Abstract
Background
Oligotropha carboxidovorans OM5 T. (DSM 1227, ATCC 49405) is a chemolithoautotrophic bacterium capable of utilizing CO (carbon monoxide) and fixing CO2 (carbon dioxide). We previously published the draft genome of this organism and recently submitted the complete genome sequence to GenBank.
Results
The genome sequence of the chemolithoautotrophic bacterium Oligotropha carboxidovorans OM5 consists of a 3.74-Mb chromosome and a 133-kb megaplasmid that contains the genes responsible for utilization of carbon monoxide, carbon dioxide, and hydrogen. To our knowledge, this strain is the first one to be sequenced in the genus Oligotropha, the closest fully sequenced relatives being Bradyrhizobium sp. BTAi and USDA110 and Nitrobacter hamburgiensis X14. Analysis of the O. carboxidovorans genome reveals potential links between plasmid-encoded chemolithoautotrophy and chromosomally-encoded lipid metabolism. Comparative analysis of O. carboxidovorans with closely related species revealed differences in metabolic pathways, particularly in carbohydrate and lipid metabolism, as well as transport pathways.
Conclusion
Oligotropha, Bradyrhizobium sp and Nitrobacter hamburgiensis X14 are phylogenetically proximal. Although there is significant conservation of genome organization between the species, there are major differences in many metabolic pathways that reflect the adaptive strategies unique to each species.
Background
OM5, the type strain of the gram-negative bacterium Oligotropha carboxidovorans [1,2] (originally called Pseudomonas carboxidovorans), was isolated from soil of waste water sewage treatment settling ponds. OM5 is capable of chemolithoautotrophic growth on CO, CO2, and H2 [3-5] and can utilize chemoorganoheterotrophic substrates under aerobic conditions, including the salts of pyruvate, formate, glyoxylate, lactate, ascorbate, malate, oxoglutarate, acetate, and oxalate.
Initial 16 S rDNA sequencing [5] showed Oligotropha carboxidovorans is phylogenetically close to two Bradyrhizobiaceae: Bradyrhizobium sp. BTAi and Nitrobacter hamburgiensis X14. All three species share some common gene(s) and operons; however, these three bacteria vary greatly in metabolism. Together they are an excellent model for understanding how closely related bacteria adapt to very different environments.
In addition to its utility for comparative genomics, O. carboxidovorans genes may have practical utility for bioenergy production. O. carboxidovorans is capable of utilizing syngas, which is a mixture of CO, CO2, and H2 that results from gasification of organic wastes, for chemolithoautotrophic growth. Microbial fermentation of this gas mixture could serve as a source for biofuels. Many of the chemolithoautotrophy genes in O. carboxidovorans are located on a megaplasmid and have been studied [2,4,6,7]. The O. carboxidovorans genome will allow further studies to understand how they are regulated and how fixed carbon is assimilated into its metabolism, which could enhance use of syngas as a biofuel source.
We previously announced the O. carboxidovorans genome sequence [8]. At the time of our previous announcement, the chromosome sequence was in two large contigs. We now report completion of the O. carboxidovorans OM5 genome and comparative genomics analysis with Bradyrhizobium sp. BTAi and N. hamburgiensis X14. Central intermediary metabolism is similar in O. carboxidovorans, Bradyrhizobium sp. BTAi, and N. hamburgiensis X14, and O. carboxidovorans shares other features such as iron transport systems. However, O. carboxidovorans also has several unique features, particularly in fatty acid oxidation and glyoxylate cycle.
Results and discussion
Genome organization
Closure of the last two gaps produced a 3,745,629 bp long circular OM5 chromosome (62.4% GC) with 3782 predicted genes. The vast majority (3722) are protein coding; 48 encode RNAs, and 12 are predicted as pseudogenes. Of the 48 RNA genes, three are rRNA genes (one rRNA operon is present), and the rest code for tRNAs. Twelve percent of the genome is predicted to be intergenic DNA. Of the OM5 protein coding genes, TIGR role categories were assigned to 87.59%. Of these, 72.01% had a definitive functional assignment, and 1.31% did not have a role category. In addition, 15.99% were identified as conserved hypothetical proteins (CHPs), and 10.67% were designated as hypothetical proteins (HPs). Transporter and enzyme functions are the most abundant major metabolic activities in the OM5 genome, together making up 10.5% of the predicted coding capacity. Table 1 shows predicted functional classifications (COG groups) of O. carboxidovorans proteins.
Table 1.
Number of genes per COG group in the O. carboxidovorans genome.
| Process | Description | Class ID | CDS No |
|---|---|---|---|
| Cellular Process and Signaling | Cell cycle control, cell division and chromosome partitioning | D | 37 |
| Cellular Process and Signaling | Cell wall/membrane/envelope biogenesis | M | 205 |
| Cellular Process and Signaling | Cell motility | N | 90 |
| Cellular Process and Signaling | Posttranslational modification, protein turnover, chaperones | O | 172 |
| Cellular Process and Signaling | Signal transduction mechanisms | T | 189 |
| Cellular Process and Signaling | Intracellular trafficking, secretion and vesicular transport | U | 117 |
| Cellular Process and Signaling | Defense mechanisms | Y | 99 |
| Cellular Process and Signaling | Extracellular structures | W | 1 |
| Information storage and processing | Chromatin structure and dynamics | B | 1 |
| Information storage and processing | Translation, ribosome structure and biogenesis | J | 193 |
| Information storage and processing | Transcription | K | 205 |
| Information storage and processing | Replication, recombination and repair | L | 175 |
| Metabolism | Energy production and conversion | C | 236 |
| Metabolism | Amino acid transport and metabolism | E | 444 |
| Metabolism | Nucleotide transport and metabolism | F | 79 |
| Metabolism | Carbohydrate transport and metabolism | G | 207 |
| Metabolism | Coenzyme transport and metabolism | H | 137 |
| Metabolism | Lipid transport and metabolism | I | 150 |
| Metabolism | Inorganic ion transport and metabolism | P | 345 |
| Metabolism | Secondary metabolites biosynthesis, transport and catabolism | Q | 112 |
| Poorly characterized | General function prediction only | R | 514 |
| Poorly characterized | Function unknown | S | 243 |
Using RfamScan http://rfam.sanger.ac.uk/[9] in Oligotroscope, we found 10 potential non-coding RNA sequences, which includes small RNA (sRNA) (Table 2). Prediction of prokaryotic sRNAs has limitations because their sequences are diverse; therefore, identification is based on conserved sequence features in the intergenic regions (promoters, terminators, and RNA secondary structures). In several bacteria, sRNAs have important regulatory functions, including regulation of outer membrane protein expression and regulating stress responses (e.g., oxidative stress, SOS/DNA damage, cold shock, and iron stress). Some sRNAs such as DsrA and RprA are translational activators [10-12]. The regulatory RNA SsrA is needed for correct high-level translation of RpoS [13].
