerus d | 4.02 (625) | 6 | 2 | 1 | 1 | 2 | 270 | trnLCUN - trnV2 |
a Expressed as a percent of the whole genome and the total number of base pairs.
b Includes all unassigned sequences ≥ 10 bp in length.
c The location and size of the UR is defined by upstream and downstream neighbouring genes.
d Data for Dendropoma maximum and Thylacodes squamigerus include pseudogene sequences found within some unassigned regions (see Tables 2 and 5).
Our analyses did uncover several interesting features within large intergenic regions of Dendropoma maximum and Thylacodes squamigerus (Figure 1). In D. maximum, we identified a 121 bp stretch within an unassigned region of 167 bp between trnR and trnY that was a perfect match to the reverse complement of a portion of rrnS. Likewise, in Thylacodes squamigerus, in an unassigned region between trnV1 and trnLCUN, we found a 140 bp stretch of sequence that was identical to a corresponding portion of rrnL. In addition, between trnLCUN and trnV2, we discovered a 120 bp remnant of nad1 (82% identical, with a 91 bp stretch differing only in 6 bases) and a 114 bp remnant of rrnS (92% identical). These pseudogene fragments in Thylacodes, as well as the extra tRNAs (trnV and trnLUUR) present in this region, are likely the result of a duplication event spanning rrnS-trnV-rrnL-trnLUUR-trnLCUN-nad1, with subsequent overwriting of some gene duplicates (Rawlings et al., in prep).
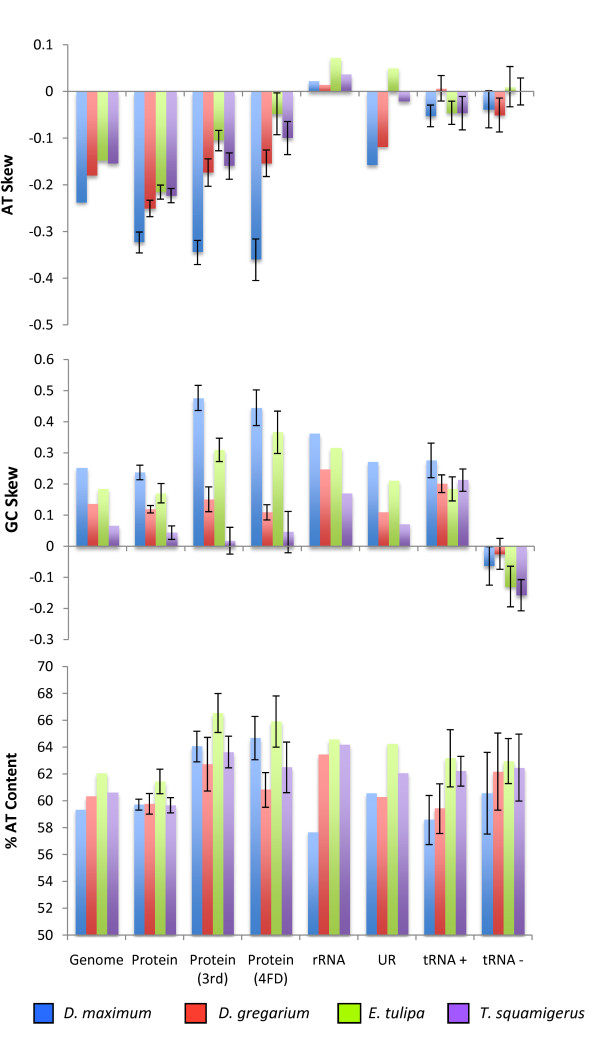
Nucleotide composition and skews
All four vermetid genomes were AT-rich, with these two nucleotides accounting for 59 - 62% of the genome (Figure 2; Additional file 7, Table S1); this trend was consistent across regions of each genome associated with protein-encoding genes, rRNA genes, tRNAs, and non-coding regions. Interestingly, however, the AT-biases of vermetid genomes were noticeably lower than those reported for other complete caenogastropod genomes (range: 65.2-70.1%, n = 14; Additional file 7, Table S1). Likewise, AT content was only moderately higher at third-codon positions and 4FD sites of protein-encoding genes, a result unusual in comparison with many other protostomes where %AT can often exceed >80% at 4FD sites [44]. Skew analyses revealed that the "+" strand was strongly biased against A (AT-skew ranging from -0.148 to -0.238) and towards G (GC-skew ranging from +0.065 to +0.251). This pattern was similar to other caenogastropods (Additional file 7, Table S1), heterobranchs [15], and the chiton, Katharina tunicata [55], but opposite to that found on the "+" strand of the abalone Haliotis [56] and the cephalopods, Nautilus and Octopus, suggestive of switches in the assignments of leading and lagging strands within the Mollusca [41].
Figure 2.
Nucleotide composition of four complete vermetid genomes. For each species, the %AT content, AT-skew, and GC-skew are shown for the "+" strand of the complete genome and its functional components, including: protein-encoding genes (complete protein, 3rd codon positions only, and 3rd positions of 4-fold degenerate (4FD) codons only), rRNA genes, unassigned regions (UR), and tRNA genes (including "+" and "-" strand encoded genes). Error bars refer to standard errors based on a sample size from each genome of: 13 protein encoding genes, 14 -15 "+" strand tRNA genes, and 7-8 "-" strand tRNA genes.
Skew patterns were different between regions of the genome depending on their function. For AT-skews, the most strongly biased areas were found associated with protein-encoding genes, but these were not necessarily most pronounced at third positions or at 4FD sites (except for D. maximum; Figure 2). The two rRNA genes showed a marked difference from protein-encoding genes, with a slight positive AT-skew, on average. Differences in skew patterns between rRNA and protein-encoding genes have been noted elsewhere [41,57], and may reflect base-pairing constraints associated with the secondary structures of these rRNAs [41]. Unassigned regions, assumed to be non-coding, were expected to experience similar selective pressures to those of 4FD third codon positions; this correspondence was not strong, however, with unassigned regions showing variation in AT-bias across taxa from slight positive AT-skews to moderate negative skews. tRNA genes regardless of whether they were coded for on the "+" or "-" strand exhibited no consistent skew patterns, with an average close to zero.
GC-skews were moderately positive for most regions of the genome, including protein-encoding genes, rRNA genes, unassigned regions, and tRNAs encoded on the "+" strand. Skews were particularly strong, however, for the third codon positions and 4FD third codon positions in Dendropoma maximum and Eualetes tulipa. On average, neutral to negative GC-skews were associated with regions of the "+" strand associated with tRNA genes encoded on the "-" strand.
