Abstract
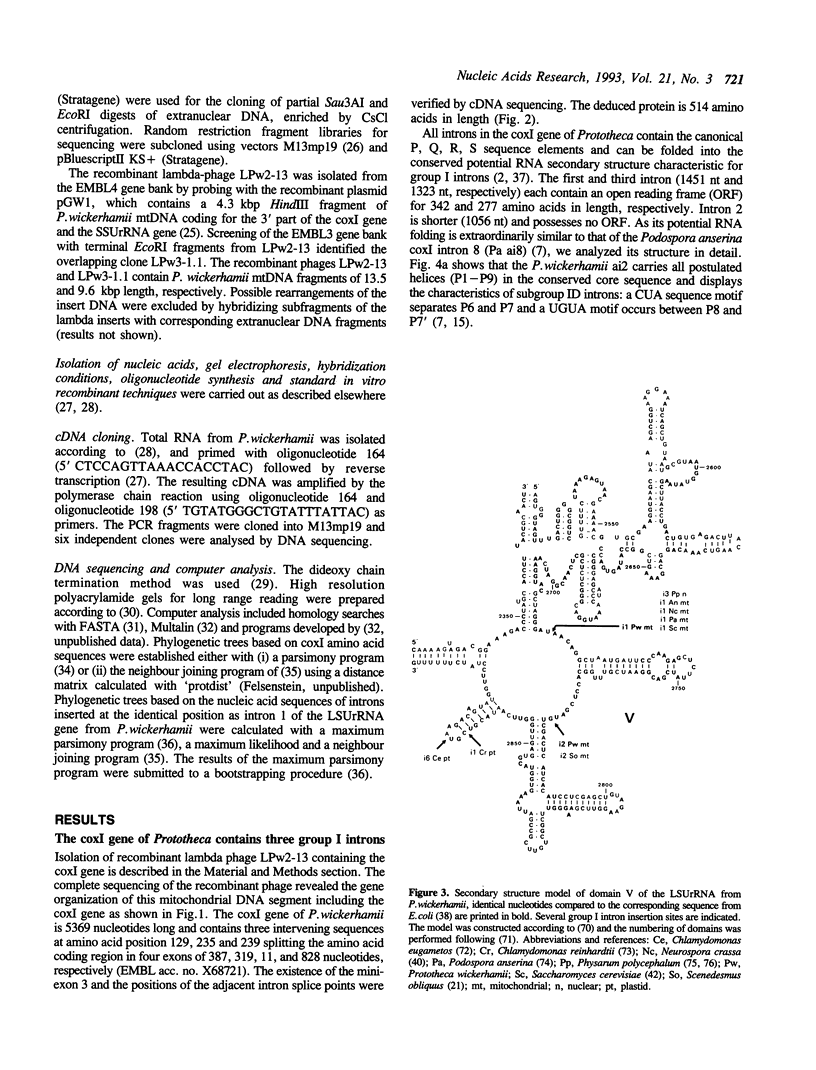
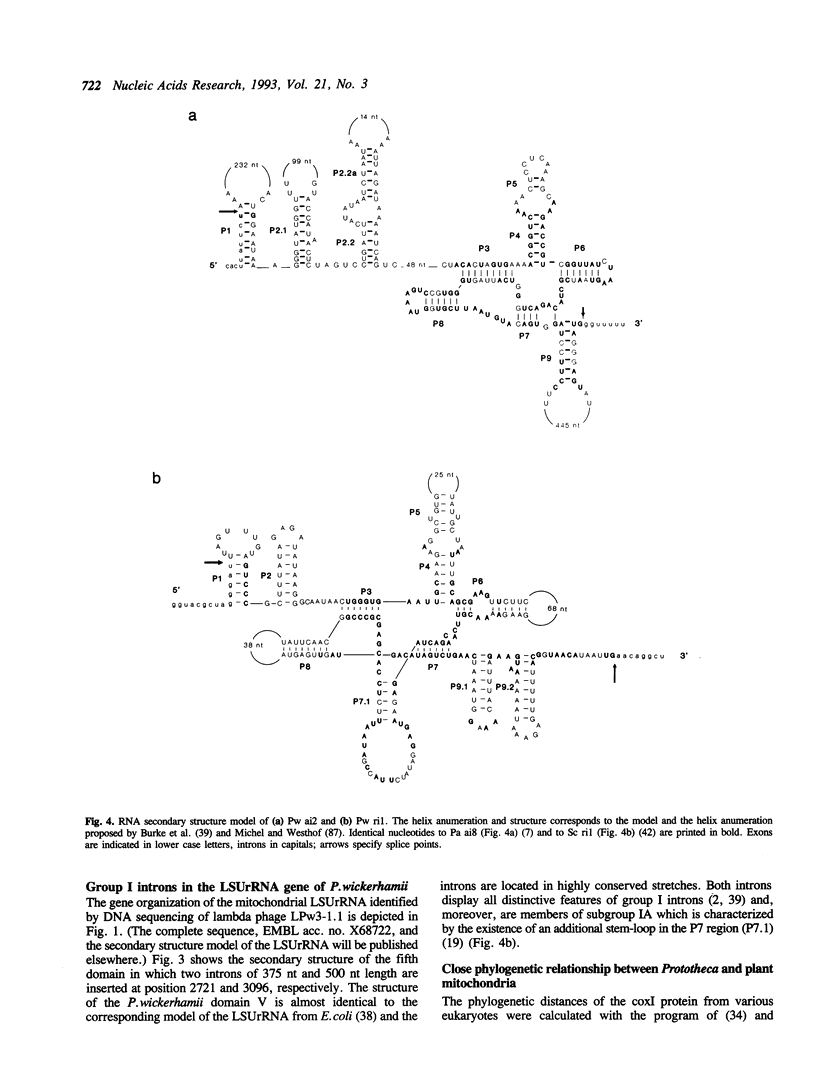
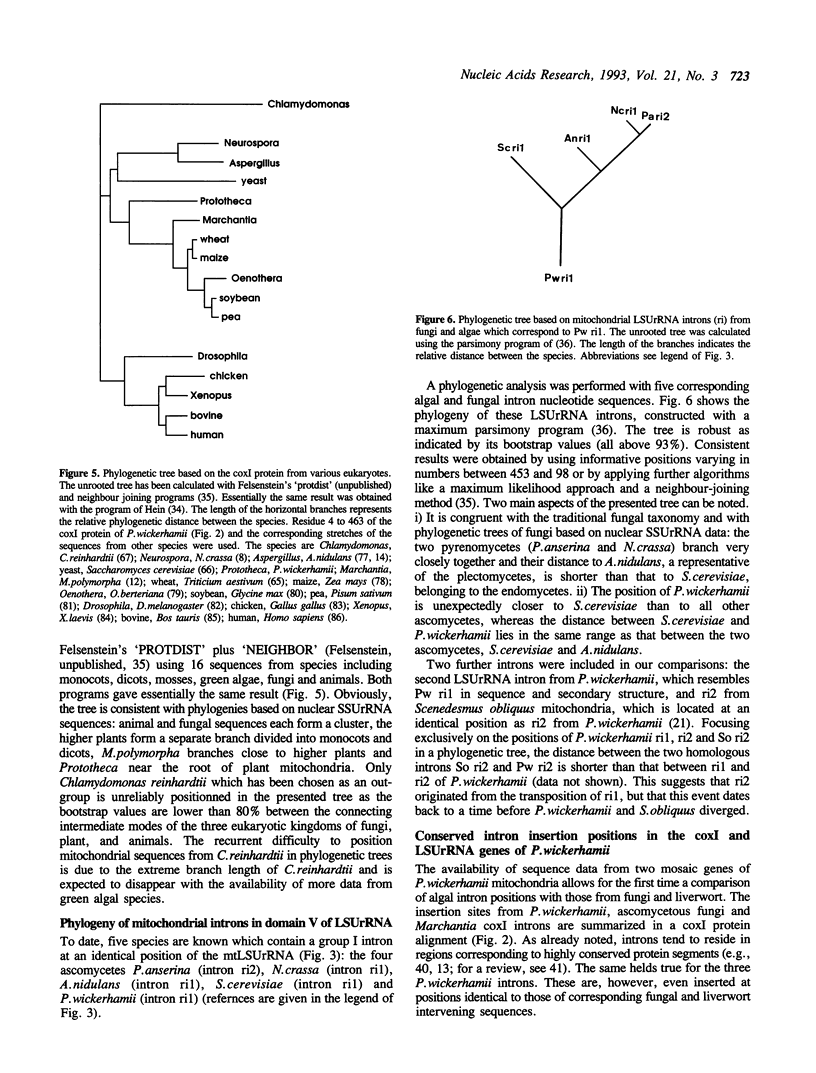
The mitochondrial DNA from the colourless alga Prototheca wickerhamii contains two mosaic genes as was revealed from complete sequencing of the circular extranuclear genome. The genes for the large subunit of the ribosomal RNA (LSUrRNA) as well as for subunit I of the cytochrome oxidase (coxI) carry two and three intronic sequences respectively. On the basis of their canonical nucleotide sequences they can be classified as group I introns. Phylogenetic comparisons of the coxI protein sequences allow us to conclude that the P.wickerhamii mtDNA is much closer related to higher plant mtDNAs than to those of the chlorophyte alga C.reinhardtii. The comparison of the intron sequences revealed several unusual features: (1) The P.wickerhamii introns are structurally related to mitochondrial introns from various ascomycetous fungi. (2) Phylogenetic analyses indicate a close relationship between fungal and algal intronic sequences. (3) The P. wickerhamii introns are located at positions within the structural genes which can be considered as preferred intron insertion sites in homologous mitochondrial genes from fungi or liverwort. In all cases, the sequences adjacent to the insertion sites are very well conserved over large evolutionary distances. Our finding of highly similar introns in fungi and algae is consistent with the idea that introns have already been present in the bacterial ancestors of present day mitochondria and evolved concomitantly with the organelles.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allet B., Rochaix J. D. Structure analysis at the ends of the intervening DNA sequences in the chloroplast 23S ribosomal genes of C. reinhardii. Cell. 1979 Sep;18(1):55–60. doi: 10.1016/0092-8674(79)90353-2. [DOI] [PubMed] [Google Scholar]
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Anderson S., de Bruijn M. H., Coulson A. R., Eperon I. C., Sanger F., Young I. G. Complete sequence of bovine mitochondrial DNA. Conserved features of the mammalian mitochondrial genome. J Mol Biol. 1982 Apr 25;156(4):683–717. doi: 10.1016/0022-2836(82)90137-1. [DOI] [PubMed] [Google Scholar]
- Belfort M. Self-splicing introns in prokaryotes: migrant fossils? Cell. 1991 Jan 11;64(1):9–11. doi: 10.1016/0092-8674(91)90201-9. [DOI] [PubMed] [Google Scholar]