Table 2.
Predicted O. carboxidovorans non-coding RNAs
| Label | Begin | End | Length | Name | Product |
|---|---|---|---|---|---|
| misc_RNA_16 | 925123 | 925197 | 75 | suhB | Antisense RNA/nc RNA |
| misc_RNA_19 | 1416092 | 1416176 | 85 | ssrA | 10Sa RNA SsrA |
| misc_RNA_18 | 1416179 | 1416292 | 114 | ssrA | 10Sa RNA SsrA |
| misc_RNA_7 | 1574853 | 1575108 | 256 | csrC | Regulatory sRNA |
| misc_RNA_8 | 2702642 | 2702856 | 215 | csrC | Regulatory sRNA |
| misc_RNA_20 | 2988271 | 2988368 | 98 | TPP | Riboswitch |
| misc_RNA_2 | 3118145 | 3118195 | 51 | serC | Attenuator |
| misc_RNA_1 | 3263978 | 3264135 | 158 | ssrS | 6 S regulatory RNA |
| misc_RNA_21 | 3502780 | 3502877 | 98 | SRP | Bacterial signal recognition particle |
| misc_RNA_9 | 3606055 | 3606129 | 75 | ctRNA | Counter-transcribed RNA |
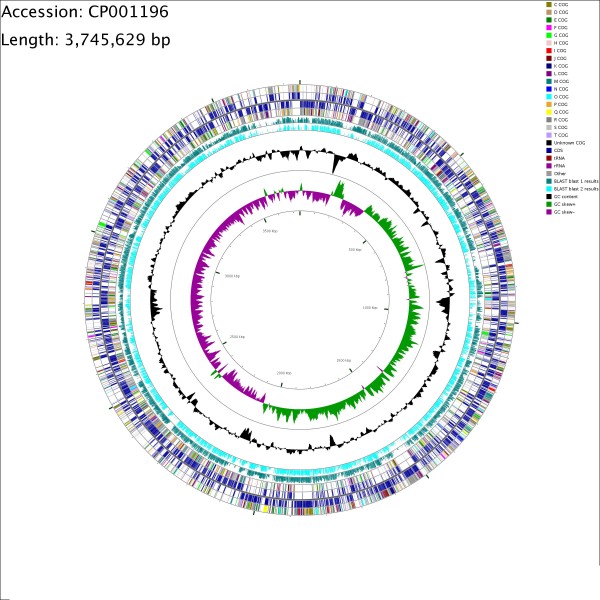
GC and AT skew analysis http://tubic.tju.edu.cn/Ori-Finder/ suggests a putative replication origin (spanning 150 nt) starting at position 357,583 [14]. The origin has a strong inflection point in the coding strand with 4 DnaA box motifs (tgtttcacg). This motif was identified using search parameters that allowed no more than one mismatch compared to the perfect E. coli DnaA box (ttatccaca) (see Figure 1; constructed using CG View) [15].
Figure 1.
Circular representation of the O. carboxidovorans OM5 genome. Circles display (from inside): genomic position in kilobases, GC-skew, GC content, BLASTX results with strain B. japonicum strain USDA110 (blast 2), BLASTX results with N. hamburgensis strain X14 (blast 1), predicted protein-coding sequences (CDS) on the reverse strand, and predicted CDS on the forward strand. Protein-coding sequences on the outer two circles are colored according to predicted COG (clusters of orthologous groups) functional categories. The single letter COG group identifications are described in Table 2.
The final sequences discussed in this paper (GenBank: NC_011386; tax ID 504832) are available at Comprehensive Microbial Resource (CMR) (cmr.jcvi.org/cgi-bin/CMR), Integrated Microbial Genomes (IMG) http://img.jgi.doe.gov/cgi-bin/pub/main.cgi, KEGG, and Genoscope (Oligotroscope).
Comparative genomics
O. carboxidovorans, N. hamburgensis, and Bradyrhizobium spp. have contrasting physiologies and ecological niches. N. hamburgensis is ubiquitous in nature; it has been isolated from soil, building sandstone, and sewage sludge. It is similar to O. carboxidovorans in that it is a facultatively lithoautotrophic bacterium, but it differs from O. carboxidovorans by using nitrite to nitrate oxidation (nitrification) as its autotrophic growth energy source. N. hamburgensis can also grow mixotrophically with NO2 and organic components or heterotrophically using only organic compounds [16,17]. Its current biotechnology applications include efficient transformation of fertilizer nitrogen into readily usable forms (such as nitrates) and nitrogen removal during wastewater treatment. B. japonicum USDA110 is a nitrogen-fixing bacterium that develops a symbiotic relationship with the soybean plant Glycine max by establishing root nodules [18]. Such symbiotic relations are agriculturally important as they obviate the need for expensive and environmentally damaging fertilizer. Bradyrhizobium sp. BTAi1 is a photosynthetic rhizobium that is also capable of nitrogen fixation, the first known example of such an organism [19]. It can form stem and root nodules.
