Abstract
Background
Genetic analysis of transcriptional profiles is a promising approach for identifying and dissecting the genetics of complex traits like meat performance. Accordingly, expression levels obtained by microarray analysis were taken as phenotypes in a linkage analysis to map eQTL. Moreover, expression levels were correlated with traits related to meat quality and principle components with high loadings of these traits. By using an up-to-date annotation and localization of the respective probe-sets, the integration of eQTL mapping data and information of trait correlated expression finally served to point to candidate genes for meat quality traits.
Results
Genome-wide transcriptional profiles of M. longissimus dorsi RNAs samples of 74 F2 animals of a pig resource population revealed 11,457 probe-sets representing genes expressed in the muscle. Linkage analysis of expression levels of these probe-sets provided 9,180 eQTL at the suggestive significance threshold of LOD > 2. We mapped 653 eQTL on the same chromosome as the corresponding gene and these were designated as 'putative cis-eQTL'. In order to link eQTL to the traits of interest, probe-sets were addressed with relative transcript abundances that showed correlation with meat quality traits at p ≤ 0.05. Out of the 653 'putative cis-eQTL', 262 transcripts were correlated with at least one meat quality trait. Furthermore, association of expression levels with composite traits with high loadings for meat quality traits generated by principle component analysis were taken into account leading to a list of 85 genes exhibiting cis-eQTL and trait dependent expression.
Conclusion
Holistic expression profiling was integrated with QTL analysis for meat quality traits. Correlations between transcript abundance and meat quality traits, combined with genetic positional information of eQTL allowed us to prioritise candidate genes for further study.
Background
Genetical genomics as a new approach which combines gene-expression data and marker genotypes in a segregating population, offers great perspectives to make a major contribution to the dissection of complex traits [1,2]. Genetical genomics aims at detecting genomic loci that control variation in gene expression, so-called expression QTL (eQTL; to distinguish them from functional QTL that affect traits at the whole-organism level, subsequently termed pheneQTL(pQTL)). The detected eQTL can represent a locus that lies close to the gene that is being controlled (cis-acting) or one or more loci that are unlinked to the gene that is being controlled (trans-acting) [1]. Expression-QTL for genes showing high correlation with the phenotype may provide the necessary information required for identifying genes that control quantitative phenotypes. Those cis-eQTLs resulting from the correlation of expression profiles with phenotypic measurements represent candidate genes for the genetic regulation underlying the variation of the physiological traits [3,4].
We have previously reported on the identification of eQTL of genes showing expression levels correlated with waterholding capacity of meat measured as drip loss [5]. Here in this study, we expand and up-dated the analysis towards global eQTL mapping of transcripts showing variable abundance in porcine muscle and using annotation and localisation data of Affymetrix microarray probe-sets based on the current porcine sequence information [6]. We focus on eQTL of genes whose expression at slaughter is significantly correlated to technological meat quality traits addressed in commercial pig breeding schemes. Taking into account own observations on the reliability of eQTL mapping we highlight eQTL located on the same chromosome as the corresponding genes themselves [7].
Results
Summary analysis of eQTL detection
Expression data were obtained from M. longissimus dorsi samples of 74 F2 animals of a resource population using Affymetrix Porcine Genome Arrays containing in total 24,123 probe-sets of which 20,689 probe-sets were assigned to a known gene [6]. MAS5 analysis revealed consistent 'present calls' for 11,457 probe-sets. This pre-selected set was further analyzed with the more sophisticated hybrid algorithm PLIER [8,9] and the expression levels were subjected to linkage analysis. Out of 11,457 probe-sets 6,117 showed at least one eQTL at the 5% chromosome-wide significance threshold (average F-value 4.92, corresponding to LOD score = 2.0 and nominal significance of p ≤ 10-3) corresponding approximately to the suggestive linkage threshold proposed by Lander & Kruglyak [10] (Table 1). In total, the 11,457 probe-sets revealed 9,180 eQTL at LOD score > 2 (Additional file 1), 1,058 eQTL at LOD score > 3, and 160 eQTL at LOD score > 4. Only 29 had eQTL with LOD score > 5. This yielded an average of 1.5 eQTL per transcript (ranging from 1 to 6 eQTLs). The eQTL were distributed over all autosomes with some regions harboring particularly many eQTL with high significance (Figure 1). Out of the 9,180 eQTL 8,168 eQTL could be assigned to the porcine genome sequence (Ensembl Sscrofa9 database, released April 2009). In total 653 eQTL were mapped on the same chromosome as the corresponding gene itself. These eQTL are putative cis-eQTL.
Table 1.
Global eQTL summary
| Total # of probe-sets | Lod score threshold | Nominal P | Total # of eQTL | # of probe-sets with at least one eQTL | # of eQTL mapped | # of eQTL with ipsi-chromosomal location* |
|---|---|---|---|---|---|---|
| 11,457 | > 2 | 4.90E-03 | 9180 | 6117 | 8168 | 653 |
| 11,457 | > 3 | 4.59E-04 | 1058 | 990 | 955 | 125 |
| 11,457 | > 4 | 4.50E-05 | 160 | 159 | 139 | 35 |
| 11,457 | > 5 | 3.52E-06 | 29 | 29 | 25 | 13 |
*, i.e. # of eQTL located on the same chromosome as the corresponding probe-set
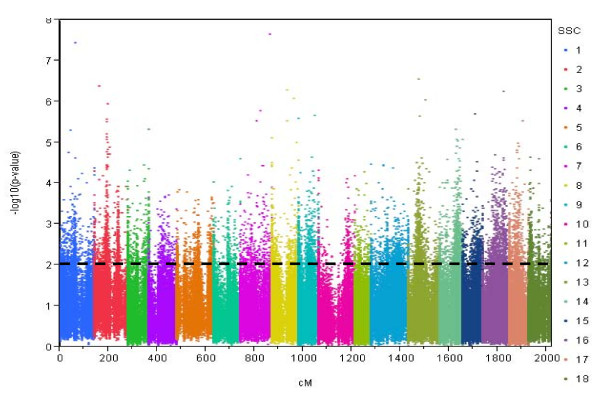
Figure 1.
Graphical scheme of eQTL across 18 chromosome obtained by eQTL analysis of 11457 probe-set. Y-axis shows P-values, expressed on the -log10 scale, and X-axis shows position of eQTL in cM though18 chromosome. The genome-wide significant threshold (p ≤ 0.05) is shown with the dot line.