- Binder S., Marchfelder A., Brennicke A., Wissinger B. RNA editing in trans-splicing intron sequences of nad2 mRNAs in Oenothera mitochondria. J Biol Chem. 1992 Apr 15;267(11):7615–7623. [PubMed] [Google Scholar]
- Bonen L., Boer P. H., McIntosh J. E., Gray M. W. Nucleotide sequence of the wheat mitochondrial gene for subunit I of cytochrome oxidase. Nucleic Acids Res. 1987 Aug 25;15(16):6734–6734. doi: 10.1093/nar/15.16.6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonitz S. G., Coruzzi G., Thalenfeld B. E., Tzagoloff A., Macino G. Assembly of the mitochondrial membrane system. Structure and nucleotide sequence of the gene coding for subunit 1 of yeast cytochrme oxidase. J Biol Chem. 1980 Dec 25;255(24):11927–11941. [PubMed] [Google Scholar]
- Burger G., Scriven C., Machleidt W., Werner S. Subunit 1 of cytochrome oxidase from Neurospora crassa: nucleotide sequence of the coding gene and partial amino acid sequence of the protein. EMBO J. 1982;1(11):1385–1391. doi: 10.1002/j.1460-2075.1982.tb01327.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke J. M., Belfort M., Cech T. R., Davies R. W., Schweyen R. J., Shub D. A., Szostak J. W., Tabak H. F. Structural conventions for group I introns. Nucleic Acids Res. 1987 Sep 25;15(18):7217–7221. doi: 10.1093/nar/15.18.7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke J. M., RajBhandary U. L. Intron within the large rRNA gene of N. crassa mitochondria: a long open reading frame and a consensus sequence possibly important in splicing. Cell. 1982 Dec;31(3 Pt 2):509–520. doi: 10.1016/0092-8674(82)90307-5. [DOI] [PubMed] [Google Scholar]
- Cedergren R., Gray M. W., Abel Y., Sankoff D. The evolutionary relationships among known life forms. J Mol Evol. 1988 Dec;28(1-2):98–112. doi: 10.1007/BF02143501. [DOI] [PubMed] [Google Scholar]
- Chapdelaine Y., Bonen L. The wheat mitochondrial gene for subunit I of the NADH dehydrogenase complex: a trans-splicing model for this gene-in-pieces. Cell. 1991 May 3;65(3):465–472. doi: 10.1016/0092-8674(91)90464-a. [DOI] [PubMed] [Google Scholar]
- Chu F. K., Maley G. F., Maley F., Belfort M. Intervening sequence in the thymidylate synthase gene of bacteriophage T4. Proc Natl Acad Sci U S A. 1984 May;81(10):3049–3053. doi: 10.1073/pnas.81.10.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu F. K., Maley G. F., West D. K., Belfort M., Maley F. Characterization of the intron in the phage T4 thymidylate synthase gene and evidence for its self-excision from the primary transcript. Cell. 1986 Apr 25;45(2):157–166. doi: 10.1016/0092-8674(86)90379-x. [DOI] [PubMed] [Google Scholar]