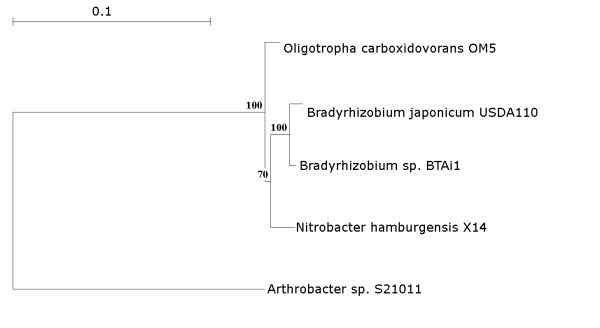
The OM5 16 S rDNA genes are most similar to those of Nitrobacter hamburgensis X14, followed by Bradyrhizobium sp. strains USDA110 and BTAi1 (Figure 2). The OM5 genome is also more syntenic to N. hamburgensis X14 than Bradyrhizobium (Table 3). Thus, overall gene synteny substantiates the rDNA phylogeny, but regional synteny between the three strains varies considerably. Synteny is well conserved between the three species in regions containing genes that encode conserved functions, such as ABC transporters, heavy metal binding and transporter proteins, ribosomal proteins, and metabolic pathways (amino acid metabolism, TCA cycle, flagellar proteins, chaperones, etc.). By contrast, synteny is less conserved in genes encoding some of the hypothetical proteins, and this difference is likely due to specific evolutionary adaption. A comparison of general genome features shows considerable variation in genome size in the three species, but otherwise the three species are similar (Table 4).
Figure 2.
Phylogenetic tree on the basis of 16 S rRNA genes for O. carboxidovorans OM5, B. japonicum USDA110, Nitrobacter hamburgensis X14, and Bradyrhizobium sp. BTAi1 where Arthrobacter was used as outgroup. The tree was created using Treecon. Bootstrapping is shown for values 70% and above. The tree is based on distance matrix where 0.1 means 10% difference between two sequences.
Table 3.
Putative orthology and synteny relations between O. carboxidovorans and other closely related genomes
| Strain | CDS Number* | Percent BBH† | Number of syntons‡ | CDS in syntons (%) |
|---|---|---|---|---|
| Bradyrhizobium japonicum USDA110 | 8317 | 29.79 | 501 | 28.9 |
| Bradyrhizobium sp. BTAi1 | 7394 | 33.35 | 467 | 31.82 |
| Bradyrhizobium sp. ORS278 | 6717 | 35.22 | 394 | 31.40 |
| Nitrobacter hamburgensis X14 | 3804 | 58.20 | 288 | 52.29 |
| Nitrobacter winogradskyi Nb-255 | 3122 | 66.18 | 264 | 60.28 |
| Rhodopseudomonas palustris CGA009 | 4813 | 47.85 | 391 | 43.74 |
| Rhodopseudomonas palustris HaA2 | 4683 | 66.18 | 365 | 60.28 |
| Azorhizobium caulinodans ORS571 | 4717 | 43.31 | 484 | 34.24 |
| Xanthobacter autotrophicus Py2 | 4746 | 42.96 | 521 | 36.98 |
| Methylobacterium populi BJ001 | 5314 | 39.14 | 457 | 27.31 |
| Oligotropha carboxidovorans OM5 | 3722 | 99.97 | 1 | 100 |
*Number of coding sequences (CDS) encoded in each respective genome
†Percent bidirectional best hits (BBH) in each respective species compared to O. carboxidovorans. BBH are defined as gene couples encoding proteins that are considered orthologous based on either reciprocal best BLAST hits or satisfying a BLASTp alignment threshold (a minimum of 35% sequence identity over 80% of the length of the smaller protein).
‡The number of syntons shared between each respective species and O. carboxidovorans. A synton is a conserved gene cluster (synteny group) shared between two bacterial genomes. All possible kinds of chromosomal rearrangements are allowed (inversion, insertion/deletion). The gap parameter, representing the maximum number of consecutive genes that are not involved in a synteny group, was set to five genes.
Table 4.
Comparison of general features between O. carboxidovorans OM5, B. japonicum USDA110, and N. hamburgensis X14
| OM5 | USDA110 | X14 | |
|---|---|---|---|
| Size (Mb) | 3745629 | 9105828 | 4406967 |
| GC% | 62.39 | 64.05 | 61.7 |
| % Coding | 87.59 | 86.59 | 83.8 |
| rRNA | 3 | 3 | 3 |
| tRNA | 48 | 51 | 55 |
| Genes not assigned role category | 1.31% | 25.87% | 1.33% |
| Genes assigned role category | 72.01% | 47.66% | 57.94% |
| Total genes | 3738 | 10740 | 4853 |
Although these three genera have different physiologies, they show significant homology at the protein level. O. carboxidovorans OM5 shares 1148 orthologous proteins (bidirectional best hits (BBHs) with identity ≥ 70%) [20] with N. hamburgensis X14, and it shares 1157 with B. japonicum USDA110. Comparing all three strains using the same criteria, there are 953 proteins that have orthologs in all three strains [Additional file 1]. Many of these are "housekeeping" proteins involved in DNA metabolism, repair, protein synthesis, central metabolic pathways, and chaperones/heat shock proteins. Many of the flagellar proteins and proteins responsible for nitrogen uptake and regulation had orthologs in all three species. Membrane proteins involved in uptake of cations and heavy metal ions (such as CusA (OCAR_4024) and CzcA-like proteins (OCAR_4741, OCAR_4871, OCAR_6437, OCAR_6658, OCAR_7479, OCAR_7726, and OCAR_7738)) also had orthologs in all three species.
On the other hand, 1090 O. carboxidovorans proteins did not have similarity with any proteins from B. japonicum and N. hamburgensis (identity < 30%) [21] [Additional file 2]. Most of these proteins are hypothetical; some are phage integrase proteins, and a few are transcriptional regulators or permease genes responsible for transport of ions across the cell membrane. These proteins appear unique to O. carboxidovorans and could be interesting for further study to determine their roles in cellular metabolism.
Mobile and extrachromosomal elements
The O. carboxidovorans genome has 49 putative phage or transposon related genes and two prophages as determined by Prophinder. No transposons are present. This number of bacteriophage genes is lower than N. hamburgensis strain X14, which has 96 predicted phage-related genes and six prophage genes. High genome plasticity or presence of a large number of mobile elements can indicate the need for rapid changes in response to the environment [22]. For example, the Pseudomonas fluorescens Pf-5 genome contains at least 8 large DNA segments that have evolved due to genetic exchanges with other bacteria [23]; these adaptations improved its ability to compete and survive in the dynamic and microbiologically complex rhizosphere. Shewanella oneidensis MR-1 also has a large number of mobile elements that provide a means of rapid adaptation [24]. Thus, O. carboxidovorans may live in an environment where there is less selective pressure compared to N. hamburgensis. By contrast, B. japonicum strain USDA110 does not have any predicted prophages (although it has 20 predicted phage-related genes). Genes coding for mobile and extrachromosomal element functions form a distinct and major category only in N. hamburgensis and not in the other two species.