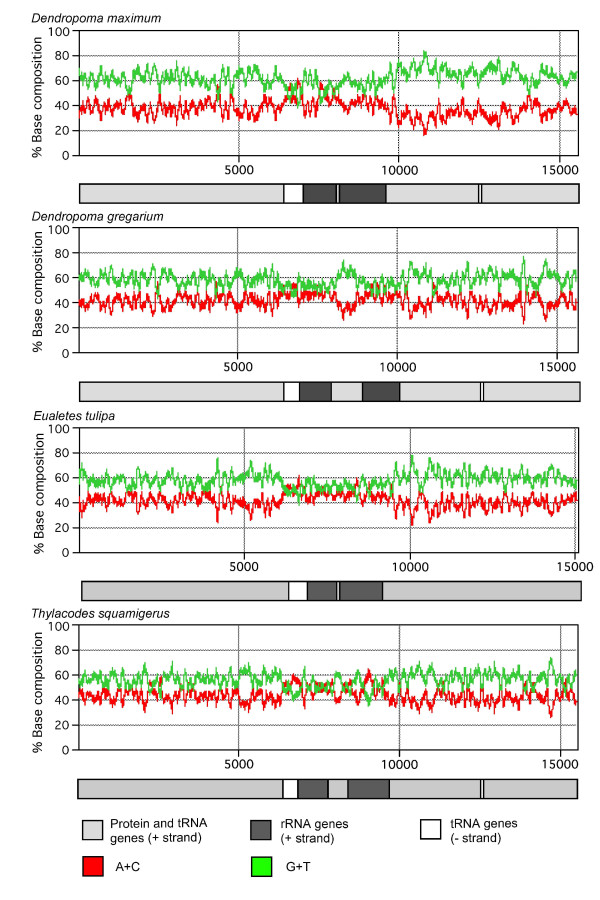
As predicted by skew patterns described above, G+T composition varied across the length of the "+" strand in all four complete genomes (Figure 3). Areas of weak or no G+T bias were typically associated with rRNAs and regions with tRNAs encoded on the "-" strand. Dendropoma maximum exhibited a particularly strong bias for G+T (>65%) in the region from 10,000 to 14,000 bp.
Figure 3.
Plots of A+C and G+T composition along the "+" strand of the mt genomes of D. maximum, D. gregarium, E. tulipa, and T. squamigerus using a sliding window of 100 nucleotides. Each plot is positioned above a linear representation of its corresponding genome highlighting regions corresponding to the presence of: 1) protein encoding genes and tRNA genes encoded on the "+" strand, 2) rRNA genes encoded on the "+" strand, and 3) tRNA genes encoded on the "-" strand. See [41] for a comparison with Nautilus and Katharina. As is the standard convention for metazoan mt genomes, cox1 has been designated the start point for the "+" strand.
Codon frequency
No evidence was found of changes to the standard genetic code employed by other molluscs (Additional file 8, Table S2). Each codon was used multiple times in the protein-encoding genes of each species (Additional file 9, Table S3), however, codon usage frequencies were not equal. Codon usage strongly reflected skew patterns at the third position for synonymous codons (both two-fold and four-fold degenerate codons; see Additional file 9, Table S3): the most abundant base at the third codon position across all four complete genomes was T (36.7-43.1%) and the least abundant base was C (9.4-17.9%). The relative ranking of G (18.5-26.5%) and of A (21.0-29.8%), however, was less consistent across genomes.
Putative origins of replication
Locating the regions of the mt genomes associated with the replication origins (ORs, also known as control regions or A+T rich regions) for both "+" and "-" strands can be challenging based on a knowledge of the nucleotide sequence of mt genomes alone. Recognition elements can be in the form of conserved sequence blocks, regions rich in A+T, stable stem-loop structures containing T-rich loops, or repetitive elements/palindromes [58]. ORs can also be identified by association with rearrangement "hotspots" [23]. Such features are not definitive evidence, however, and can differ in utility across taxonomic groups. For instance, only 35% of tentatively identified molluscan control regions have been found to have palindromes and the average %AT content of these regions is in the range of 65-70%, close to the average genomic %AT content for molluscs (based on 9 molluscan genomes analyzed) [59]. Consequently, looking for palindromes or A+T rich regions may not be helpful in identifying the OR of molluscs. In contrast, 85% of insect control regions have palindromes and CR regions are associated with >80% AT [59].
Because none of the recognition elements described above were useful in locating putative ORs within the complete genomes of the four vermetids examined here, we attempted to locate ORs by examining the relationship between nucleotide composition and the position of protein-encoding genes within these genomes. Reyes et al. [45] determined that the AT and GC-skews at 4FD sites of protein-encoding genes of 25 mammalian mt genomes were significantly correlated with the duration of single stranded state of heavy-strand genes during replication, with increased duration reflecting, in part, the proximity of protein encoding genes to the origin of replication of the heavy strand (ORH), as well as their position relative to the OR of the light strand (ORL). Longer durations in the single stranded condition increase the vulnerability of DNAs to hydrolytic and oxidative damage creating the compositional asymmetries between the heavy and light strands (or leading and lagging strands). In vertebrates, the ORL is typically located two-thirds of the way around the genome from the ORH. In insects, the only protostomes examined in detail, however, the replication origins for both strands (leading and lagging) appear to be in close proximity, near the ends of a conserved block within the A+T rich region, with the lagging strand beginning after 95% of the leading strand has been replicated [60]. Comparative studies are lacking for molluscs.
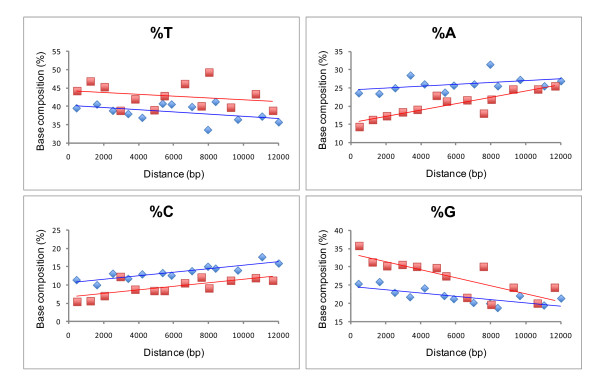
If the ORs for both strands are located close to one another in molluscs, then examining nucleotide composition in relation to the position of each protein-encoding gene could reveal putative sites for the OR. We tested this by plotting nucleotide composition at 4FD third codon positions as in [45] versus the midpoint position of each protein-encoding gene (Figure 4). By moving the putative OR between different protein-encoding genes, we found a compelling linear relationship, with a negative slope for %T and %G and a positive slope for %A and %C, when we positioned the OR between nad2 and nad1 (or between nad2 and nad6 for D. gregarium). This region generally encompasses the cassette of 7 tRNA genes encoded on the "-" strand, two rRNA genes, as well as additional tRNAs, albeit with some notable differences amongst taxa (Figure 1). This relationship is shown based on averaged values across all four complete vermetid genomes (Figure 4: diamonds), and separately for the genome of Dendropoma maximum (Figure 4: squares) which exhibited the strongest pattern across all four taxa. Reyes et al. [45] found an increase in the frequency of A and C at 4FD sites in the light (sense) strand in direct relation to the single-stranded duration of the heavy strand. They speculated that this was the result of spontaneous deamination of adenine into hypoxanthine, which base pairs with C rather than T, and cytosine into uracil, which base pairs with A rather than G, along the heavy strand during its single-stranded state in replication (with associated increases in nucleotides A and C in the light, sense strand). In vermetid genomes, where genes are encoded on the opposite strand to vertebrates (i.e. the heavy strand is the sense or "+" strand for all vermetid protein-encoding genes, see Figure 3), genes with lower frequencies of G and T (or higher frequencies of C and A) at 4FD sites should be those experiencing shorter durations in the single-stranded condition. The marked difference in nucleotide composition of 4FD sites between nad2 (low %G and %T) and nad1 (high %G and %T) is thus suggestive of substantial differences in exposure of these two genes to the single-stranded condition. Consequently, the OR may lie between these two protein-encoding genes. Comparisons of nucleotide composition patterns along the genomes of protostome taxa with well defined ORs are now necessary to confirm or refute the predictive power of such analyses in identifying control regions.