- Colleaux L., Michel-Wolwertz M. R., Matagne R. F., Dujon B. The apocytochrome b gene of Chlamydomonas smithii contains a mobile intron related to both Saccharomyces and Neurospora introns. Mol Gen Genet. 1990 Sep;223(2):288–296. doi: 10.1007/BF00265065. [DOI] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988 Nov 25;16(22):10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings D. J., Domenico J. M., Nelson J. DNA sequence and secondary structures of the large subunit rRNA coding regions and its two class I introns of mitochondrial DNA from Podospora anserina. J Mol Evol. 1989 Mar;28(3):242–255. doi: 10.1007/BF02102482. [DOI] [PubMed] [Google Scholar]
- Cummings D. J., Domenico J. M. Sequence analysis of mitochondrial DNA from Podospora anserina. Pervasiveness of a class I intron in three separate genes. J Mol Biol. 1988 Dec 20;204(4):815–839. doi: 10.1016/0022-2836(88)90044-7. [DOI] [PubMed] [Google Scholar]
- Cummings D. J., Michel F., McNally K. L. DNA sequence analysis of the 24.5 kilobase pair cytochrome oxidase subunit I mitochondrial gene from Podospora anserina: a gene with sixteen introns. Curr Genet. 1989 Dec;16(5-6):381–406. doi: 10.1007/BF00340719. [DOI] [PubMed] [Google Scholar]
- Cummings D. J., Michel F., McNally K. L. DNA sequence analysis of the apocytochrome b gene of Podospora anserina: a new family of intronic open reading frame. Curr Genet. 1989 Dec;16(5-6):407–418. doi: 10.1007/BF00340720. [DOI] [PubMed] [Google Scholar]
- Desjardins P., Morais R. Sequence and gene organization of the chicken mitochondrial genome. A novel gene order in higher vertebrates. J Mol Biol. 1990 Apr 20;212(4):599–634. doi: 10.1016/0022-2836(90)90225-B. [DOI] [PubMed] [Google Scholar]
- Dujon B. Group I introns as mobile genetic elements: facts and mechanistic speculations--a review. Gene. 1989 Oct 15;82(1):91–114. doi: 10.1016/0378-1119(89)90034-6. [DOI] [PubMed] [Google Scholar]
- Dujon B. Sequence of the intron and flanking exons of the mitochondrial 21S rRNA gene of yeast strains having different alleles at the omega and rib-1 loci. Cell. 1980 May;20(1):185–197. doi: 10.1016/0092-8674(80)90246-9. [DOI] [PubMed] [Google Scholar]