Integration of trait, expression, and mapping data to identify genes related to the meat quality
In order to link the eQTL to the genetic background of a classical phenotypic trait of interest, it is necessary to establish a relationship between the variation of that classical phenotypic trait, the expression levels of various transcripts, and the mapping position of the eQTL and the corresponding transcripts. Therefore, we selected transcripts whose expression level showed significant correlation with individual meat quality traits and with the composite traits, PC2 and PC3 (Tables 2, 3, 4) (p ≤ 0.05, corresponding to correlation coefficients ranging between |0.24-0.50|) and we focussed on those transcripts with eQTL on their own chromosome as the structurally fixed linkage group (collateral transcript and eQTL position). Out of 653 eQTL that were mapped on the chromosome of their corresponding genes 262 transcripts showed correlation with at least one meat quality trait. These are displayed in Figure 2, 3, 4, 5 and 6. The linkage map showed accurate match of microsatellites marker order with the marker arrangement in the current porcine genome sequence assembly (Sscrofa9 assembly, April 2009). A total of 73 out 115 microsatellites marker were found to align on the porcine genome sequence assembly map position by BLAST analysis. Reflecting this alignment, the position of 262 transcripts in the porcine physical map was assigned to the corresponding eQTL position in the genetic linkage maps. Most of the eQTL of transcripts whose expression level correlated with meat quality traits were found on SSC1, 2, 3, 6, 9 and 14. Table 2 summarizes the number of eQTL related to various meat quality traits.
Table 2.
The number of eQTL of probe-sets exhibiting expression levels that are correlated with meat quality traits
| No. of eQTL with trait correlated express gene | |||
|---|---|---|---|
| Meat quality parameters | Traits | All eQTL | eQTL located on the same chromosome as the corresponding probe-set |
| routine parameters | |||
| - pH | pH1ld | 425 | 35 |
| pH24ld | 384 | 34 | |
| pH24sm | 271 | 28 | |
| - conductivity | LF1h loin | 253 | 16 |
| LF24ld | 793 | 50 | |
| LF24ld | 1012 | 64 | |
| meat texture | DL | 298 | 26 |
| TL | 285 | 28 | |
| CL | 76 | 40 | |
| technological meat quality parameters | QPTO | 273 | 12 |
| SF | 439 | 35 | |
Trait abbreviations:
pH1ld pH-value in Mld at 13th/14th rib 45 min post mortem
pH24ld pH-value in Mld at 13th/14th rib 24 h post mortem
pH24sm pH-value in M. semimembranosus (Msm) at 24 h post mortem
LF1ld conductivity in Mld at 13th/14th rib45 min post mortem
LF24ld conductivity in Mld at 13th/14th rib post mortem
LF24sm conductivity in Msm at 24 h post mortem
DL Drip loss: percentage of weight loss after 48 h of Mld samples collected at 24 h post mortem
TL Thaw loss: percentage of weight loss of Mld samples frozen at -20°C.
CL Cooking loss: percentage of weight loss of Mld samples incubated in water at 75°C for 50 min
OPTO meat colour 24 h post mortem in M. longissimus dorsi (Mld)at 13th/14th rib; OPTO star
SF Shear force was measured by the Instron-4310 equipment
Table 3.
Probe-sets with expression levels that are correlated with principal component 2 and information about their eQTL.
| Annotation | eQTL | individual trait correlation* | |||
|---|---|---|---|---|---|
| Prob_set_IDs | Gene Symbols | SSC | cM | F | traits |
| Ssc.25132.3.S1 | RAD23B | 1 | 130 | 5.11 | DL, -pH1ld, LF1ld, LF24sm |
| Ssc.11839.1.S1 | ZNF462 | 1 | 138 | 8.12 | SF |
| Ssc.21636.1.A1 | FILIP1 | 1 | 142 | 5.4 | pH24ld |
| Ssc.21225.1.S1 | RNASEH2C | 2 | 1 | 5.2 | LF24sm |
| Ssc.27245.1.S1 | RTN3 | 2 | 18 | 10.8 | -LF24sm |
| Ssc.5869.1.S1 | SART1 | 2 | 42 | 8.7 | LF24sm |
| Ssc.11130.2.A1 | 2 | 50 | 5.4 | LF24sm | |
| Ssc.16460.1.S1 | EGR1 | 2 | 52 | 6.2 | -LF24sm |
| Ssc.28435.1.A1 | EHD1 | 2 | 56 | 8.21 | CL, -pH24ld, -pH24sm, LF24sm |
| Ssc.6529.1.A1 | AFF4 | 2 | 132 | 9.3 | pH24ld, pH24sm, -LF24ld |
| Ssc.16877.1.S1 | LOC728816 | 3 | 0 | 6.4 | OPTO |
| Ssc.15224.3.A1 | FOSL2 | 3 | 0 | 5.8 | pH24ld, pH24sm |
| Ssc.1051.1.S1 | TGOLN2 | 3 | 86 | 5.9 | pH24ld |
| Ssc.15224.2.S1 | FOSL2 | 3 | 86 | 6.2 | -DL, pH24ld, pH24sm |
| Ssc.15224.1.S1 | FOSL2 | 3 | 86 | 6.3 | -DL, -SF, -TL, pH24ld, pH24sm |
| Ssc.24035.1.S1 | EIF2C2 | 4 | 45 | 7.4 | -DL, -TL |
| Ssc.5112.2.S1 | LMNA | 4 | 67 | 9.54 | pH24ld, pH24sm |
| Ssc.3935.1.S1 | SLC48A1 | 5 | 89 | 5.2 | LF24sm |
| Ssc.26780.1.S1 | BCL2L12 | 6 | 0 | 6.9 | DL, -pH24ld, -pH24sm, LF24sm, |
| Ssc.1545.1.A1 | TCF25 | 6 | 0 | 5.7 | LF24sm |
| Ssc.20488.1.A1 | RPS9 | 6 | 65 | 8.4 | DL, TL |
| Ssc.14436.1.S1 | USP14 | 6 | 92 | 5.4 | pH24ld |
| Ssc.10297.3.S1 | CAPZB | 6 | 95 | 6.2 | DL, CL, TL, -pH24ld, LF24ld, LF24sm |
| Ssc.10297.1.S1 | CAPZB | 6 | 99 | 6 | CL, LF24sm |
| Ssc.21139.2.S1 | CLIC5 | 7 | 14 | 6 | pH1ld |
| Ssc.10148.1.S1 | MTHFD1 | 7 | 81 | 16.1 | -OPTO, LF1ld |
| Ssc.22107.1.A1 | LOC100131693 | 8 | 110 | 5.8 | pH24ld, LF24sm |
| Ssc.16983.1.S1 | TRAPPC4 | 9 | 62 | 7.7 | -OPTO |
| Ssc.22376.1.A1 | 12 | 63 | 5.5 | OPTO | |
| Ssc.4217.1.S1 | ITIH4 | 13 | 47 | 7.3 | -DL, pH1ld, -LF24sm |
| Ssc.20319.1.S1 | TMEM115 | 13 | 47 | 5.2 | LF24sm, |
| Ssc.31206.3.S1 | CGGBP1 | 13 | 52 | 5.9 | -pH24ld, |
| Ssc.10429.1.S1 | ANKRD1 | 14 | 0 | 6 | -DL, pH24ld, -LF24sm |
| Ssc.10911.1.A1 | ADRBK2 | 14 | 0 | 6 | -CL, pH24ld, -LF24ld |
| Ssc.18050.1.S1 | ZRANB1 | 14 | 87 | 5.6 | -LF24sm |
| Ssc.11397.1.A1 | MTG1 | 14 | 96 | 6.7 | LF24sm |
| Ssc.6058.1.S1 | LOC100129026 | 16 | 55 | 4.9 | -DL, -SF, -LF24sm |
| Ssc.18640.3.S1 | UBE2D1 | 18 | 34 | 4.9 | DL, LF24sm |
"*"the probe-sets listed here represent a subset of probe-sets correlated with individual meat quality traits as indicated in this column naming the traits with which the corresponding probe-sets show correlated expression in addition to the correlated expression with PC2; "-" indicates negative correlation
Trait abbreviations are given at table 2.