Figure 4.
Variation in nucleotide composition at 4FD sites of protein-coding genes in relation to the position of each gene within the genome. Data are presented for: 1) the mean of all four vermetid genomes (diamonds) and 2) Dendropoma maximum alone (squares). The position of each protein encoding gene was calculated as the number of nucleotides from the beginning of nad1 (or nad6 for D. gregarium) to the midpoint of the target gene. This different start point for D. gregarium was based on the derived translocation of nad6 to a position directly upstream from nad1 in this taxon [18]. Regression equations are as follows. All four genomes: Y = -2.92 × 10-4 X + 40.20, r2 = 0.21[%T]; Y = 2.59 × 10-4 X + 24.47, r2 = 0.18 [%A]; Y = 4.84 × 10-4 X + 10.60, r2 = 0.79 [%C]; Y = -4.51 × 10-4 X + 24.73, r2 = 0.56 [%G]; D. maximum alone: Y = -2.45 × 10-4 X + 44.22, r2 = 0.07 [%T]; Y = 8.66 × 10-4 X + 15.47, r2 = 0.78 [%A]; Y = 4.83 × 10-4 X + 6.63, r2 = 0.55 [%C]; Y = -1.10 × 10-3 X + 33.67, r2 = 0.65 [%G].
Two other observations support the general location for the OR between nad2 and nad1. First, this region is associated with a major change in base compositional bias reflecting the presence of rRNA genes and a cassette of tRNAs encoded on the "-" strand (Figure 3). Such changes are thought to be associated with ORs [41,58]. Second, this region of the mt genome appears to be involved in a number of gene order rearrangements, possibly reflecting a rearrangement "hotspot" (see Gene order rearrangements, below). Hotspots have been associated with ORs in previous studies [23]. In many other caenogastropods, however, ORs have been tentatively identified in a different region of the genome, despite the presence of a similar cassette of tRNA genes located on the "-" strand [48,52,53]. In these taxa, the OR is thought to be present between trnF and coxIII within an unassigned stretch of sequence of variable length (from 15 - 848 bp) associated with inverted repeats and secondary structure elements. This gene boundary is not present in the Vermetidae. Direct examination of mRNAs is now needed to demonstrate conclusively the presence of ORs within vermetid and other caenogastropod mt genomes.
Gene order rearrangements
Each of the vermetid genomes examined here possessed a unique gene arrangement (Figure 5). The four complete vermetid genomes differed primarily in the position of tRNA genes, the most mobile elements within the mt genome [16]. While the position of many tRNA genes was conserved, the location of trnA, trnK, trnP, trnT, trnLCUN, trnLUUR, and trnV was more variable across the four genomes. Only D. gregarium differed in the order of protein-encoding genes, with nad6 changing position relative to other protein encoding and rRNA genes, as described in [18]. The two partial genomes of Vermetus erectus and Thylaeodus sp. provided additional evidence of the extent of gene rearrangement within this family, however, with novel arrangements both tRNA and protein-encoding genes.
Figure 5.
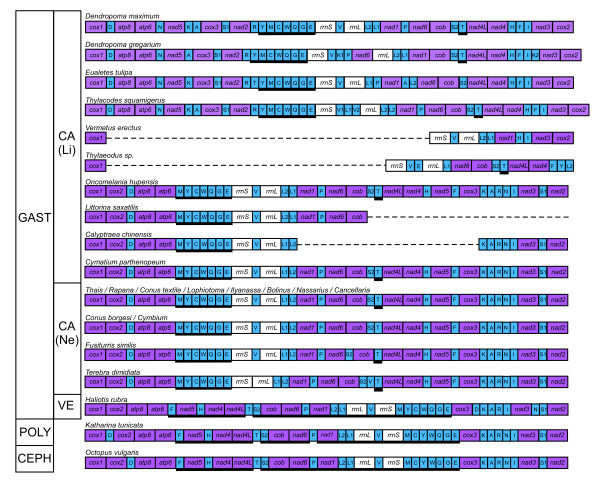
Linear arrangement of protein-encoding, rRNA and tRNA genes within the complete mt genomes of vermetid gastropods, caenogastropods, and select representatives of other molluscan groups. Partial genomes of Thylaeodus sp, Vermetus erectus, Littorina saxatilis and Calyptraea chinensis are also included. As is the standard convention for metazoan mt genomes, cox1 has been designated the start point for the "+" strand for all genomes. The dashed line indicates missing genes in the representation of partial genomes. Genes are transcribed left-to-right as depicted except for those underlined to signify opposite orientation. Gene names are as in Figure 1 except that L1 and L2 refer to trnLCUN and trnLUUR, respectively, and S1 and S2 refer to trnSAGN and trnSUCN, respectively. Pseudogenes identified in the genomes of Dendropoma maximum and Thylacodes squamigerus are not shown. In some instances, gene orders differ from their original publications (e.g., [53]); this is based on revised annotations listed in MitoZoa [63]. Abbreviations are as follows. GAST: Class Gastropoda; POLY: Class Polyplacophora; CEPH: Class Cephalopoda; CA: Caenogastropoda; Li: Littorinimorpha; Ne: Neogastropoda; VE: Vetigastropoda.
Given the extent of shared gene boundaries with representatives of other molluscan classes, Grande et al. [15] have suggested that the mt genome of the abalone, Haliotis rubra [56], represents the ancestral gene order of gastropods, albeit with two derived changes. The position of trnD and trnN are likely autapomorphies of Haliotis, since many caenogastropods share a different (and presumed ancestral) position for trnD with Octopus and trnN with Octopus and Katharina (Figure 5). Based on this inferred plesiomorphic gastropod condition, vermetids exhibit a number of derived gene order changes, some of which are shared with other caenogastropods. All published caenogastropod mt genomes, and the four complete vermetid genomes examined here, share an inversion of a block of 23 genes spanning from trnF to trnE in Haliotis, with a reversion (to the original strand) of genes spanning trnM to trnE within this block. As discussed by [48], this rearrangement must have occurred before the divergence of caenogastropods, but after their separation from the Vetigastropoda (Haliotis).
Vermetid mt genomes also shared an interesting gene order change with two other members of the superfamily Littorinimorpha, Littorina saxatilis and Oncomelania hupensis, associated with the position of the two leucine tRNA genes, trnLUUR and trnLCUN (Figure 5). Within the mt genome of Haliotis, Octopus, and Katharina, these two leucine tRNAs are sandwiched between rrnL and nad1 in the following arrangement: rrnL-trnLCUN-trnLUUR-nad1. This gene order is retained in all caenogastropod mt genomes sampled to date, except for Littorina, Oncomelania, Dendropoma maximum, D. gregarium, and Vermetus erectus, where the relative position of these two genes has switched, such that trnLUUR is located directly upstream from trnLCUN. (Note: this gene order is incorrectly annotated in [48] (pg 40), and this gene order change is not recognized in [15](page 11) or [53](page 4), where it is stated that there are no differences in gene order between the partial genome of Littorina and complete genome of neogastropods). This difference in the order of these two tRNAs is also present in several other vermetid taxa (data not shown), but is not present in two additional members of the Littorinimorpha: Cymatium parthenopeum and Calyptraea chinensis [53]. While this shared gene rearrangement may be a synapomorphy defining a clade within the Littorinimorpha to which the Littorinidae (Littorina), Pomatiopsidae (Oncomelania) and Vermetidae belong, changes in tRNA positions involving neighbouring leucine tRNA genes should be treated with caution. Gene translocations among neighbouring genes appear to occur with increased frequency in mt genomes and thus may be more likely to arise independently [23]. Such "position switches" between neighbours may therefore be less reliable phylogenetic characters for addressing deeper level phylogenetic questions. In addition, simple changes in position of these two leucine tRNA genes can mask more complicated dynamics that may involve gene duplications and tRNA remolding events [28] (see Evidence for tRNA remolding and recruitment, below). Uncovering the dynamics that have occurred within this region of the genome can be essential to interpreting gene identity correctly and accurately reconstructing past gene rearrangement events. The discovery of two trnLUUR genes side by side between rrnL and nad1 of Thylacodes squamigerus further suggests that such tRNA remolding events may be at play within vermetid mt genomes.