- Erickson J. M., Rahire M., Rochaix J. D. Chlamydomonas reinhardii gene for the 32 000 mol. wt. protein of photosystem II contains four large introns and is located entirely within the chloroplast inverted repeat. EMBO J. 1984 Dec 1;3(12):2753–2762. doi: 10.1002/j.1460-2075.1984.tb02206.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gass D. A., Makaroff C. A., Palmer J. D. Variable intron content of the NADH dehydrogenase subunit 4 gene of plant mitochondria. Curr Genet. 1992 Apr;21(4-5):423–430. doi: 10.1007/BF00351705. [DOI] [PubMed] [Google Scholar]
- Goodrich-Blair H., Scarlato V., Gott J. M., Xu M. Q., Shub D. A. A self-splicing group I intron in the DNA polymerase gene of Bacillus subtilis bacteriophage SPO1. Cell. 1990 Oct 19;63(2):417–424. doi: 10.1016/0092-8674(90)90174-d. [DOI] [PubMed] [Google Scholar]
- Gray M. W., Cedergren R., Abel Y., Sankoff D. On the evolutionary origin of the plant mitochondrion and its genome. Proc Natl Acad Sci U S A. 1989 Apr;86(7):2267–2271. doi: 10.1073/pnas.86.7.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M. W. Organelle origins and ribosomal RNA. Biochem Cell Biol. 1988 May;66(5):325–348. doi: 10.1139/o88-042. [DOI] [PubMed] [Google Scholar]
- Hein J. Unified approach to alignment and phylogenies. Methods Enzymol. 1990;183:626–645. doi: 10.1016/0076-6879(90)83041-7. [DOI] [PubMed] [Google Scholar]
- Hiesel R., Schobel W., Schuster W., Brennicke A. The cytochrome oxidase subunit I and subunit III genes in Oenothera mitochondria are transcribed from identical promoter sequences. EMBO J. 1987 Jan;6(1):29–34. doi: 10.1002/j.1460-2075.1987.tb04714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac P. G., Jones V. P., Leaver C. J. The maize cytochrome c oxidase subunit I gene: sequence, expression and rearrangement in cytoplasmic male sterile plants. EMBO J. 1985 Jul;4(7):1617–1623. doi: 10.1002/j.1460-2075.1985.tb03828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier A., Dujon B. The intron of the mitochondrial 21S rRNA gene: distribution in different yeast species and sequence comparison between Kluyveromyces thermotolerans and Saccharomyces cerevisiae. Mol Gen Genet. 1983;192(3):487–499. doi: 10.1007/BF00392195. [DOI] [PubMed] [Google Scholar]
- Kan N. C., Gall J. G. The intervening sequence of the ribosomal RNA gene is highly conserved between two Tetrahymena species. Nucleic Acids Res. 1982 May 11;10(9):2809–2822. doi: 10.1093/nar/10.9.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsch T., Kuck U., Esser K. Mitochondrial group I introns from the filamentous fungus Podospora anserina code for polypeptides related to maturases. Nucleic Acids Res. 1987 Aug 25;15(16):6743–6744. doi: 10.1093/nar/15.16.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmerer E. C., Kao T. H., Deng G. R., Wu R. Isolation and nucleotide sequence of the pea cytochrome oxidase subunit I gene. Plant Mol Biol. 1989 Jul;13(1):121–124. doi: 10.1007/BF00027340. [DOI] [PubMed] [Google Scholar]