Table 4.
Probe-sets with expression levels that are correlated with principal component 3 and information about their eQTL
| Annotation | eQTL | individual trait correlation* | |||
|---|---|---|---|---|---|
| Prob_set_IDs | Gene Symbols | SSC | cM | F | traits |
| Ssc.16910.1.S1 | ROD1 | 1 | 13 | 5.92 | CL, LF24ld |
| Ssc.26338.1.S1 | PRKAG1 | 1 | 30 | 6.69 | -LF24ld |
| Ssc.29222.1.S1 | UBE2CBP | 1 | 31 | 5.72 | pH1ld, -LF1ld, -LF24ld |
| Ssc.3853.1.S1 | C9orf61 | 1 | 54 | 7.98 | -DL, pH1ld, -LF24sm |
| Ssc.20392.1.S1 | BVES | 1 | 63 | 22.1 | -pH1ld |
| Ssc.7558.1.A1 | 1 | 127 | 7.96 | -SF, pH1ld, -LF24ld | |
| Ssc.25132.3.S1 | RAD23B | 1 | 130 | 5.11 | DL, -pH1ld, LF1ld, LF24sm |
| Ssc.16460.1.S1 | EGR1 | 2 | 52 | 6.24 | -LF24sm |
| Ssc.29893.1.A1 | RHOBTB3 | 2 | 94 | 7.37 | LF24ld |
| Ssc.14340.3.S1 | LITAF | 3 | 37 | 6.69 | -pH1ld |
| Ssc.7947.1.A1 | PREPL | 3 | 61 | 5.13 | -pH1ld |
| Ssc.2441.2.A1 | SLC3A1 | 3 | 64 | 6.08 | -pH1ld, LF24ld |
| Ssc.24213.2.S1 | UXS1 | 3 | 74 | 5.71 | LF24ld, LF24sm |
| Ssc.5415.1.S1 | DDX1 | 3 | 86 | 4.96 | pH1ld, -pH24ld, -LF24ld |
| Ssc.8547.1.A1 | 3 | 86 | 7.11 | pH1ld, LF24sm | |
| Ssc.19258.1.S1 | KCNJ8 | 5 | 0 | 5.98 | -LF1ld |
| Ssc.5769.1.S1 | Rassf3 | 5 | 3 | 7.01 | pH1ld, -LF24ld |
| Ssc.3935.1.S1 | SLC48A1 | 5 | 89 | 5.17 | LF24sm |
| Ssc.10297.3.S1 | CAPZB | 6 | 95 | 6.17 | DL, CL, TL, -pH24ld, LF24ld, LF24sm |
| Ssc.10297.1.S1 | CAPZB | 6 | 99 | 6 | CL, LF24sm |
| Ssc.2152.2.S1 | BSDC1 | 6 | 112 | 4.92 | TL |
| Ssc.25358.1.S1 | GRAMD2 | 7 | 5 | 5.41 | -CL, pH1ld, -LF24ld |
| Ssc.21139.2.S1 | CLIC5 | 7 | 14 | 6 | pH1ld |
| Ssc.17920.2.S1 | C15orf17 | 7 | 44 | 7.21 | LF24sm |
| Ssc.30959.1.A1 | LOC100153449 | 7 | 55 | 6.11 | pH1ld |
| Ssc.17920.1.A1 | C15orf17 | 7 | 91 | 7.56 | -LF24sm |
| Ssc.8611.2.S1 | MLL5 | 9 | 0 | 6.06 | LF24ld, LF24sm |
| Ssc.28945.3.S1 | FAM133B | 9 | 0 | 5.15 | LF24sm |
| Ssc.23527.1.A1 | C11orf57 | 9 | 0 | 7.79 | LF24sm |
| Ssc.8415.1.A1 | BACE1 | 9 | 2 | 5.28 | LF24sm |
| Ssc.16454.1.S1 | DNAJC2 | 9 | 7 | 5.51 | LF24sm |
| Ssc.6155.1.S1 | CTSC | 9 | 13 | 8.95 | pH24ld, pH24sm |
| Ssc.28609.1.S1 | SNHG1 | 9 | 45 | 5.21 | -LF24sm |
| Ssc.25107.1.S1 | BAT2D1 | 9 | 51 | 6.56 | LF24ld, LF24sm |
| Ssc.1537.1.S2 | YKT6 | 9 | 82 | 5.23 | LF24sm |
| Ssc.8563.2.S1 | RBM27 | 11 | 67 | 5.11 | LF24ld |
| Ssc.30812.1.S1 | SPAG7 | 12 | 0 | 5.02 | pH1ld, LF24sm |
| Ssc.6130.1.S1 | FBXW11 | 13 | 46 | 8.08 | pH24sm |
| Ssc.4217.1.S1 | ITIH4 | 13 | 47 | 7.31 | -DL, pH1ld, -LF24sm |
| Ssc.6767.1.S1 | dJ196E23.2 | 13 | 53 | 5.05 | LF24sm |
| Ssc.7384.1.A1 | ZFYVE20 | 13 | 53 | 7.69 | LF24sm |
| Ssc.30107.1.S1 | C10orf59 | 14 | 50 | 6.38 | -LF1ld |
| Ssc.11118.1.S1 | ATP5B | 14 | 86 | 5.44 | -LF24ld |
| Ssc.24012.1.S1 | TSN | 15 | 10 | 5.26 | -LF24ld |
| Ssc.9483.1.A1 | Osbpl6 | 15 | 28 | 7.28 | -LF24sm |
| Ssc.13904.1.S1 | TM2D2 | 15 | 51 | 6.73 | -pH1ld, LF24sm |
| Ssc.13358.1.A1 | Agbl3 | 18 | 4 | 5.67 | DL,LF24sm |
"*"the probe-sets listed here represent a subset of probe-sets correlated with individual meat quality traits as indicated in this column naming the traits with which the corresponding probe-sets show correlated expression in addition to the correlated expression with PC3; "-" indicates negative correlation
Trait abbreviations are given at table 2.