Some gene order arrangements were unique to the Vermetidae (Figure 5). All four complete vermetid genomes shared a block of three protein-encoding genes and 3 - 4 tRNA genes: trnN-nad5-trnK-trnA-cox3-trnS1-nad2, with trnA and trnK absent from this block in Eualetes and D. gregarium, respectively. This gene rearrangement was associated with the movement of nad5 from its conserved position between trnF and trnH in Katharina, Octopus, Haliotis, and other caenogastropods, and with the break-up of cox3, trnS1, and nad2 from their conserved association within a block of genes including cox3 - 5 tRNAs - nad3 - trnS1 - nad2 in these same taxa. TrnD also shared a unique derived position between cox1 and atp8 in the Vermetidae. While trnD in Haliotis has differing neighbouring genes, the gene order cox1-trnD is shared with the chiton, Katharina, and trnD-atp8 is shared with Octopus, as well as all other caenogastropods. The derived vermetid arrangement thus appears to be associated with a switch in the position of cox2 relative to cox1, trnD, and atp8, along a lineage leading to the Vermetidae. In addition, the vermetids also shared a derived change in the relative positions of trnY and trnM within the cassette of 7 tRNA genes encoded on the "-" strand. Such distinctions between the mt genome of vermetids and other littorinimorphs are particularly relevant here given that there is both molecular and morphological support for the inclusion of littorines and vermetids in a clade of lower caenogastropods [28,37,38]. Gene rearrangements shared by vermetids to the exclusion of other Littorinimorpha (Littorina, Oncomelania, Calyptraea, and Cymatium), thus can be inferred to have occurred following the divergence of the ancestral vermetid from the common ancestor of these taxa.
Numerous gene order changes have also occurred within the family Vermetidae (Figure 5). The Vermetidae has a fossil record extending to the Late Cretaceous, suggesting that this gene reshuffling has happened within the past 65 million years, and based on molecular dating, perhaps within the past 38 million years [18]. Preliminary evidence suggests that the mt genome of Dendropoma maximum represents the ancestral gene order of the Vermetidae. The translocation of trnK1 and trnP-nad6 found in Dendropoma gregarium has also been shown to be a derived gene order change within this genus [18]. Relative to D. maximum, most gene rearrangements evident in the three other complete genomes have occurred between trnR and trnT. Eualetes differs from D. maximum in the position of four tRNAs: trnA, trnP, trnLUUR and trnT, with gene remolding events between trnLCUN and trnLUUR likely associated with gene order changes involving these two isoaccepting tRNA genes (see Evidence for tRNA remolding and recruitment, below). Differences between the gene arrangement of Dendropoma maximum and Thylacodes appear related to a gene duplication event in Thylacodes spanning rrnS to nad1, as inferred from gene vestiges of rrnS, rrnL, and nad1 and extra tRNA genes (trnV, trnLUUR). Details of this rearrangement will be examined further in another paper (Rawlings et al. in prep). Many of the conserved gene boundaries described above for the four complete vermetid genomes sequenced, even those shared with other caenogastropods, Haliotis, Katharina, and Octopus, were not evident in the partial genomes of V. erectus and Thylaeodus (Figures 1 &5). The scale of these rearrangements is impressive given that these changes have happened within a family of gastropods, and suggests that further sampling of the Vermetidae should uncover more changes and perhaps intermediate stages that may reveal the dynamics underlying the gene rearrangements shown here. The genome of Thylacodes squamigerus provides hope of this as vestiges of once functional genes discovered between annotated genes are helping to understand the mechanism of gene order change and the appearance of duplicated tRNAs. This expectation has also been borne out by additional data collected as part of a phylogenetic analysis of the Vermetidae: numerous gene order changes have now been uncovered within vermetid taxa based on short sequences (<3.5 kb) extending from rrnS to nad1 (Rawlings et al., in prep; data not shown). With further sampling of the Vermetidae, we anticipate that these gene order rearrangements will provide a suite of robust characters that can be used, in addition to morphological and nucleotide sequence characters, to build a well-supported phylogenetic hypothesis for this family and to firmly place the Vermetidae within the context of caenogastropod evolution.
Evidence for tRNA remolding and recruitment
Implicit in the use of secondary structure characteristics and anticodon triplets to recognize tRNAs is the assumption that tRNA genes cannot change identity by simple nucleotide substitutions in their anticodon. For the most part this seems to be true within animal mt genomes, likely because of the presence of specific recognition elements that are required by tRNA synthetases to identify and correctly charge their associated tRNAs. However, evidence is accumulating that tRNAs do occasionally change identities. Through a process known as tRNA remolding, identity change does occur between the two isoaccepting leucine tRNA genes [25,26,28] and possibly also between the two isoaccepting serine tRNA genes [61]. Cases of tRNA identity change across non-isoaccepting tRNAs, referred to as tRNA recruitment, are now also coming to light [27]. The strongest evidence of tRNA remolding is the discovery of unexpectedly high levels of sequence similarity between two tRNA leucine genes [28]. Consequently, these dynamics are most easily recognized when they have happened recently, before mutational changes can obscure the common history of the duplicated tRNAs. Although gene remolding and recruitment events are often associated with changes in gene order, the pattern of duplicate loss may result in maintenance of the original gene order. In fact, recognizing gene remolding events can often help to uncover genome dynamics that are hidden at the level of gene order alone [28].