- Knoop V., Schuster W., Wissinger B., Brennicke A. Trans splicing integrates an exon of 22 nucleotides into the nad5 mRNA in higher plant mitochondria. EMBO J. 1991 Nov;10(11):3483–3493. doi: 10.1002/j.1460-2075.1991.tb04912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhsel M. G., Strickland R., Palmer J. D. An ancient group I intron shared by eubacteria and chloroplasts. Science. 1990 Dec 14;250(4987):1570–1573. doi: 10.1126/science.2125748. [DOI] [PubMed] [Google Scholar]
- Kück U., Godehardt I., Schmidt U. A self-splicing group II intron in the mitochondrial large subunit rRNA (LSUrRNA) gene of the eukaryotic alga Scenedesmus obliquus. Nucleic Acids Res. 1990 May 11;18(9):2691–2697. doi: 10.1093/nar/18.9.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kück U. The intron of a plastid gene from a green alga contains an open reading frame for a reverse transcriptase-like enzyme. Mol Gen Genet. 1989 Aug;218(2):257–265. doi: 10.1007/BF00331276. [DOI] [PubMed] [Google Scholar]
- Lambowitz A. M. Infectious introns. Cell. 1989 Feb 10;56(3):323–326. doi: 10.1016/0092-8674(89)90232-8. [DOI] [PubMed] [Google Scholar]
- Lang B. F., Burger G. A collection of programs for nucleic acid and protein analysis, written in FORTRAN 77 for IBM-PC compatible microcomputers. Nucleic Acids Res. 1986 Jan 10;14(1):455–465. doi: 10.1093/nar/14.1.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang B. F., Burger G. A rapid, high resolution DNA sequencing gel system. Anal Biochem. 1990 Jul;188(1):176–180. doi: 10.1016/0003-2697(90)90548-n. [DOI] [PubMed] [Google Scholar]
- Lang B. F., Cedergren R., Gray M. W. The mitochondrial genome of the fission yeast, Schizosaccharomyces pombe. Sequence of the large-subunit ribosomal RNA gene, comparison of potential secondary structure in fungal mitochondrial large-subunit rRNAs and evolutionary considerations. Eur J Biochem. 1987 Dec 15;169(3):527–537. doi: 10.1111/j.1432-1033.1987.tb13641.x. [DOI] [PubMed] [Google Scholar]