Figure 2.
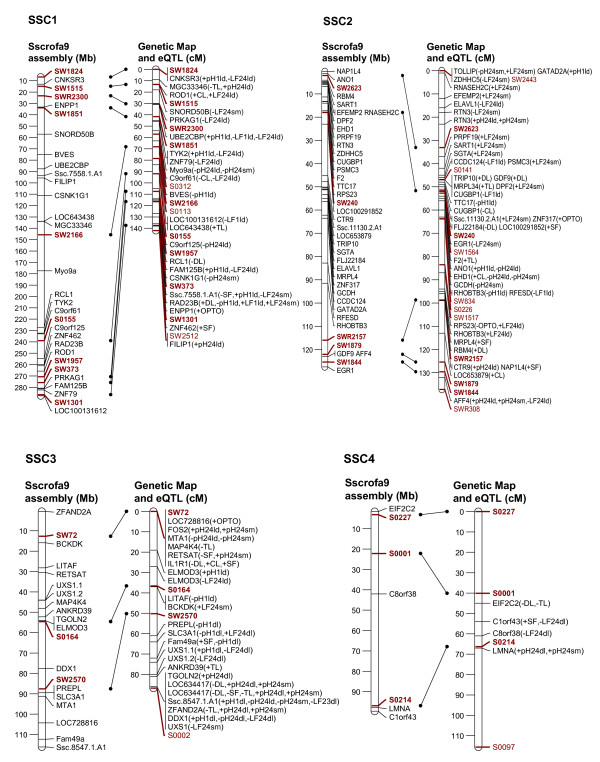
Position of eQTL of probe-sets that show trait-correlated expression and that are located on the same chromosome (SSC1 to 4) as the corresponding genes. Left: physical map with positions of microsatellite markers (red) and genes (black) represented by the probe-sets in Mb according to the Sscrofa9 genome sequence; Right: sex-averaged linkage maps of SSC 1 to 4 with positions of microsatellite markers and eQTL of corresponding probe-sets in cM; Both maps are linked based on the microsatellite marker positions (red and bold). Meat quality traits, with which the expression levels of probe-sets are correlated, are presented in brackets. Trait abbreviations are given at table 2.
Figure 3.
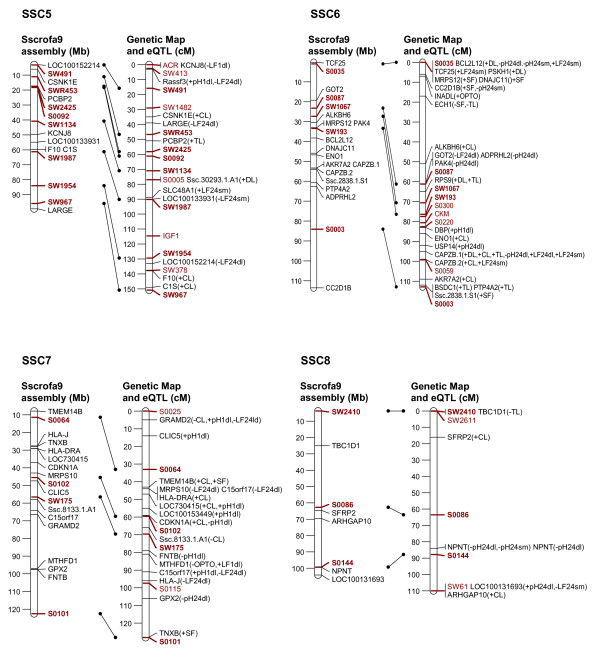
Position of eQTL of probe-sets that show trait-correlated expression and that are located on the same chromosome (SSC5 to 8) as the corresponding genes. Left: physical map with positions of microsatellite markers (red) and genes (black) represented by the probe-sets Mb according to the Sscrofa9 genome sequence; Right: sex-averaged linkage maps of SSC 1 to 4 with positions of microsatellite markers and eQTL of corresponding probe-sets in cM; Both maps are linked based on the microsatellite marker positions (red and bold). Meat quality traits, with which the expression levels of probe-sets are correlated, are presented in brackets. Trait abbreviations are given at table 2.
Figure 4.
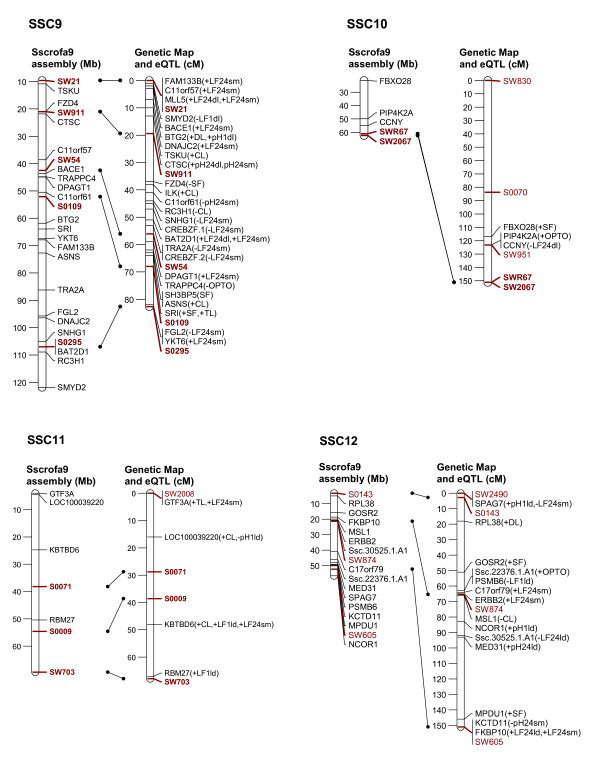
Position of eQTL of probe-sets that show trait-correlated expression and that are located on the same chromosome (SSC9 to 12) as the corresponding genes. Left: physical map with positions of microsatellite markers (red) and genes (black) represented by the probe-sets Mb according to the Sscrofa9 genome sequence; Right: sex-averaged linkage maps of SSC 1 to 4 with positions of microsatellite markers and eQTL of corresponding probe-sets in cM; Both maps are linked based on the microsatellite marker positions (red and bold). Meat quality traits, with which the expression levels of probe-sets are correlated, are presented in brackets. Trait abbreviations are given at table 2.