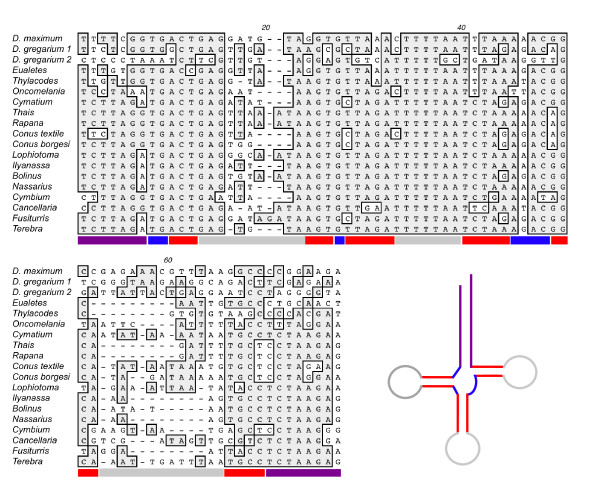
Given that leucine tRNAs seem particularly susceptible to remolding events [26,28], we investigated the sequence similarity between both leucine tRNAs within each vermetid taxon as well as other select caenogastropods, Haliotis, Octopus, and Katharina (Table 9). High sequence similarities (80% or greater) between leucine tRNA genes were found in five taxa (Table 9) suggesting that one tRNA has taken over the role of the other through a process of gene duplication, mutation in the anticodon triplet and the eventual loss of the original gene [26,28]. The T. squamigerus genome represents a particularly interesting case of this. Within this genome, the two copies of LUUR only share 67.6% sequence identity; in comparison there is 98.4% sequence similarity between trnLCUN and trnLUUR2. Consequently, we can assume that trnLUUR2 is a duplicated copy of trnLCUN that has subsequently undergone a mutation in the third position of the anticodon to assume the identity of trnLUUR. Such events argue for the exclusion of tRNA leucine genes in gene-order based phylogenetic analyses at high taxonomic levels; at low taxonomic levels, such as within the Vermetidae, however, they offer the promise of new phylogenetic characters and the discovery of gene dynamics that may not be evident at the level of gene order alone. Given the importance of identifying remolding events, it is surprising that so little attention is often paid to the dynamics of these two leucine tRNA genes. Failure to distinguish between these two genes correctly can also lead to mistakes in interpretation, where two genomes are considered to have identical gene orders, but differ in the position of these two leucine tRNAs (see comparisons between Littorina saxatilis and other caenogastropods in [15,48,53]).
Table 9.
Percentage similarity in nucleotide sequences between isoaccepting leucine tRNA genes (trnLUUR and trnLCUN) within the mt genomes of six vermetids, sixteen additional caenogastropods, and three other molluscs for comparison
| Species | tRNA comparisonab | Percent similarityc |
|---|---|---|
| Dendropoma maximum | LUUR-LCUN | 60.9% |
| Dendropoma gregarium | LUUR-LCUN | 69.0% |
| Eualetes tulipa | LCUN-/-LUUR | 98.4% |
| Thylacodes squamigerusd | LCUN-/-LUUR1 | 66.2% |
| LCUN-/-LUUR2 | 98.4% | |
| LUUR1-LUUR2 | 67.6% | |
| Vermetus erectus | LUUR-LCUN | 76.6% |
| Thylaeodus sp. | LCUN-/-LUUR | 87.0% |
| Oncomelania | LUUR-LCUN | 91.2% |
| Littorina | LUUR-LCUN | 80.3% |
| Calyptraea | LCUN-LUUR | 58.0% |
| Cymatium | LCUN-LUUR | 61.8% |
| Thais | LCUN-LUUR | 58.0% |
| Rapana | LCUN-LUUR | Insufficient datae |
| Conus textile | LCUN-LUUR | 60.9% |
| Conus borgesi | LCUN-LUUR | 60.9% |
| Lophiotoma | LCUN-LUUR | 62.3% |
| Ilyanassa | LCUN-LUUR | 64.7% |
| Nassarius | LCUN-LUUR | 64.7% |
| Bolinus | LCUN-LUUR | 62.3% |
| Cancellaria | LCUN-LUUR | 52.7% |
| Cymbium | LCUN-LUUR | 58.2% |
| Fusiturris | LCUN-LUUR | 66.7% |
| Terebra | LCUN-LUUR | 59.4% |
| Haliotis rubra | LCUN-LUUR | 64.2% |
| Katharina tunicata | LCUN-LUUR | 48.5% |
| Octopus vulgaris | LCUN-LUUR | 66.7% |
a tRNA genes were aligned using Clustal X with subsequent adjustments made according to secondary structure.
b Leucine tRNAs are presented according to relative order and position: one dash indicates that the two tRNAs are adjacent to each other; two dashes separated by a backslash indicate that these two genes are separated by at least one other gene.
c Percent similarity reflects the number of nucleotide matches over the total length of the alignment, excluding the third base of the anticodon triplet.
d Because Thylacodes had two copies of trnLUUR, three pairwise comparisons between leucine tRNA genes were made for this species.
e Sequence contained ambiguous nucleotides.
The presence of a second trnK within the genome of Dendropoma gregarium is suggestive of a past duplication event within this genome, likely the result of slipped-strand mispairing during replication. The sequence similarity between the two lysine tRNAs (trnK) is not high (38.0%). Comparisons between these sequences and the sequences of presumed trnK orthologs from other taxa revealed strong similarities between these and the trnK1 located between trnV and trnP (Figure 6). The second trnK (trnK2), located between trnI and nad3 lacked many of the conserved sequence elements present in other trnKs (Figure 6). We compared trnK2 with other tRNAs within the genome of D. gregarium to determine if this might represent a case of tRNA recruitment [27], but did not uncover any strong matches with any other gene. Consequently, the origin and function of this putative tRNA are currently unclear.
Figure 6.
Structural alignments of lysine tRNA genes sampled from four vermetid gastropods and other select caenogastropods. Two tRNA genes are included from Dendropoma gregarium, one located between trnV and trnP (trnK1), the other located between trnI and nad3 (trnK2). Nucleotides appearing in >50% of taxa at a given position in the gene are shaded grey. Colors below each region of the alignment are used to show the associated position in the colored tRNA secondary structure model.
Conclusions
Here we have presented gene order and sequence characteristics for six gastropods within the family Vermetidae, including the complete mt genomes of four species (Dendropoma maximum, D. gregarium, Eualetes tulipa, and Thylacodes squamigerus) and the partial mt genomes of two others (Vermetus erectus and Thylaeodus sp.). The publication of these genomes increases the number of complete caenogastropod mt genomes sequenced from 16 to 20 and presents the first direct evidence of major gene order rearrangements within the Littorinimorpha.
While molluscs have long been recognized as having unusually dynamic mt genomes, preliminary sampling of the Caenogastropoda represented by genomes from 16 species has suggested a model of highly conserved mt gene order. Our results reverse this trend and provide further evidence of the lability of the molluscan mt genome by describing extensive gene order changes that have occurred within one family of caenogastropod molluscs, the Vermetidae. Based on these results, we anticipate that the mt genomes of caenogastropods exhibit a variety of gene order arrangements, which, if explored and exploited, could provide a wealth of phylogenetically-informative characters to enhance our understanding of the evolutionary radiation of this diverse clade of gastropods.
Despite the extent of mt gene rearrangement within the Vermetidae, their genomes exhibit similar characteristics to other caenogastropods. Each complete genome contained the full complement of 37 genes, although additional tRNA genes were evident in the mt genomes of D. gregarium (trnK) and Thylacodes squamigerus (trnV, trnLUUR). Protein-encoding and rRNA genes were all encoded on the same strand (i.e. the "+" strand), not unusual for caenogastropods, whereas tRNA genes, the most mobile mt genomic components, were distributed between the "+" and "-" strands. Nucleotide skews patterns (i.e. AT and GC-skews) of vermetid genomes and their various components were similar to those described for other caenogastropods, although the AT bias was less pronounced than other caenogastropods described to date. Although no control regions were definitively identified, a compelling trend in the relationship between nucleotide composition and position from nad1 was uncovered, which appears worthy of further investigation.