Energy metabolism
Many of the unique features of OM5 energy metabolism enabling it to use CO (carboxidotrophy) or H2 (hydrogenotrophy) under aerobic chemolithoautotrophic conditions [25,26] are encoded on the pHCG3 megaplasmid. OM5 central energy metabolism genes are similar to N. hamburgensis and B. japonicum (as well as other members of Bradyrhizobiaceae). For energy production, OM5 mainly uses NADH-quinone oxidoreductases by oxidative phosphorylation. The chromosome encodes a F-type ATPase that is typical in bacteria and cytochrome c oxidase (cbb3 type), which would be used in various other carbon metabolism pathways. Gene(s) responsible for glycolysis and gluconeogenesis are located on the chromosome; they allow heterotrophic growth on nutrient rich media (tryptic soy broth) and on minimal medium containing acetate [8].
Carbohydrate metabolism
Genes enabling CO2 fixation via the Calvin-Benson-Bassham (CBB) reductive pentose phosphate cycle are located on the megaplasmid [4]. The OM5 chromosome encodes a complete Krebs cycle [Additional file 3] and a complete anaplerotic glyoxylate cycle [Additional file 3]. The glyoxylate cycle functions similar to the Krebs cycle but lacks enzyme steps that generate CO2. It is primarily an oxidative pathway that allows synthesis of carbohydrates from acetyl~SCoA, which usually is derived from the oxidation of fatty acids by the β-oxidation pathway. The glyoxylate cycle is found in many β-proteobacteria, including OM5, particularly in organisms that grow well in media in which acetate and other Krebs cycle dicarboxylic acid intermediates are the sole carbon growth source. Compared to N. hamburgensis, the OM5 chromosome encodes many gene(s) that participate in glyoxylate and dicarboxylate metabolism alone [Additional file 3]. The difference may reflect the inability of N. hamburgenesis to utilize dicarboxylates. B. japonicum is capable of catabolizing dicarboxylic acids [16,18]; not surprisingly, it has nearly the same number of gene(s) as O. carboxidovorans.
Fatty acid biosynthesis and oxidation
All three species have enzymes participating in saturated and unsaturated fatty acid and phospholipid synthesis. In OM5, of the 2.54% genes responsible for fatty acid and phospholipid metabolism, 61 code for fatty acid biosynthesis, and 32 code for fat degradation. Genes present in OM5 involved in fatty acid biosynthesis encode orthologs for Acp, AccA, AccB, AccC, AccD, FabD, FabH, FabF, FabG, FabA, FabZ, and FabI [27]. OM5 is capable of generating malonyl-CoA via acetyl-CoA carboxylase (Acc) (OCAR_5979, 5980, 4079) and transferring malonyl-CoA to acyl carrier protein by FabD. It also has genes necessary for fatty acid biosynthesis initiation (fabH, fabB and fabF), keto group reduction (fabG), dehydration (fabA or fabZ), and enoyl reduction (fabI). As expected, because O. carboxidovorans, N. hamburgensis, or B. japonicum have fabI, they do not have fabK and fabL.
Compared with N. hamburgensis or B. japonicum, O. carboxidovorans has fewer fatty acid oxidation enzymes [Additional file 3]. In particular, OM5 lacks the genes that degrade long chain fatty acids. For example, O. carboxidovorans does not have a gene encoding bll2324 (long-chain-fatty-acid--[acyl-carrier-protein] ligase), which is involved in degradation of long chain fatty acids. This gene is present in B. japonicum. Fatty acid methyl ester analysis on strain OM5 indicated an accumulation of some long chain fatty acids when it is grown under different environmental conditions (unpublished results). We believe the absence of these genes in O. carboxidovorans may contribute to accumulation of long chain fatty acids under certain conditions.
Chemolithoautotrophic growth
O. carboxidovorans is an autotrophic organism that can grow at the expense of inorganic compounds and use CO2 as a carbon source. Its nitrogen comes from inorganic compounds such as NH3, NO3-, or N2. During chemolithoautotrophic growth, it derives energy from oxidation of CO to CO2. This function is encoded on a megaplasmid encoding the enzyme CO dehydrogenase (cox gene cluster). Plasmid-cured mutants (or mutants in which deletions have been introduced into the plasmid) either lose all chemolithoautotrophic growth capabilities or cannot utilize H2 plus CO2 but retain the ability of using CO plus CO2 [4,25,28].
The N. hamburgensis genome contains genes that are found in carboxidotrophic bacteria. For example, it has several genes encoding cytochrome b561, an important electron transfer component in aerobic carboxidotrophic bacteria [29]. The N. hamburgensis genome also has four gene clusters and a lone CDS encoding multiple homologs of molybdopterin-containing carbon monoxide dehydrogenase (Mo-CODH). The largest of these clusters (Nham_2601 to Nham_2608) has high similarity to B. japonicum USDA110 and few other species [16]. In fact, the N. hamburgensis genome has more copies of Mo-CODH-like genes than it does of nitrite oxidoreductase (membrane bound Mo-dependent enzyme required for converting nitrite to nitrate) [16]. This suggests that these proteins have a vital, but poorly understood, role in the lifestyle of N. hamburgensis. B. japonicum USDA110 also encodes a Mo-CODH, and it can oxidize CO at the expense of nitrate reduction under anaerobic conditions. However, it does not grow under these conditions [30]. In strain USDA110, anaerobic CO uptake only occurs in the presence of nitrate, which is not a substrate or electron acceptor for anaerobic CODH. Therefore, no carbon fixation occurs [16,31]. Thus, although these two species differ from O. carboxidovorans in their chemolithoautotrophic lifestyle, they have similar enzyme(s) for this purpose, particularly CODH.