Figure 5.
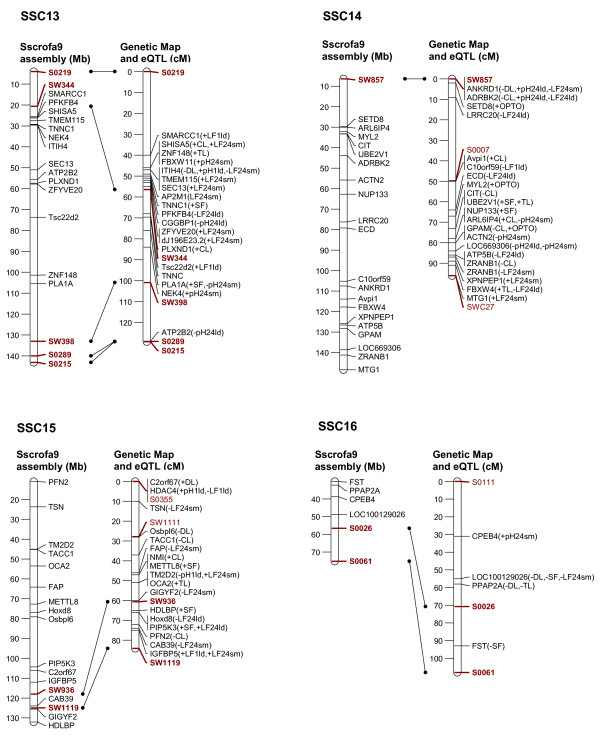
Position of eQTL of probe-sets that show trait-correlated expression and that are located on the same chromosome (SSC12 to 16) as the corresponding genes. Left: physical map with positions of microsatellite markers (red) and genes (black) represented by the probe-sets Mb according to the Sscrofa9 genome sequence; Right: sex-averaged linkage maps of SSC 1 to 4 with positions of microsatellite markers and eQTL of corresponding probe-sets in cM; Both maps are linked based on the microsatellite marker positions (red and bold). Meat quality traits, with which the expression levels of probe-sets are correlated, are presented in brackets. Trait abbreviations are given at table 2.
Figure 6.
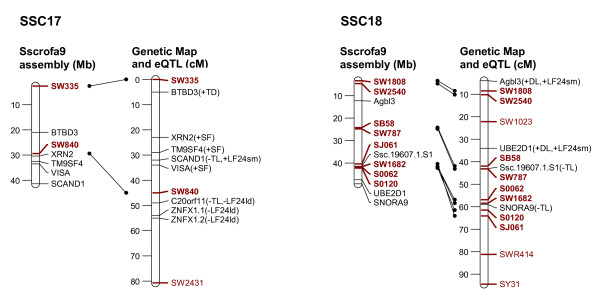
Position of eQTL of probe-sets that show trait-correlated expression and that are located on the same chromosome (SSC17 and 18) as the corresponding genes. Left: physical map with positions of microsatellite markers (red) and genes (black) represented by the probe-sets Mb according to the Sscrofa9 genome sequence; Right: sex-averaged linkage maps of SSC 1 to 4 with positions of microsatellite markers and eQTL of corresponding probe-sets in cM; Both maps are linked based on the microsatellite marker positions (red and bold). Meat quality traits, with which the expression levels of probe-sets are correlated, are presented in brackets. Trait abbreviations are given at table 2.
Most of these meat quality parameters were correlated or dependent on each other. Such correlations were reported by many studies [11-13]. In a previous study we used principle components (PCs) to reduce the multi dimensional data sets into lower dimensions [13]. The principal component (PC2) is a meat quality vector and has high loadings for pH24ld, pH24st and meat color which are inversely correlated with drip loss. The principal component (PC3) is also a meat quality vector with large positive contribution of conductivity (LF1ld, LF24ld, LF24sm) and negative contribution of pH1ld. In order to scale down the list of 262 putative cis-eQTL for meat quality, transcript levels of genes correlated with the two PCs with high loadings of meat quality traits were considered. The level of expression of 38 and 47 probe-sets, respectively, were correlated with the new composite traits that were generated by principal component analysis PC2 and PC3 (Tables 3 and 4). In order to unravel the composite traits, the correlation of these 38 and 47 transcripts with each single meat quality trait contributing to the PC was screened and traits with significant correlations with the respective genes are given in tables 3 and 4. Thus the probe-sets represent a subset of probe-sets correlated with individual meat quality traits. As previously shown, analyses of enrichment of functional annotation groups as defined in the Gene Ontology (GO), the Kyoto Encyclopedia of Genes and Genomes (KEGG) databases, or in the Ingenuity Pathways Analysis library, highlight ubiquitination, phosphorylation, mitochondrion dysfunction, actin-, integrin-, PDGF-, EGF-, VEGF-, and Ca-signalling pathways [13]. Most of the eQTL related to PC2 were found on chromosomes 2, 3, and 6; for PC3 on chromosomes 1, 3, 7, and 9.
Discussion
Complex traits are genetically controlled by many loci. The identification of candidate genes for these traits is widely based on the principle of 'collecting evidences' in order to prioritize genes for further analysis from the huge lists of functional and positional candidate genes. The detection of eQTL provides information about the positional aspects of gene expression regulation. Together with the growing knowledge of genome sequences and gene annotation, this gives insight into the architecture of regulatory networks. In order to relate eQTL analysis to the genetic background of any complex trait, either a positional link to QTL for the complex trait of interest (pQTL) and/or a functional link to the trait expression is required. Here we report on the detection of eQTL for some 11,000 transcripts found in porcine skeletal muscle by microarray analysis. We demonstrate that the global microarray eQTL analysis can serve for narrowing down the candidate genes for quantitative traits related to meat quality when it is integrated with the analysis of the correlation of expression levels with the traits of interest.