- Lang B. F. The mitochondrial genome of the fission yeast Schizosaccharomyces pombe: highly homologous introns are inserted at the same position of the otherwise less conserved cox1 genes in Schizosaccharomyces pombe and Aspergillus nidulans. EMBO J. 1984 Sep;3(9):2129–2136. doi: 10.1002/j.1460-2075.1984.tb02102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazowska J., Claisse M., Gargouri A., Kotylak Z., Spyridakis A., Slonimski P. P. Protein encoded by the third intron of cytochrome b gene in Saccharomyces cerevisiae is an mRNA maturase. Analysis of mitochondrial mutants, RNA transcripts proteins and evolutionary relationships. J Mol Biol. 1989 Jan 20;205(2):275–289. doi: 10.1016/0022-2836(89)90341-0. [DOI] [PubMed] [Google Scholar]
- Lippok B., Brennicke A., Wissinger B. The coxII gene in carrot mitochondria contains two introns. Mol Gen Genet. 1992 Mar;232(2):322–327. doi: 10.1007/BF00280012. [DOI] [PubMed] [Google Scholar]
- Michaelis G., Vahrenholz C., Pratje E. Mitochondrial DNA of Chlamydomonas reinhardtii: the gene for apocytochrome b and the complete functional map of the 15.8 kb DNA. Mol Gen Genet. 1990 Sep;223(2):211–216. doi: 10.1007/BF00265056. [DOI] [PubMed] [Google Scholar]
- Michel F., Dujon B. Conservation of RNA secondary structures in two intron families including mitochondrial-, chloroplast- and nuclear-encoded members. EMBO J. 1983;2(1):33–38. doi: 10.1002/j.1460-2075.1983.tb01376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Dujon B. Genetic exchanges between bacteriophage T4 and filamentous fungi? Cell. 1986 Aug 1;46(3):323–323. doi: 10.1016/0092-8674(86)90651-3. [DOI] [PubMed] [Google Scholar]
- Michel F., Jacquier A., Dujon B. Comparison of fungal mitochondrial introns reveals extensive homologies in RNA secondary structure. Biochimie. 1982 Oct;64(10):867–881. doi: 10.1016/s0300-9084(82)80349-0. [DOI] [PubMed] [Google Scholar]
- Michel F., Lang B. F. Mitochondrial class II introns encode proteins related to the reverse transcriptases of retroviruses. Nature. 1985 Aug 15;316(6029):641–643. doi: 10.1038/316641a0. [DOI] [PubMed] [Google Scholar]
- Michel F., Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990 Dec 5;216(3):585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- Mota E. M., Collins R. A. Independent evolution of structural and coding regions in a Neurospora mitochondrial intron. Nature. 1988 Apr 14;332(6165):654–656. doi: 10.1038/332654a0. [DOI] [PubMed] [Google Scholar]
- Muscarella D. E., Vogt V. M. A mobile group I intron in the nuclear rDNA of Physarum polycephalum. Cell. 1989 Feb 10;56(3):443–454. doi: 10.1016/0092-8674(89)90247-x. [DOI] [PubMed] [Google Scholar]