In O. carboxidovorans, the cox genes are specifically and coordinately transcribed under chemolithoautotrophic conditions in the presence of CO [26]; therefore, sensing CO is an important function. CO sensors are typically heme-based [32]. Two O. carboxidovorans genes encode proteins (CoxC and CoxH) that are putatively associated with CO sensing based on sequence motifs similar to two component regulatory systems [4]. However, the O. carboxidovorans sequence upstream of the cox cluster does not have genes with unambiguous functions in the sensing of CO or the regulation of cox genes. Other species, such as Azotobacter vinelandii and Bradyrhizobium, contain a gene encoding CooA transcriptional receptor that may be involved in CO sensing and regulating the function of CO dehydrogenase [4]. BLASTP results suggest this protein belongs to the ACAD superfamily, and it shows similarity to acyl-CoA dehydrogenases involved in lipid metabolism. O. carboxidovorans has chromosomally encoded acyl-CoA dehydrogenase genes; some of them are directly involved in fatty acid metabolism (OCAR_5221, OCAR_5223, OCAR_5308, OCAR_5791, OCAR_6069, OCAR_6878, and OCAR_6891). Therefore, it is possible that chromosomally encoded lipid metabolism in O. carboxidovorans and plasmid-encoded CO fixation are linked, but further studies are needed to confirm this.
Transport and binding
One tenth of O. carboxidovorans genes are responsible for cell motility, binding, and membrane transport. By contrast, 6% of N. hamburgensis genes are in this category. This difference likely reflects their contrasting lifestyles. As an environmental species, O. carboxidovorans likely has increased need for a diversity of these functions, while the symbiotic species N. hamburgensis is in a relatively constant environment.
Iron transport and related proteins are responsible for specific uptake of ferric-siderophore complexes, which are high affinity iron chelators in gram-negative bacteria. O. carboxidovorans codes for 3 Fhu gene(s), OCAR_4564, OCAR_4560, and OCAR_4561, that are periplasmic proteins and may function as an ABC-type transport mechanism driven by ATP hydrolysis. The E. coli fhu system consists of four genes, designated fhuA, fhuC, fhuD and fhuB, that are arranged in one operon [33]. Periplasmic FhuD (31 kDa) and cytoplasmic-membrane-associated FhuC (29 kDa) and FhuB (41 kDa) are proteins necessary for the transport of ferrichrome and other Fe3+-hydroxamate compounds (Fe3+-aerobactin, Fe3+-coprogen) across the cytoplasmic membrane from the periplasm into the cytoplasm [34-36]. The protein complex TonB-ExbB-ExbD [37,38] provides energy for this process. The presence of genes encoding these proteins suggests a functional iron siderophore transport complex in O. carboxidovorans.
Iron acquisition is important in bacteria, both as a requirement for growth and as an environmental signal that regulates gene expression [39]. Iron transport is especially important in nitrogen fixing bacteria [40] because it increases fitness in the competitive rhizosphere [41,42]. Rhizobium leguminosarum (a symbiotic nitrogen fixing species) contains a fhuDCB operon for siderophore uptake [43]. Bradyrhizobium also has iron transport systems, but unlike O. carboxidovorans, it does not utilize TonB iron transport. The presence of Fe siderophores and transporter genes in non pathogenic, non-nitrogen fixing O. carboxidovorans reflects its similarity to nitrogen fixing symbiotic bacteria. The presence of these genes in O. carboxidovorans also indicates that its natural environment, like most natural environments, is restricted in iron [40].
Unlike N. hamburgensis, O. carboxidovorans and B. japonicum have nickel/peptide transporting genes. Other O. carboxidovorans transport related proteins (e.g,. oligosaccharide/lipoprotein/amino acid transport related proteins) are also present in N. hamburgensis and B. japonicum.
Conclusion
The previously reported sequence of the O. carboxidovorans megaplasmid revealed mechanisms for its chemolithoautotrophic growth. The complete O. carboxidovorans genome reveals metabolic pathways used during heterotrophic growth and how chemolithoautotrophic growth is linked with central metabolism. Further research is needed to identify genes involved in the regulation of chemolithoautotrophic growth versus heterotrophic growth.
O. carboxidovorans appears to be adapted to a relatively stable microenvironment because its genome contains remarkably few phage- or transposon-related genes in comparison to N. hamburgensis and other soil bacteria. The lack of any typical Gram-negative communication system based on acylated homoserine lactones (AHLs) also suggests an exclusive microhabitat, similar to Azoarcus BH72 [44]. Production of AHLs is not uncommon in environmental bacteria, but it only occurs in some microhabitats under appropriate conditions [45]. Another important characteristic of the O. carboxidovorans genome is the presence of TonB-dependent receptors for iron transport across the cell membrane, unlike N. hamburgensis and B. japonicum. The role of iron acquisition in O. carboxidovorans metabolism is another very interesting area for future research.
O. carboxidovorans, N. hamburgensis, and B. japonicum each have unique genetic pathways that enable them to have specialized functions such as CO oxidation, nitrite oxidation, and N2 fixation. These genetic pathways have historically been the most well studied from these species. However, comparison of their genomes reveals that there are many metabolic pathways that are conserved between these closely related species. Comparison of these three genomes also revealed previously unknown differences in the "conserved" metabolic pathways. The cumulative effects of these metabolic differences, along with the unique pathways, allow these phylogenetically related species to adapt to diverse environments [46]. Therefore, the unique pathways for these organisms must be understood in the context of their respective genomes. This current analysis is only a beginning, and further work on the function of O. carboxidovorans proteins will yield valuable information on how this species survives in its unique environment and how it adapts to both heterotrophic and chemolithoautotrophic lifestyles.
Methods
Sequencing methods
O. carboxidovorans OM5 (ATCC no. 49405) was grown on carboxydobacterium medium (ATCC no 1789) using acetate as a substrate at 30°C with continuous shaking for 72 h under aerobic conditions. DNA isolation, sequencing, and assembly were conducted as described previously [8]. The assembly at this point had two large contigs. The two gaps were closed by PCR amplification and primer walking. Sequencing reactions were performed to correct a misassembly in the genome. The gaps had G+C rich regions that resulted in early signal loss during sequencing. To circumvent this problem, we used SequenceRx Enhancer Solution A (Invitrogen), which enables longer reads on templates containing regions with secondary structure caused by high G+C content, small nucleotide repeats, hairpins, direct repeats, and inverted repeats. Other techniques were used by MWG Biotech (Eurofins) to resolve hard sequencing stops to complete the reads. The complete genome was submitted to NCBI.