The genetic architecture of transcript level variation in the porcine F2 resource population was highly variable and complex. eQTL were detected for 53% of the 11,457 probe-sets, with many of them exhibiting more than one eQTL summing up to more than 9000 eQTL in total (Table 1). The expression levels of the majority of the transcripts showed quantitative variation. So the transcript levels are probably quantitatively controlled. The link between the linkage maps and porcine assembly sequence enable discriminating cis or trans-eQTL. All 23,934 probe-sets sequence represented on the porcine microarray and the microsatellites sequences were BLASTed against the Ensembl porcine BAC sequence (Sscrofa9, April 2009). Our microsatellite order showed high accuracy compared to the Ensembl porcine BAC sequence.
Correlations between transcript abundance and meat quality, combined with genetic positional information of eQTL allowed us to prioritize a small number of candidate genes. We considered transcripts, which exhibited expression levels correlated with meat quality traits, and which had eQTL on the same chromosome as the transcripts itself.
Our previous studies showed that cis regulation is a stable characteristic of individual transcripts. Consequently, a global microarray eQTL analysis of a limited number of samples can be used for exploring functional and regulatory gene networks and scanning for cis-eQTL. In particular, the assignment of eQTL to chromosomes is reliable; though some cis-eQTL change their position, they were consistently assigned to the same chromosome when comparing analyses based on 74 microarrays or 276 real time RT-PCRs [7]. Based on this observation, we decided to highlight only eQTL which were located on the gene itself or the same chromosome. Moreover, this relaxed measure was chosen in order to avoid the exclusion of any true cis-acting eQTL that are not precisely mapped, because of the limited resolution of the genetic map. The observation that cis-eQTL were more consistently detected than trans-eQTL was also made by comparing the results from different studies of eQTL with different numbers of animals and different tissue types [14-17].
Expression QTL mapping, with its potential to categorize cis and trans-effects, provides the mean to discriminate between "effect" and "cause" with respect to trait-associated differential expression. Though a relaxed window of cis-eQTLs was used, a low proportion of 10% of putative cis-eQTL was found in this study, compared to other previous studies [18,19]. Cis-regulated genes are of interest, because the underlying genes are expected to harbour genetic variants that influence their own expression level, which may also influence the physiological traits of interest, if transcript abundance is correlated with the target phenotype [17]. This provides a function-related evidence of the candidacy of a particular gene.
The usefulness of eQTL for identification of quantitative trait genes was demonstrated [3,5,20,21]. The causal link between sequence variation, gene expression, and phenotype arises because the polymorphism might be responsible for both a cis-eQTL and the QTL contributing to a quantitative phenotype, so called pQTL. QTL for traits related to meat quality were previously mapped in the population used here [12,22]. The matching of pQTL, eQTL and the localization of the corresponding genes with trait-correlated expression provides positional and functional evidence for the potential role of the respective genes and strongly promote them as candidate genes. Mapping of eQTL enables displaying regulatory networks and localizing genomic variation affecting the mRNA expression of a gene either within the genes itself (cis) or distant from the gene (trans). The key advantage of eQTL mapping is that it connects variation at the level of RNA expression to variation at the level of DNA. Only the latter provides versatile tools for breeding whereas the first reveals information on the biology of a trait and directs to new candidate genes. Moreover, integration of (1) information on QTL for a trait of interest in breeding (pheneQTL = pQTL) with analysis of (2) trait correlated expression and with (3) mapping of expression QTL (eQTL) for the corresponding trait-dependent-regulated genes facilitates the identification of genes and pathways with cumulative evidence of their involvement in the biology of the traits of interest and enable to built priority lists of candidate genes [23]. However, there are also some issues that limit of the use of the genetical genomics approach, in particular the resolution of the genetic maps that is depending on the number of markers and animals used and the structure of the population used and artifacts caused by the limited sensitivity and specificity of microarray experiments [24].
In a previous study we used the principle components with high loadings of meat quality traits to identify functional networks of genes and to gain knowledge of biological and physiological processes taking place during the conversion of muscle to meat [13]. Here in this study, we combined principle component analysis of traits related to meat quality with eQTL to scale down the list of candidate genes for the traits.
Conclusion
Holistic expression profiling was integrated with QTL analyses for meat quality traits. This is, to our knowledge, the first report of a comprehensive scan for cis-eQTL associated with meat quality traits in the pig. Correlations between transcript abundance and meat quality traits, combined with genetic positional information of eQTL allowed us to prioritise candidate genes for further study. Accordingly, a list of candidate genes for meat quality was set up. The further identification of the causative polymorphisms and the determination of their functional role are even more challenging, since there are several different molecular mechanisms through which mRNA levels in cells can be regulated.
Methods
Animals and tissue collection
This study was based on trait measurements, genotyping procedures, expression profiling, and linkage analysis performed in the three-generation resource family (DuPi) founded by crossbreeding Duroc and Pietrain and described in detail by Liu et al. [12,22]. All animals were free of the mutation at the ryanodine receptor locus, RYR1, which is responsible for malignant hyperthermia syndrome. A total of 572 F2 animals comprising 31 full-sib families were used for recording of meat performance traits and construction of a linkage map comprising genotypes of 115 microsatellite markers. Expression profiling and eQTL analysis were done with 74 F2 animals of this resource population that represented a subset of the population covering 25 full-sib families derived from all five F1 boars of the population and 18 out of 27 F1 sows. The experimental research on animals was done according to the German Animal Welfare Act and approved by the animal welfare board of the Leibniz Institute of Farm Animal Biology, FBN Dummerstorf.
Traits and phenotypes
Phenotypic data of F2 animals were collected following the guidelines of the German performance test (ZDS 2003) [25]. After slaughter technological parameters of meat quality, i.e. pH-value, conductivity and colour, were measured by using Star-series equipment (Rudolf Matthaeus Company, Germany). Measures of pH and conductivity were at 45 min post mortem (pH1) and 24 h post mortem (pH24), respectively; both in M. longissimus dorsi between 13th/14th rib (pH1ld, pH24ld, LF1ld, LF24ld) and in the ham (M. semimembranosus) (pH24sm, LF24sm), respectively. Muscle colour was measured at 24 h post mortem by Opto-Star. Drip loss was scored based on a bag-method with a size-standardized sample from the M. longissimus dorsi collected at 24 hours post mortem that was weighted, suspended in a plastic bag, held at 4°C for 48 h, and thereafter re-weighed [26,27]. To obtain cooking loss, a loin cube was taken from the M. longissimus dorsi, weighed, placed in a polyethylene bag and incubated in water at 75°C for 50 minutes. The bag was then immersed in flowing water at room temperature for 30 minutes and the solid portion was re-weighed. Thawing loss was determined similarly after at least 24 h of freezing at -20°C. Drip loss, cooking loss, and thawing loss were calculated as a percentage of weight loss based on the start weight of a sample. Shear force was measured by the Instron-4310 equipment and the average values of four replicates were used for analyses. The procedure 'Factor' of the SAS software package (SAS version 9.1 SAS Institute, Cary, NC) was used to derive four principle components (PCs) based on 19 traits related to muscle and carcass properties of 572 DUPI animals; i.e. the dimensionality of the data was reduced from the original 19 traits to 4 principal components (PCs), where each PC is a linear combination of all traits without a significant loss of information. PC1 and PC4 represent variation in carcass traits, whereas PC2 and PC3 represent aspects of meat quality, consequently PC2 and PC3 were used to correlate with expression profiles. The principal component analysis of meat quality traits is discussed in Ponsuksili et al., 2009 [13].