- Netzker R., Köchel H. G., Basak N., Küntzel H. Nucleotide sequence of Aspergillus nidulans mitochondrial genes coding for ATPase subunit 6, cytochrome oxidase subunit 3, seven unidentified proteins, four tRNAs and L-rRNA. Nucleic Acids Res. 1982 Aug 11;10(15):4783–4794. doi: 10.1093/nar/10.15.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller H. F. Structure of ribosomal RNA. Annu Rev Biochem. 1984;53:119–162. doi: 10.1146/annurev.bi.53.070184.001003. [DOI] [PubMed] [Google Scholar]
- Nomiyama H., Kuhara S., Kukita T., Otsuka T., Sakaki Y. Nucleotide sequence of the ribosomal RNA gene of Physarum polycephalum: intron 2 and its flanking regions of the 26S rRNA gene. Nucleic Acids Res. 1981 Nov 11;9(21):5507–5520. doi: 10.1093/nar/9.21.5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomiyama H., Sakaki Y., Takagi Y. Nucleotide sequence of a ribosomal RNA gene intron from slime mold Physarum polycephalum. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1376–1380. doi: 10.1073/pnas.78.3.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent J. M., Palmer J. D. RNA-mediated transfer of the gene coxII from the mitochondrion to the nucleus during flowering plant evolution. Cell. 1991 Aug 9;66(3):473–481. doi: 10.1016/0092-8674(81)90011-8. [DOI] [PubMed] [Google Scholar]
- Oda K., Yamato K., Ohta E., Nakamura Y., Takemura M., Nozato N., Akashi K., Kanegae T., Ogura Y., Kohchi T. Gene organization deduced from the complete sequence of liverwort Marchantia polymorpha mitochondrial DNA. A primitive form of plant mitochondrial genome. J Mol Biol. 1992 Jan 5;223(1):1–7. doi: 10.1016/0022-2836(92)90708-r. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe B. A., Ma D. P., Wilson R. K., Wong J. F. The complete nucleotide sequence of the Xenopus laevis mitochondrial genome. J Biol Chem. 1985 Aug 15;260(17):9759–9774. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster W., Brennicke A. Plastid, nuclear and reverse transcriptase sequences in the mitochondrial genome of Oenothera: is genetic information transferred between organelles via RNA? EMBO J. 1987 Oct;6(10):2857–2863. doi: 10.1002/j.1460-2075.1987.tb02587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz A., Gubbins E. J., Bogorad L. The anticodon of the maize chloroplast gene for tRNA Leu UAA is split by a large intron. Nucleic Acids Res. 1982 May 25;10(10):3027–3037. doi: 10.1093/nar/10.10.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkl H., Wolf K. The mosaic cox1 gene in the mitochondrial genome of Schizosaccharomyces pombe: minimal structural requirements and evolution of group I introns. Gene. 1986;45(3):289–297. doi: 10.1016/0378-1119(86)90027-2. [DOI] [PubMed] [Google Scholar]