Once the complete genome was submitted to NCBI, Comprehensive Microbial Resource (CMR) and Integrated Microbial Genomes (IMG) were used to compare OM5 against all of the publicly available, complete prokaryotic genomes. Orifinder, Prophinder [47]http://aclame.ulb.ac.be/Tools/Prophinder/, and CGVIEW were used to determine the origin of the circular genome, identify prophages, and to obtain a circular graphical representation of the genome.
Comparative genomics
The O. carboxidovorans genome sequence was submitted to Magnifying Genomes Microbial Genome Annotation System (MaGe) [48], which benevolently created Oligotroscope. This website allowed manual annotation as well as orthology and synteny detection of microbial genomes available in NCBI. Putative orthologous relationships between genomes were defined as gene couples satisfying the bidirectional best hit (BBH) criterion or a BLASTP alignment threshold (a minimum of 70% sequence identity over 80% of the length of the smallest protein) [48]. Unique proteins were identified as proteins having <30% sequence identity over 80% of the length. These relationships were subsequently used to search for conserved gene clusters (synteny groups or syntons) among several bacterial genomes using the Syntonizer function. All possible kinds of chromosomal rearrangements were allowed (inversion, insertion/deletion). The gap parameter, representing the maximum number of consecutive genes that are not involved in a synteny group, was set to five genes. Orthology relations were strengthened by synteny detection (conservation of chromosomal co-localization between pairs of orthologous genes from different genomes) using the Phyloprofile synteny option in Oligotroscope. The method used was not restricted to the BBH definition; therefore, it allowed for multiple correspondences between genes (fusion/fission, duplication) and chromosomal rearrangements (inversion, insertion/deletion). Oligotroscope was also used to determine the misc_RNAs in the genome (Table 1).
We determined O. carboxidovorans-specific genes (genes encoding proteins where no orthologs were detected in the compared species and not in any synteny group) using the two most closely related species with fully sequenced genomes for comparison, Bradyrhizobium sp. (strain USDA110 and BTAi1) and N. hamburgensis X14. Cartographic representations in the MaGe web interface allowed us to confirm the manual annotations and detect specific gene(s).
Phylogenetic tree construction
For tree construction, 16 S rRNA sequences were aligned in ClustalX [49], followed by tree construction using Treecon [50]. Distance estimations were performed in Treecon by Jukes and Cantor method [51]. Tree topology for the 16 S rRNA nucleotide sequences were inferred by the distance matrix using Neighbour Joining method [52].
Pathway analysis
For pathway analysis, we used the Kyoto Encyclopedia of Genes and Genomes (KEGG) PATHWAY Database, which includes the completed sequence of strain OM5 http://www.genome.jp/kegg-bin/show_organism?org=oca. The KEGG database was also used to trace the enzymes/pathways in Bradyrhizobium USDA110 and N. hamburgensis X14 and compare with strain OM5.
Authors' contributions
DP conducted sequencing and closure of the genome as well as annotation and analysis. SMB, SCB, YSD, and MLL conceived and directed the project. SMB and YSD oversaw computational analyses, SCB and MLL oversaw sequencing aspects, and MLL oversaw microbiological aspects. DP wrote the manuscript draft, and all authors edited and approved the final manuscript.
Supplementary Material
Orthologous proteins in O. carboxidovorans OM5, N. hamburgensis X14, and Bradyrhizobium spp. USDA110.
Proteins unique to O. carboxidovorans OM5 as compared to N. hamburgensis X14, and Bradyrhizobium spp. USDA110.
O. carboxidovorans enzyme complexes involved in some metabolic pathways.
Contributor Information
Debarati Paul, Email: dp307@msstate.edu.
Susan M Bridges, Email: bridges@cse.msstate.edu.
Shane C Burgess, Email: burgess@cvm.msstate.edu.
Yoginder S Dandass, Email: yogi@cse.msstate.edu.
Mark L Lawrence, Email: lawrence@cvm.msstate.edu.
Acknowledgements
This research was supported by the U.S. Department of Energy through the MSU Sustainable Energy Research Center. We are grateful to the Mississippi State University Life Sciences and Biotechnology Institute for hosting Manatee. We thank Todd French for technical advice and Michelle Banes for technical assistance. We thank Ranjit Kumar and Philippe Chouvarine for bioinformatics support. We gratefully acknowledge the assistance from all the web-based analysis tools that have been used for preparing the manuscript.
References
- Meyer O, Schlegel HG. Reisolation of the carbon monoxide utilizing hydrogen bacterium Pseudomonas carboxydovorans (Kistner) comb. nov. Archives of microbiology. 1978;118(1):35–43. doi: 10.1007/BF00406071. [DOI] [PubMed] [Google Scholar]
- Pelzmann A, Ferner M, Gnida M, Meyer-Klaucke W, Maisel T, Meyer O. The CoxD Protein of Oligotropha carboxidovorans Is a Predicted AAA+ ATPase Chaperone Involved in the Biogenesis of the CO Dehydrogenase [CuSMoO2] Cluster. The Journal of biological chemistry. 2009;284(14):9578–9586. doi: 10.1074/jbc.M805354200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergey DH, Holt JG. Bergey's manual of determinative bacteriology. 9. Baltimore: Williams & Wilkins; 1994. [Google Scholar]
- Fuhrmann S, Ferner M, Jeffke T, Henne A, Gottschalk G, Meyer O. Complete nucleotide sequence of the circular megaplasmid pHCG3 of Oligotropha carboxidovorans: function in the chemolithoautotrophic utilization of CO, H(2) and CO(2) Gene. 2003;322:67–75. doi: 10.1016/j.gene.2003.08.027. [DOI] [PubMed] [Google Scholar]
- Guerrero R. Bergey's manuals and the classification of prokaryotes. Int Microbiol. 2001;4(2):103–109. doi: 10.1007/s101230100021. [DOI] [PubMed] [Google Scholar]