Whole genome expression profiling
Immediately post mortem tissue samples were collected and snap frozen that were taken between the 13th and 14th rib from the center of M. longissimus dorsi of 74 animals. Total RNA was isolated using TRI Reagent (Sigma, Taufkirchen, Germany) according to the manufacturer's protocol. After DNaseI treatment the RNA was cleaned up using the RNeasy Kit (Qiagen, Hilden, Germany). The quantity of RNA was established using the NanoDrop ND-1000 spectrophotometer (Peqlab, Erlangen, Germany) and the integrity was checked by running 1 μg of RNA on 1% agarose gel. In addition absence of DNA contamination was checked using the RNA as a template in standard PCR amplifying fragments of RPL32 and HPRT1. Muscle expression pattern were assessed using 74 Porcine Genome Array which contains 24,123 probe-sets that interrogate 20,689 transcripts that were assigned to known genes [6]. Preparation of target products, hybridization and scanning using the GeneChip scanner 3000 were performed according to Affymetrix protocols using 5 μg of total RNA to prepare antisense biotinylated RNA. The quality of hybridization was assessed in all samples following the manufacturer's recommendations. Data were analysed with Affymetrix GCOS 1.1.1 software using global scaling to a target signal of 500. Data were then imported into Arrays Assist software (Stratagene Europe, Amsterdam, The Netherlands) for subsequent analysis. The data were processed with MAS5.0 to generate cell intensity files (using default settings with detection P-values of < 0.04 for 'present', ≥0.04 and ≤ 0.06 for 'marginal', < 0.06 for 'absent'; only 'present' calls were used). Quantitative expression levels of the present transcripts were estimated using PLIER (Probe Logarithmic Intensity Error) for normalization. The microarray data related to all samples have been deposited in the Gene Expression Omnibus (GEO, [28]) public repository (GEO accession number: GSE10204).
Correlation between phenotype and gene expression
Phenotypic data, i.e. expression levels and meat quality data as well as PC traits, were adjusted for systematic effects by analysis of variance performed with the procedure 'Mixed' of the SAS software package (SAS System for Windows, Release 9.02) before analysing their correlation and performing eQTL analysis. Full-sib family and sex were used as fixed effects, carcass weight as a covariate and slaughter date as random effect. Based on observations from 74 animals Pearson correlation coefficients were calculated between the residuals of the log2 transformed expression intensities of all 11,453 probes and each individual meat quality trait as well as between the expression levels and both composite traits, PC2 and PC3. For each pair of transcript level and phenotype, Pearson correlation together with the P-value was computed (significance threshold p ≤ 0.05). The correlation coefficients at p ≤ 0.05 ranged between |0.24-0.50|. The observed false discovery rates were 0.08-0.34 which is reasonable for a microarray study, in particular, considering the relatively relaxed p-value (p ≤ 0.05).
eQTL analysis
In order to map eQTL adjusted expression values of 11,457 probe-sets were regressed onto the additive and dominance coefficients in intervals of 1 cM using the F2 option of QTL express [29]. Chromosome-wide significance levels were estimated by permutation tests using 5000 permutations [30]. The 5% chromosome-wide threshold corresponds approximately to the suggestive linkage threshold proposed by Lander & Kruglyak (1995) [10]. Average significance thresholds were 4.92 corresponding to LOD = 2.0 with nominal p ≤ 10-3. Annotations and localization of probe-sets was based on assembly Sscrofa9 (April 2009) [6]. In order to link the genetic map to Ensembl Sscrofa9, all 115 microsatellites sequences, which were used in the linkage map, were BLASTed against the Ensembl porcine BAC sequence (Sscrofa9, April 2009). Only 73 microsatellites sequences could be localized on Sscrofa 9 as shown on Figure 2, 3, 4, 5, 6.
Authors' contributions
SP analyzed the microarray data and drafted the manuscript; EM, MS, and KS aided in data analysis and helped in drafting the manuscript; KW conceived and designed the study, contributed to data interpretation and helped in drafting the manuscript. All authors read and approved the final manuscript.
Supplementary Material
eQTL detected for probe-sets representing transcripts that are expressed in porcine M. longissimus dorsi. All 9,180 eQTL (at LOD score > 2) estimated for probe-sets of the Affymetrix Porcine Genome Array, their position in cM on linkage groups, corresponding gene names and chromosomal positions according to the current porcine genome sequence assembly (Sscrofa9 assembly, April 2009) [6]
Contributor Information
Siriluck Ponsuksili, Email: s.wimmers@fbn-dummerstorf.de.
Eduard Murani, Email: murani@fbn-dummerstorf.de.
Manfred Schwerin, Email: schwerin@fbn-dummerstorf.de.
Karl Schellander, Email: ksch@itz.uni-bonn.de.
Klaus Wimmers, Email: wimmers@fbn-dummerstorf.de.