Our results also demonstrate that focused sampling of mt genomes at low taxonomic levels can be extremely productive, both in terms of uncovering characters useful in phylogeny and in understanding more fully the evolutionary dynamics and mechanics of mt gene rearrangements. Each of the six vermetid mt genomes examined had a unique gene order, with evidence of gene rearrangements involving translocations of tRNA and protein-encoding genes, and one gene inversion, occurring within this family. Additional rearrangements have also been uncovered amongst other representatives of this family suggesting that further sampling of complete mt genomes within the Vermetidae will also prove useful. The extent of gene rearrangement within such an evolutionarily young group offers the opportunity to explore gene dynamics such as tRNA remolding, which appears to have been rife within this family, and to investigate its consequences for phylogenetic analyses based on gene orders. The sampling of mt genomes, such as that of Thylacodes squamigerus, in the intermediate stages of a gene rearrangement, may also help to illuminate mechanisms of gene translocation and inversion and to interpret past events that have shaped an organism's current mt gene order. Continued studies of the mt genome dynamics within the Vermetidae will improve our understanding of this family's phylogeny, its phylogenetic placement within the Littorinimorpha, and the mechanisms and processes that have acted to shape the mt genomes of this family and other metazoans.
Authors' contributions
RB and TAR obtained the samples. TAR and TMC extracted the DNA and amplified the genomes which were then sequenced by JLB. TAR and MJM annotated the genome, MJM undertook the descriptive analyses and TAR and MJM wrote the first draft of the manuscript. All authors participated in subsequent revisions of the manuscript.
Supplementary Material
Figure S1. Inferred tRNA secondary structures based on the nucleotide sequences of 22 mitochondrial tRNA genes identified from the complete mt genome of Dendropoma maximum. tRNA genes are labeled according to their amino acid specificity and are arranged alphabetically.
Figure S2. Inferred tRNA secondary structures based on the nucleotide sequences of 23 mitochondrial tRNA genes identified from the complete mt genome of Dendropoma gregarium. tRNA genes are labeled according to their amino acid specificity and are arranged alphabetically.
Figure S3. Inferred tRNA secondary structures based on the nucleotide sequences of 22 mitochondrial tRNA genes identified from the complete mt genome of Eualetes tulipa. tRNA genes are labeled according to their amino acid specificity and are arranged alphabetically.
Figure S4. Inferred tRNA secondary structures based on the nucleotide sequences of 24 mitochondrial tRNA genes identified from the complete mt genome of Thylacodes squamigerus. tRNA genes are labeled according to the amino acid specificity and are arranged alphabetically.
Figure S5. Inferred tRNA secondary structures based on the nucleotide sequences of eight mitochondrial tRNA genes identified from the partial mt genome of Thylaeodus sp. tRNA genes are labeled according to their amino acid specificity and are arranged alphabetically.
Figure S6. Inferred tRNA secondary structures based on the nucleotide sequences of five mitochondrial tRNA genes identified from the partial mt genome of Vermetus erectus. tRNA genes are labeled according to their amino acid specificity and are arranged alphabetically.
Table S1. Base compositions and nucleotide skews for new vermetid mt genomes, existing caenogastropod mt genomes, and other select molluscs.
Table S2. The conservation of amino acid identity in select codons from cox1, cox2, and cox3 genes from the mt genomes of Dendropoma maximum, D. gregarium, Eualetes tulipa, and Thylacodes squamigerus.
Table S3. Summary of codon usage across all protein-encoding genes in the mitochondrial genomes of Dendropoma maximum, D. gregarium, Eualetes tulipa, and Thylacodes squamigerus.
Contributor Information
Timothy A Rawlings, Email: Timothy_Rawlings@cbu.ca.
Martin J MacInnis, Email: mmacinni@interchange.ubc.ca.
Rüdiger Bieler, Email: rbieler@fieldmuseum.org.
Jeffrey L Boore, Email: JLBoore@GenomeProjectSolutions.com.
Timothy M Collins, Email: CollinsT@fiu.edu.
Acknowledgements
We thank Drs. Michael Hadfield, Isabella Kappner, Jon Norenburg, and Richard and Megumi Strathmann for vermetid samples, and Dr. Mónica Medina for assistance with genome sequencing. MJM was funded in part by CBU RP Grant 8341 to TAR. Vermetid research was supported by NSF DEB(REVSYS)-0841760/0841777 to RB, TMC, and TAR.
References
- Boore JL. Animal mitochondrial genomes. Nucleic Acids Research. 1999;27(8):1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MJ, Arndt A, Gorski S, Fajber E. The phylogeny of echinoderm classes based on mitochondrial gene arrangements. Journal of Molecular Evolution. 1993;36:545–554. doi: 10.1007/BF00556359. [DOI] [PubMed] [Google Scholar]
- Boore JL, Brown WM. Complete sequence of the mitochondrial DNA of the annelid worm Lumbricus terrestris. Genetics. 1995;141:305–319. doi: 10.1093/genetics/141.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavrov DV, Brown WM, Boore JL. Phylogenetic position of the Pentastomida and (pan)crustacean relationships. Proceedings of the Royal Society of London Series B-Biological Sciences. 2004;271(1538):537–544. doi: 10.1098/rspb.2003.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore JL, Brown WM. Mitochondrial genomes of Galathealinum, Helobdella, and Platynereis: sequence and gene arrangement comparisons indicate that Pogonophora is not a phylum and Annelida and Arthropoda are not sister taxa. Molecular Biology and Evolution. 2000;17(1):87–106. doi: 10.1093/oxfordjournals.molbev.a026241. [DOI] [PubMed] [Google Scholar]
- Lavrov DV, Lang BF. Poriferan mtDNA and animal phylogeny based on mitochondrial gene arrangements. Systematic Biology. 2005;54(4):651–659. doi: 10.1080/10635150500221044. [DOI] [PubMed] [Google Scholar]
- Boore JL, Macey JR, Medina M. Sequencing and comparing whole mitochondrial genomes of animals. Methods in Enzymology. 2005;395:311–348. doi: 10.1016/S0076-6879(05)95019-2. full_text. [DOI] [PubMed] [Google Scholar]
- Simison WB, Lindberg DR, Boore JL. Rolling circle amplification of metazoan mitochondrial genomes. Molecular Phylogenetics and Evolution. 2006;39(2):562–567. doi: 10.1016/j.ympev.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Hudson ME. Sequencing breakthroughs for genomic ecology and evolutionary biology. Molecular Ecology Resources. 2007;8:3–17. doi: 10.1111/j.1471-8286.2007.02019.x. [DOI] [PubMed] [Google Scholar]
- Blanchette M, Kunisawa T, Sankoff D. Gene order breakpoint evidence in animal mitochondrial phylogeny. Journal of Molecular Evolution. 1999;49(2):193–203. doi: 10.1007/PL00006542. [DOI] [PubMed] [Google Scholar]
- Moret BME, Tang JJ, Wang LS, Warnow T. Steps toward accurate reconstructions of phylogenies from gene-order data. Journal of Computer and System Sciences. 2002;65(3):508–525. doi: 10.1016/S0022-0000(02)00007-7. [DOI] [Google Scholar]