- Hugendieck I, Meyer O. The structural genes encoding CO dehydrogenase subunits (cox L, M and S) in Pseudomonas carboxydovorans OM5 reside on plasmid pHCG3 and are, with the exception of Streptomyces thermoautotrophicus, conserved in carboxydotrophic bacteria. Archives of microbiology. 1992;157(3):301–304. doi: 10.1007/BF00245166. [DOI] [PubMed] [Google Scholar]
- Santiago B, Meyer O. Purification and molecular characterization of the H2 uptake membrane-bound NiFe-hydrogenase from the carboxidotrophic bacterium Oligotropha carboxidovorans. Journal of bacteriology. 1997;179(19):6053–6060. doi: 10.1128/jb.179.19.6053-6060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D, Bridges S, Burgess SC, Dandass Y, Lawrence ML. Genome sequence of the chemolithoautotrophic bacterium Oligotropha carboxidovorans OM5T. Journal of bacteriology. 2008;190(15):5531–5532. doi: 10.1128/JB.00614-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, Rfam: updates to the RNA families database. Nucleic acids research. 2009. pp. D136–140. [DOI] [PMC free article] [PubMed]
- Vogel J, Argaman L, Wagner EG, Altuvia S. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr Biol. 2004;14(24):2271–2276. doi: 10.1016/j.cub.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jager JG, Huttenhofer A, Wagner EG. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic acids research. 2003;31(22):6435–6443. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Wagner EG. Target identification of small noncoding RNAs in bacteria. Current opinion in microbiology. 2007;10(3):262–270. doi: 10.1016/j.mib.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Ranquet C, Gottesman S. Translational regulation of the Escherichia coli stress factor RpoS: a role for SsrA and Lon. Journal of bacteriology. 2007;189(13):4872–4879. doi: 10.1128/JB.01838-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Zhang CT. Ori-Finder: a web-based system for finding oriCs in unannotated bacterial genomes. BMC bioinformatics. 2008;9:79. doi: 10.1186/1471-2105-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stothard P, Wishart DS. Circular genome visualization and exploration using CGView. 2005. pp. 537–539. [DOI] [PubMed]
- Starkenburg SR, Larimer FW, Stein LY, Klotz MG, Chain PS, Sayavedra-Soto LA, Poret-Peterson AT, Gentry ME, Arp DJ, Ward B. et al. Complete genome sequence of Nitrobacter hamburgensis X14 and comparative genomic analysis of species within the genus Nitrobacter. Applied and environmental microbiology. 2008;74(9):2852–2863. doi: 10.1128/AEM.02311-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirstein K, Bock E. Close genetic relationship between Nitrobacter hamburgensis nitrite oxidoreductase and Escherichia coli nitrate reductases. Archives of microbiology. 1993;160(6):447–453. doi: 10.1007/BF00245305. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Nakamura Y, Sato S, Minamisawa K, Uchiumi T, Sasamoto S, Watanabe A, Idesawa K, Iriguchi M, Kawashima K. et al. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 2002;9(6):189–197. doi: 10.1093/dnares/9.6.189. [DOI] [PubMed] [Google Scholar]
- Cytryn EJ, Jitacksorn S, Giraud E, Sadowsky MJ. Insights learned from pBTAi1, a 229-kb accessory plasmid from Bradyrhizobium sp. strain BTAi1 and prevalence of accessory plasmids in other Bradyrhizobium sp. strains. The ISME journal. 2008;2(2):158–170. doi: 10.1038/ismej.2007.105. [DOI] [PubMed] [Google Scholar]
- Hiller NL, Janto B, Hogg JS, Boissy R, Yu S, Powell E, Keefe R, Ehrlich NE, Shen K, Hayes J. et al. Comparative genomic analyses of seventeen Streptococcus pneumoniae strains: insights into the pneumococcal supragenome. Journal of bacteriology. 2007;189(22):8186–8195. doi: 10.1128/JB.00690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Ouzounis C, Sander C, Tuparev G, Vriend G. A database of protein structure families with common folding motifs. Protein Sci. 1992;1(12):1691–1698. doi: 10.1002/pro.5560011217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerlin P, Reid-Smith RJ. Antimicrobial resistance: its emergence and transmission. Animal health research reviews/Conference of Research Workers in Animal Diseases. 2008;9(2):115–126. doi: 10.1017/S146625230800159X. [DOI] [PubMed] [Google Scholar]
- Mavrodi DV, Loper JE, Paulsen IT, Thomashow LS. Mobile genetic elements in the genome of the beneficial rhizobacterium Pseudomonas fluorescens Pf-5. BMC microbiology. 2009;9:8. doi: 10.1186/1471-2180-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romine MF, Carlson TS, Norbeck AD, McCue LA, Lipton MS. Identification of mobile elements and pseudogenes in the Shewanella oneidensis MR-1 genome. Applied and environmental microbiology. 2008;74(10):3257–3265. doi: 10.1128/AEM.02720-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraut M, Hugendieck I, Herwig S, Meyer O. Homology and distribution of CO dehydrogenase structural genes in carboxydotrophic bacteria. Archives of microbiology. 1989;152(4):335–341. doi: 10.1007/BF00425170. [DOI] [PubMed] [Google Scholar]
- Santiago B, Schubel U, Egelseer C, Meyer O. Sequence analysis, characterization and CO-specific transcription of the cox gene cluster on the megaplasmid pHCG3 of Oligotropha carboxidovorans. Gene. 1999;236(1):115–124. doi: 10.1016/S0378-1119(99)00245-0. [DOI] [PubMed] [Google Scholar]
- Heath RJ. Bacterial fatty-acid biosynthesis: an antibacterial drug target waiting to be exploited. Drug discovery today. 2001;6(14):715.. doi: 10.1016/S1359-6446(01)01881-5. [DOI] [PubMed] [Google Scholar]
- Kraut M, Meyer O. Plasmids in carboxydotrophic bacteria: physical and restriction analysis. Archives of microbiology. 1988;149(6):540–546. doi: 10.1007/BF00446758. [DOI] [Google Scholar]
- Meyer O, Gremer L, Ferner R, Ferner M, Dobbek H, Gnida M, Meyer-Klaucke W, Huber R. The role of Se, Mo and Fe in the structure and function of carbon monoxide dehydrogenase. Biological chemistry. 2000;381(9-10):865–876. doi: 10.1515/BC.2000.108. [DOI] [PubMed] [Google Scholar]