- Turmel M., Boulanger J., Schnare M. N., Gray M. W., Lemieux C. Six group I introns and three internal transcribed spacers in the chloroplast large subunit ribosomal RNA gene of the green alga Chlamydomonas eugametos. J Mol Biol. 1991 Mar 20;218(2):293–311. doi: 10.1016/0022-2836(91)90713-g. [DOI] [PubMed] [Google Scholar]
- Wahleithner J. A., MacFarlane J. L., Wolstenholme D. R. A sequence encoding a maturase-related protein in a group II intron of a plant mitochondrial nad1 gene. Proc Natl Acad Sci U S A. 1990 Jan;87(2):548–552. doi: 10.1073/pnas.87.2.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring R. B., Brown T. A., Ray J. A., Scazzocchio C., Davies R. W. Three variant introns of the same general class in the mitochondrial gene for cytochrome oxidase subunit 1 in Aspergillus nidulans. EMBO J. 1984 Sep;3(9):2121–2128. doi: 10.1002/j.1460-2075.1984.tb02100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring R. B., Davies R. W., Scazzocchio C., Brown T. A. Internal structure of a mitochondrial intron of Aspergillus nidulans. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6332–6336. doi: 10.1073/pnas.79.20.6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissinger B., Schuster W., Brennicke A. Trans splicing in Oenothera mitochondria: nad1 mRNAs are edited in exon and trans-splicing group II intron sequences. Cell. 1991 May 3;65(3):473–482. doi: 10.1016/0092-8674(91)90465-b. [DOI] [PubMed] [Google Scholar]
- Wolf K., Del Giudice L. The variable mitochondrial genome of ascomycetes: organization, mutational alterations, and expression. Adv Genet. 1988;25:185–308. doi: 10.1016/s0065-2660(08)60460-5. [DOI] [PubMed] [Google Scholar]
- Wolff G., Kück U. The structural analysis of the mitochondrial SSUrRNA implies a close phylogenetic relationship between mitochondria from plants and from the heterotrophic alga Prototheca wickerhamii. Curr Genet. 1990 Apr;17(4):347–351. doi: 10.1007/BF00314883. [DOI] [PubMed] [Google Scholar]
- Xu M. Q., Kathe S. D., Goodrich-Blair H., Nierzwicki-Bauer S. A., Shub D. A. Bacterial origin of a chloroplast intron: conserved self-splicing group I introns in cyanobacteria. Science. 1990 Dec 14;250(4987):1566–1570. doi: 10.1126/science.2125747. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]