- Larget B, Simon DL, Kadane JB, Sweet D. A Bayesian analysis of metazoan mitochondrial genome arrangements. Molecular Biology and Evolution. 2005;22(3):486–495. doi: 10.1093/molbev/msi032. [DOI] [PubMed] [Google Scholar]
- Simison WB, Boore JL. In: Phylogeny and Evolution of the Mollusca. Ponder W, Lindberg DR, editor. Berkeley, CA: University of California Press; 2008. Molluscan evolutionary genomics; pp. 447–461. [Google Scholar]
- Vallès Y, Boore JL. Lophotrochozoan mitochondrial genomes. Integrative and Comparative Biology. 2006;46(4):544–557. doi: 10.1093/icb/icj056. [DOI] [PubMed] [Google Scholar]
- Grande C, Templado J, Zardoya R. Evolution of gastropod mitochondrial genome arrangements. BMC Evolutionary Biology. 2008;8 doi: 10.1186/1471-2148-8-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gissi C, Iannelli F, Pesole G. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity. 2008;101(4):301–320. doi: 10.1038/hdy.2008.62. [DOI] [PubMed] [Google Scholar]
- Milbury CA, Gaffney PM. Complete mitochondrial DNA sequence of the eastern oyster Crassostrea virginica. Marine Biotechnology. 2005;7(6):697–712. doi: 10.1007/s10126-005-0004-0. [DOI] [PubMed] [Google Scholar]
- Rawlings TA, Collins TM, Bieler R. A major mitochondrial gene rearrangement among closely related species. Molecular Biology and Evolution. 2001;18(8):1604–1609. doi: 10.1093/oxfordjournals.molbev.a003949. [DOI] [PubMed] [Google Scholar]
- Serb JM, Lydeard C. Complete mtDNA Sequence of the North American Freshwater Mussel, Lampsilis ornata (Unionidae): an examination of the evolution and phylogenetic utility of mitochondrial genome organization in Bivalvia (Mollusca) Molecular Biology and Evolution. 2003;20(11):1854–1866. doi: 10.1093/molbev/msg218. [DOI] [PubMed] [Google Scholar]
- Dowton M, Castro LR, Austin AD. Mitochondrial gene rearrangements as phylogenetic characters in the invertebrates: the examination of genome 'morphology'. Invertebrate Systematics. 2002;16(3):345–356. doi: 10.1071/IS02003. [DOI] [Google Scholar]
- Ballard JWO, Rand DM. The population biology of mitochondrial DNA and its phylogenetic implications. Annual Review of Ecology Evolution and Systematics. 2005;36:621–642. doi: 10.1146/annurev.ecolsys.36.091704.175513. [DOI] [Google Scholar]
- Levinson G, Gutman GA. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Molecular Biology and Evolution. 1987;4(3):203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- Boore JL, Brown WM. Big trees from little genomes: mitochondrial gene order as a phylogenetic tool. Current Opinion in Genetics & Development. 1998;8(6):668–674. doi: 10.1016/S0959-437X(98)80035-X. [DOI] [PubMed] [Google Scholar]
- Lunt DH, Hyman BC. Animal mitochondrial DNA recombination. Nature. 1997;387:247. doi: 10.1038/387247a0. [DOI] [PubMed] [Google Scholar]
- Cantatore P, Gadaleta MN, Roberti M, Saccone C, Wilson AC. Duplication and remoulding of tRNA genes during the evolutionary rearrangement of mitochondrial genomes. Nature. 1987;329:853–855. doi: 10.1038/329853a0. [DOI] [PubMed] [Google Scholar]
- Higgs PG, Jameson D, Jow H, Rattray M. The evolution of tRNA-Leu genes in animal mitochondrial genomes. Journal of Molecular Evolution. 2003;57(4):435–445. doi: 10.1007/s00239-003-2494-6. [DOI] [PubMed] [Google Scholar]
- Lavrov DV, Lang BF. Transfer RNA gene recruitment in mitochondrial DNA. Trends in Genetics. 2005;21(3):129–133. doi: 10.1016/j.tig.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Rawlings TA, Collins TM, Bieler R. Changing identities: tRNA duplication and remolding within animal mitochondrial genomes. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(26):15700–15705. doi: 10.1073/pnas.2535036100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowton M, Austin AD. Evolutionary dynamics of a mitochondrial rearrangement "hot spot" in the Hymenoptera. Molecular Biology and Evolution. 1999;16(2):298–309. doi: 10.1093/oxfordjournals.molbev.a026111. [DOI] [PubMed] [Google Scholar]
- San Mauro D, Gower DJ, Zardoya R, Wilkinson M. A hotspot of gene order rearrangement by tandem duplication and random loss in the vertebrate mitochondrial genome. Molecular Biology and Evolution. 2006;23(1):227–234. doi: 10.1093/molbev/msj025. [DOI] [PubMed] [Google Scholar]
- Lavrov DV, Boore JL, Brown WM. Complete mtDNA sequences of two millipedes suggest a new model for mitochondrial gene rearrangements: Duplication and nonrandom loss. Molecular Biology and Evolution. 2002;19(2):163–169. doi: 10.1093/oxfordjournals.molbev.a004068. [DOI] [PubMed] [Google Scholar]
- Flook P, Rowell H, Gellissen G. Homoplastic rearrangements of insect mitochondrial transfer-RNA genes. Naturwissenschaften. 1995;82(7):336–337. doi: 10.1007/BF01131531. [DOI] [Google Scholar]
- Macey JR, Papenfuss TJ, Kuehl JV, Fourcade HM, Boore JL. Phylogenetic relationships among amphisbaenian reptiles based on complete mitochondrial genomic sequences. Molecular Phylogenetics and Evolution. 2004;33(1):22–31. doi: 10.1016/j.ympev.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Dowton M, Cameron SL, Dowavic JI, Austin AD, Whiting MF. Characterization of 67 mitochondrial tRNA gene rearrangements in the Hymenoptera suggests that mitochondrial tRNA gene position is selectively neutral. Molecular Biology and Evolution. 2009;26(7):1607–1617. doi: 10.1093/molbev/msp072. [DOI] [PubMed] [Google Scholar]
- Iannelli F, Griggio F, Pesole G, Gissi C. The mitochondrial genome of Phallusia mammillata and Phallusia fumigata (Tunicata, Ascidiacea): high genome plasticity at intra-genus level. BMC Evolutionary Biology. 2007;7 doi: 10.1186/1471-2148-7-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponder WF, Lindberg DR. Towards a phylogeny of gastropod molluscs: an analysis using morphological characters. Zoological Journal of the Linnean Society. 1997;119:83–265. doi: 10.1111/j.1096-3642.1997.tb00137.x. [DOI] [Google Scholar]
- Colgan DJ, Ponder WF, Beacham E, Macaranas J. Molecular phylogenetics of Caenogastropoda (Gastropoda: Mollusca) Molecular Phylogenetics and Evolution. 2007;42(3):717–737. doi: 10.1016/j.ympev.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Healy JM. Sperm morphology in Serpulorbis and Dendropoma and its relevance to the systematic position of the Vermetidae. Journal of Molluscan Studies. 1988;54:295–308. doi: 10.1093/mollus/54.3.295. [DOI] [Google Scholar]
- Collins TM, Frazer KS, Palmer AR, Vermeij GJ, Brown WM. Evolutionary history of Northern Hemisphere Nucella: molecules, morphology, ecology, and fossils. Evolution. 1996;50:2287–2304. doi: 10.2307/2410698. [DOI] [PubMed] [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology. 1994;3(5):294–299. [PubMed] [Google Scholar]
- Boore JL. The complete sequence of the mitochondrial genome of Nautilus macromphalus (Mollusca: Cephalopoda) BMC Genomics. 2006;7 doi: 10.1186/1471-2164-7-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Research. 1997;25(5):955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslett D, Canback B. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 2008;24(2):172–175. doi: 10.1093/bioinformatics/btm573. [DOI] [PubMed] [Google Scholar]