- Lorite MJ, Tachil J, Sanjuan J, Meyer O, Bedmar EJ. Carbon monoxide dehydrogenase activity in Bradyrhizobium japonicum. Applied and environmental microbiology. 2000;66(5):1871–1876. doi: 10.1128/AEM.66.5.1871-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King GM. Nitrate-dependent anaerobic carbon monoxide oxidation by aerobic CO-oxidizing bacteria. FEMS microbiology ecology. 2006;56(1):1–7. doi: 10.1111/j.1574-6941.2006.00065.x. [DOI] [PubMed] [Google Scholar]
- Chan MK. CooA, CAP and allostery. Nature structural biology. 2000;7(10):822–824. doi: 10.1038/79559. [DOI] [PubMed] [Google Scholar]
- Burkhardt R, Braun V. Nucleotide sequence of the fhuC and fhuD genes involved in iron (III) hydroxamate transport: domains in FhuC homologous to ATP-binding proteins. Mol Gen Genet. 1987;209(1):49–55. doi: 10.1007/BF00329835. [DOI] [PubMed] [Google Scholar]
- Braun V, Gunter K, Hantke K. Transport of iron across the outer membrane. Biology of metals. 1991;4(1):14–22. doi: 10.1007/BF01135552. [DOI] [PubMed] [Google Scholar]
- Coulton JW, Mason P, Allatt DD. fhuC and fhuD genes for iron (III)-ferrichrome transport into Escherichia coli K-12. Journal of bacteriology. 1987;169(8):3844–3849. doi: 10.1128/jb.169.8.3844-3849.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mademidis A, Killmann H, Kraas W, Flechsler I, Jung G, Braun V. ATP-dependent ferric hydroxamate transport system in Escherichia coli: periplasmic FhuD interacts with a periplasmic and with a transmembrane/cytoplasmic region of the integral membrane protein FhuB, as revealed by competitive peptide mapping. Molecular microbiology. 1997;26(5):1109–1123. doi: 10.1046/j.1365-2958.1997.6592008.x. [DOI] [PubMed] [Google Scholar]
- Gunter K, Braun V. In vivo evidence for FhuA outer membrane receptor interaction with the TonB inner membrane protein of Escherichia coli. FEBS letters. 1990;274(1-2):85–88. doi: 10.1016/0014-5793(90)81335-L. [DOI] [PubMed] [Google Scholar]
- Postle K, Kadner RJ. Touch and go: tying TonB to transport. Molecular microbiology. 2003;49(4):869–882. doi: 10.1046/j.1365-2958.2003.03629.x. [DOI] [PubMed] [Google Scholar]
- Braun V. Iron uptake mechanisms and their regulation in pathogenic bacteria. Int J Med Microbiol. 2001;291(2):67–79. doi: 10.1078/1438-4221-00103. [DOI] [PubMed] [Google Scholar]
- Barton L, Johnson G, Bishop Y. The Metabolism of Iron by Nitrogen-Fixing Rhizospheric Bacteria. Iron Nutrition in Plants and Rhizospheric Microorganisms. 2006. pp. 199–214. full_text.
- Yan Y, Yang J, Dou Y, Chen M, Ping S, Peng J, Lu W, Zhang W, Yao Z, Li H. et al. Nitrogen fixation island and rhizosphere competence traits in the genome of root-associated Pseudomonas stutzeri A1501. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(21):7564–7569. doi: 10.1073/pnas.0801093105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilands JB. Siderophores: structure and function of microbial iron transport compounds. The Journal of biological chemistry. 1995;270(45):26723–26726. doi: 10.1074/jbc.270.45.26723. [DOI] [PubMed] [Google Scholar]
- Stevens JB, Carter RA, Hussain H, Carson KC, Dilworth MJ, Johnston AW. The fhu genes of Rhizobium leguminosarum, specifying siderophore uptake proteins: fhuDCB are adjacent to a pseudogene version of fhuA. Microbiology (Reading, England) 1999;145(Pt 3):593–601. doi: 10.1099/13500872-145-3-593. [DOI] [PubMed] [Google Scholar]
- Krause A, Ramakumar A, Bartels D, Battistoni F, Bekel T, Boch J, Bohm M, Friedrich F, Hurek T, Krause L. et al. Complete genome of the mutualistic, N2-fixing grass endophyte Azoarcus sp. strain BH72. Nature biotechnology. 2006;24(11):1385–1391. doi: 10.1038/nbt1243. [DOI] [PubMed] [Google Scholar]
- Burmølle M, Hansen LH, Oregaard G, Sørensen SJ. Presence of N-Acyl Homoserine Lactones in Soil Detected by a Whole-Cell Biosensor and Flow Cytometry. Microbial Ecology. 2003;45(3):226–236. doi: 10.1007/s00248-002-2028-6. [DOI] [PubMed] [Google Scholar]
- Kelley BP, Sharan R, Karp RM, Sittler T, Root DE, Stockwell BR, Ideker T. Conserved pathways within bacteria and yeast as revealed by global protein network alignment. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(20):11394–11399. doi: 10.1073/pnas.1534710100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima-Mendez G, Van Helden J, Toussaint A, Leplae R. Prophinder: a computational tool for prophage prediction in prokaryotic genomes. Bioinformatics (Oxford, England) 2008;24(6):863–865. doi: 10.1093/bioinformatics/btn043. [DOI] [PubMed] [Google Scholar]
- Vallenet D, Labarre L, Rouy Z, Barbe V, Bocs S, Cruveiller S, Lajus A, Pascal G, Scarpelli C, Medigue C. MaGe: a microbial genome annotation system supported by synteny results. Nucleic acids research. 2006;34(1):53–65. doi: 10.1093/nar/gkj406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic acids research. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, De Wachter R. Construction of evolutionary distance trees with TREECON for Windows: accounting for variation in nucleotide substitution rate among sites. Comput Appl Biosci. 1997;13(3):227–230. doi: 10.1093/bioinformatics/13.3.227. [DOI] [PubMed] [Google Scholar]
- Som A. Theoretical foundation to estimate the relative efficiencies of the Jukes-Cantor+gamma model and the Jukes-Cantor model in obtaining the correct phylogenetic tree. Gene. 2006;385:103–110. doi: 10.1016/j.gene.2006.03.027. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular biology and evolution. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Orthologous proteins in O. carboxidovorans OM5, N. hamburgensis X14, and Bradyrhizobium spp. USDA110.
Proteins unique to O. carboxidovorans OM5 as compared to N. hamburgensis X14, and Bradyrhizobium spp. USDA110.
O. carboxidovorans enzyme complexes involved in some metabolic pathways.