Acknowledgements
The authors thank Annette Jugert and Joana Bittner for excellent technical help. This research was supported by German Research Foundation (Deutsche Forschungs-gemeinschaft, DFG; Forschergruppe 'DRIP', FOR 753)
References
- Jansen RC, Nap JP. Genetical genomics: the added value from segregation. Trends Genet. 2001;17:388–391. doi: 10.1016/S0168-9525(01)02310-1. [DOI] [PubMed] [Google Scholar]
- Jansen RC. Studying complex biological systems using multifactorial perturbation. Nat Rev Genet. 2003;4:145–151. doi: 10.1038/nrg996. [DOI] [PubMed] [Google Scholar]
- Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, Guhathakurta D, Sieberts SK, Monks S, Reitman M, Zhang C, Lum PY, Leonardson A, Thieringer R, Metzger JM, Yang L, Castle J, Zhu H, Kash SF, Drake TA, Sachs A, Lusis AJ. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doss S, Schadt EE, Drake TA, Lusis AJ. Cis-acting expression quantitative trait loci in mice. Genome Res. 2005;15:681–691. doi: 10.1101/gr.3216905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponsuksili S, Jonas E, Murani E, Phatsara C, Srikanchai T, Walz C, Schwerin M, Schellander K, Wimmers K. Trait correlated expression combined with expression QTL analysis reveals biological pathways and candidate genes affecting water holding capacity of muscle. BMC Genomics. 2008;9:367. doi: 10.1186/1471-2164-9-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naraballobh W, Chomdej S, Murani E, Wimmers K, Ponsuksili S. Annotation and in silico localization of the Affymetrix GeneChip Porcine Genome Array. Arch Tierz. 2010;53:230–238. [Google Scholar]
- Ponsuksili S, Murani E, Phatsara C, Schwerin M, Schellander K, Wimmers K. Expression quantitative trait loci analysis of genes in porcine muscle by quantitative real time RT-PCR compared to microarray data. Heredity. 2010. in press . [DOI] [PubMed]
- Affymetrix Technical Manual. Affymetrix GeneChip Expression Analysis. Affymetrix, Santa Clara, CA; 2001. [Google Scholar]
- Affymetrix Technical Note. Guide to Probe Logarithmic Intensity Error (PLIER) Estimation. Affymetrix, Santa Clara, CA; 2005. [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Hodgson RR, Belk KE, Savell JW, Cross HR, Williams FL. Development of a quantitative quality grading system for mature cow carcasses. J Anim Sci. 1992;70:1840–1847. doi: 10.2527/1992.7061840x. [DOI] [PubMed] [Google Scholar]
- Liu G, Jennen DG, Tholen E, Juengst H, Kleinwächter T, Hölker M. et al. A genome scan reveals QTL for growth, fatness, leanness and meat quality in a Duroc-Pietrain resource population. Anim Genet. 2007;38:241–252. doi: 10.1111/j.1365-2052.2007.01592.x. [DOI] [PubMed] [Google Scholar]
- Ponsuksili S, Murani E, Phatsara C, Schwerin M, Schellander K, Wimmers K. Porcine muscle sensory attributes associate with major changes in gene networks involving CAPZB, ANKRD1, and CTBP2. Funct Integr Genomics. 2009;9:455–471. doi: 10.1007/s10142-009-0131-1. [DOI] [PubMed] [Google Scholar]
- Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- Yvert G, Brem RB, Whittle J, Akey JM, Foss E, Smith EN. et al. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat Genet. 2003;35:57–64. doi: 10.1038/ng1222. [DOI] [PubMed] [Google Scholar]
- Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS. et al. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göring HH, Curran JE, Johnson MP, Dyer TD, Charlesworth J, Cole SA. et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, Ruff TG, Milligan SB, Lamb JR, Cavet G, Linsley PS, Mao M, Stoughton RB, Friend SH. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–301. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- Hubner N, Wallace CA, Zimdahl H, Petretto E, Schulz H, Maciver F, Mueller M, Hummel O, Monti J, Zidek V, Musilova A, Kren V, Causton H, Game L, Born G, Schmidt S, Müller A, Cook SA, Kurtz TW, Whittaker J, Pravenec M, Aitman TJ. Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nat Genet. 2005;37:243–253. doi: 10.1038/ng1522. [DOI] [PubMed] [Google Scholar]
- Bystrykh L, Weersing E, Dontje B, Sutton S, Pletcher MT, Wiltshire T. et al. Uncovering regulatory pathways that affect hematopoietic stem cell function using 'genetical genomics'. Nat Genet. 2005;37:225–232. doi: 10.1038/ng1497. [DOI] [PubMed] [Google Scholar]
- Mehrabian M, Allayee H, Stockton J, Lum PY, Drake TA, Castellani LW, Suh M, Armour C, Edwards S, Lamb J, Lusis AJ, Schadt EE. Integrating genotypic and expression data in a segregating mouse population to identify 5-lipoxygenase as a susceptibility gene for obesity and bone traits. Nat Genet. 2005;37:1224–1233. doi: 10.1038/ng1619. [DOI] [PubMed] [Google Scholar]
- Liu G, Kim JJ, Jonas E, Wimmers K, Ponsuksili S, Murani E, Phatsara C, Tholen E, Jüngst H, Tesfaye D, Chen JL, Schellander K. Combined line-cross and half-sib QTL analysis in Duroc-Pietrain population. Mamm Genome. 2008;6:429–438. doi: 10.1007/s00335-008-9132-y. [DOI] [PubMed] [Google Scholar]
- Wimmers K, Murani E, Ponsuksili S. Functional genomics and genetical genomics approaches towards elucidating networks of genes affecting meat performance in pigs. Brief Funct Genomics. 2010;9:251–258. doi: 10.1093/bfgp/elq003. [DOI] [PubMed] [Google Scholar]
- Verdugo A, Farber CR, Warden CH, Medrano JF. Serious limitations of the QTL/microarray approach for QTL gene discovery. BMC Biology. 2010;8:96. doi: 10.1186/1741-7007-8-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZDS, Zentral Verband der Deutschen Schweineproduktion e. V. Richtlinie fuer die Stationspruefung auf Mastleistung, Schlachtkoerperwert und Fleischbeschaffenheit beim Schwein. Bonn, Germany; 2003. [Google Scholar]
- Honikel KO. Wasserbindungsvermögen von Fleisch. Mitteilungsblatt der BAFF. 1986;6:7150–7154. [Google Scholar]
- Kauffman RG, Eikelenboom G, Wal van der PG, Merkus G, Zaar M. The use of filter paper to estimate drip loss of porcine musculature. Meat Sci. 1986;18:191–200. doi: 10.1016/0309-1740(86)90033-1. [DOI] [PubMed] [Google Scholar]
- Gene Expression Omnibus. http://www.ncbi.nlm.nih.gov/geo/
- Seaton G, Haley CS, Knott SA, Kearsey M, Visscher PM. QTL Express: mapping quantitative trait loci in simple and complex pedigrees. Bioinformatics. 2002;18:339–340. doi: 10.1093/bioinformatics/18.2.339. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eQTL detected for probe-sets representing transcripts that are expressed in porcine M. longissimus dorsi. All 9,180 eQTL (at LOD score > 2) estimated for probe-sets of the Affymetrix Porcine Genome Array, their position in cM on linkage groups, corresponding gene names and chromosomal positions according to the current porcine genome sequence assembly (Sscrofa9 assembly, April 2009) [6]