- Perna NT, Kocher TD. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. Journal of Molecular Evolution. 1995;41(3):353–358. doi: 10.1007/BF01215182. [DOI] [PubMed] [Google Scholar]
- Reyes A, Gissi C, Pesole G, Saccone C. Asymmetrical directional mutation pressure in the mitochondrial genome of mammals. Molecular Biology and Evolution. 1998;15(8):957–966. doi: 10.1093/oxfordjournals.molbev.a026011. [DOI] [PubMed] [Google Scholar]
- Santos MAS, Moura G, Massey SE, Tuite MF. Driving change: the evolution of alternative genetic codes. Trends in Genetics. 2004;20(2):95–102. doi: 10.1016/j.tig.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Yokobori S, Suzuki T, Watanabe K. Genetic code variations in mitochondria: tRNA as a major determinant of genetic code plasticity. Journal of Molecular Evolution. 2001;53:(4–5):314-326. doi: 10.1007/s002390010221. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay PK, Stevenson BJ, Cady MT, Olivera BM, Wolstenholme DR. Complete mitochondrial DNA sequence of a Conoidean gastropod, Lophiotoma (Xenuroturris) cerithiformis: gene order and gastropod phylogeny. Toxicon. 2006;48(1):29–43. doi: 10.1016/j.toxicon.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Boore JL, Daehler LL, Brown WM. Complete sequence, gene arrangement, and genetic code of mitochondrial DNA of the cephalochordate Branchiostoma floridae (Amphioxus) Molecular Biology and Evolution. 1999;16(3):410–418. doi: 10.1093/oxfordjournals.molbev.a026122. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology: 1994; Menlo Park, California. AAAI Press; 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers; pp. 28–36. [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay PK, Stevenson BJ, Ownby JP, Cady MT, Watkins M, Olivera BM. The mitochondrial genome of Conus textile, coxI-coxII intergenic sequences and conoidean evolution. Molecular Phylogenetics and Evolution. 2008;46(1):215–223. doi: 10.1016/j.ympev.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha RL, Grande C, Zardoya R. Neogastropod phylogenetic relationships based on entire mitochondrial genomes. BMC Evolutionary Biology. 2009;9:210. doi: 10.1186/1471-2148-9-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang KH, Hwang UW. Complete mitochondrial genome of Bugula neritina (Bryozoa, Gymnolaemata, Cheilostomata): phylogenetic position of Bryozoa and phylogeny of lophophorates within the Lophotrochozoa. BMC Genomics. 2009;10 doi: 10.1186/1471-2164-10-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore JL, Brown WM. Complete DNA-sequence of the mitochondrial genome of the black chiton, Katharina tunicata. Genetics. 1994;138(2):423–443. doi: 10.1093/genetics/138.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard BT, Kerr LJ, McKiernan JM, Jansen ES, Hanna PJ. Mitochondrial DNA sequence and gene organization in the Australian Blacklip Abalone Haliotis rubra (Leach) Marine Biotechnology. 2005;7:645–658. doi: 10.1007/s10126-005-0013-z. [DOI] [PubMed] [Google Scholar]
- Wang X, Lavrov DV. Seventeen new complete mtDNA sequences reveal extensive mitochondrial genome evolution within the Demospongiae. Public Library of Science One. 2008;3(7):e2723. doi: 10.1371/journal.pone.0002723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugler MR, France SC. The mitochondrial genome of a deep-sea bamboo coral (Cnidaria, Anthozoa, Octocorallia, Isididae): genome structure and putative origins of replication are not conserved among octocorals. Journal of Molecular Evolution. 2008;67(2):125–136. doi: 10.1007/s00239-008-9116-2. [DOI] [PubMed] [Google Scholar]
- Arunkumar KP Nagaraju J Unusually long palindromes are abundant in mitochondrial control regions of insects and nematodes Public Library of Science One 200611e11017205114 [Google Scholar]
- Saito S, Tamura K, Aotsuka T. Replication origin of mitochondrial DNA in insects. Genetics. 2005;171(4):1695–1705. doi: 10.1534/genetics.105.046243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavrov DV, Brown WM. Trichinella spiralis mtDNA: a nematode mitochondrial genome that encodes a putative ATP8 and normally structures tRNAs and has a gene arrangement relatable to those of coelomate metazoans. Genetics. 2001;157(2):621–637. doi: 10.1093/genetics/157.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieler R, Petit RE. Thylacodes - Thylacodus - Tulaxodus: Worm-snail name confusion and the status of Serpulorbis (Gastropoda: Vermetidae) Malacologia. 2010;52(1):183–187. doi: 10.4002/040.052.0114. [DOI] [Google Scholar]
- Lupi R, D'Onorio de Meo P, Picardi E, D'Antonio M, Paoletti D, Castrignano T, Pesole G, Gissi C. MitoZoa: A curated mitochondrial genome database of metazoans for comparative genomics studies. Mitochondrion. 2010;10(2):192–199. doi: 10.1016/j.mito.2010.01.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Inferred tRNA secondary structures based on the nucleotide sequences of 22 mitochondrial tRNA genes identified from the complete mt genome of Dendropoma maximum. tRNA genes are labeled according to their amino acid specificity and are arranged alphabetically.
Figure S2. Inferred tRNA secondary structures based on the nucleotide sequences of 23 mitochondrial tRNA genes identified from the complete mt genome of Dendropoma gregarium. tRNA genes are labeled according to their amino acid specificity and are arranged alphabetically.
Figure S3. Inferred tRNA secondary structures based on the nucleotide sequences of 22 mitochondrial tRNA genes identified from the complete mt genome of Eualetes tulipa. tRNA genes are labeled according to their amino acid specificity and are arranged alphabetically.
Figure S4. Inferred tRNA secondary structures based on the nucleotide sequences of 24 mitochondrial tRNA genes identified from the complete mt genome of Thylacodes squamigerus. tRNA genes are labeled according to the amino acid specificity and are arranged alphabetically.
Figure S5. Inferred tRNA secondary structures based on the nucleotide sequences of eight mitochondrial tRNA genes identified from the partial mt genome of Thylaeodus sp. tRNA genes are labeled according to their amino acid specificity and are arranged alphabetically.
Figure S6. Inferred tRNA secondary structures based on the nucleotide sequences of five mitochondrial tRNA genes identified from the partial mt genome of Vermetus erectus. tRNA genes are labeled according to their amino acid specificity and are arranged alphabetically.
Table S1. Base compositions and nucleotide skews for new vermetid mt genomes, existing caenogastropod mt genomes, and other select molluscs.
Table S2. The conservation of amino acid identity in select codons from cox1, cox2, and cox3 genes from the mt genomes of Dendropoma maximum, D. gregarium, Eualetes tulipa, and Thylacodes squamigerus.
Table S3. Summary of codon usage across all protein-encoding genes in the mitochondrial genomes of Dendropoma maximum, D. gregarium, Eualetes tulipa, and Thylacodes squamigerus.