Abstract
Background
Lantibiotics are lanthionine-containing, post-translationally modified antimicrobial peptides. These peptides have significant, but largely untapped, potential as preservatives and chemotherapeutic agents. Type 1 lantibiotics are those in which lanthionine residues are introduced into the structural peptide (LanA) through the activity of separate lanthionine dehydratase (LanB) and lanthionine synthetase (LanC) enzymes. Here we take advantage of the conserved nature of LanC enzymes to devise an in silico approach to identify potential lantibiotic-encoding gene clusters in genome sequenced bacteria.
Results
In total 49 novel type 1 lantibiotic clusters were identified which unexpectedly were associated with species, genera and even phyla of bacteria which have not previously been associated with lantibiotic production.
Conclusions
Multiple type 1 lantibiotic gene clusters were identified at a frequency that suggests that these antimicrobials are much more widespread than previously thought. These clusters represent a rich repository which can yield a large number of valuable novel antimicrobials and biosynthetic enzymes.
Background
Bacteriocins are bacterially produced peptide antibiotics. Two major classes of gram-positive bacteriocins have been recognised, Class I undergo significant post-translationally modifications while the Class II are unmodified [1,2]. The majority of the class I bacteriocins are lantibiotics; small peptides containing internal bridges resulting from the formation of (β-methyl)lanthionine residues. The structural gene encodes a ribosomally synthesised precursor prepeptide which is generically named LanA. This prepeptide contains a leader sequence at the N-terminus, which is ultimately cleaved, and a propeptide at the C-terminus. Many or all of the serine and threonine residues within the propeptide are dehydrated to form dehydroalanine (Dha) and dehydrobutyrine (Dhb), respectively. When these modified residues interact with an intrapeptide cysteine, a thioether bond is formed resulting in the formation of lanthionine (Lan, from Dha) or β-methyl lanthionine (meLan, from Dhb).
The lantibiotics and lantipeptides (lanthionine containing peptides which lack antimicrobial activity) can be divided into four groups according to the nature of the enzymes which catalyse (me)Lan formation [3]. In the case of type 1 lantibiotics two enzymes are involved; LanB, the lanthionine dehydratase which catalyses the dehydration of the amino acids, and LanC, the lanthionine synthetase which catalyses thioether formation. Type 2 lantibiotics contain a single LanM enzyme which performs both functions. Type 3 and 4 are lantipeptides which are also catalysed by distinct enzymes such as the RamC-like and LanL enzymes [4,5]. The type 1 and 2 lantibiotics can also be further subdivided on the basis of the amino acid sequence of the unmodified propeptide. In the case of the type 1 lantibiotics, five such subgroups have been identified, each of which is named after the corresponding prototypical lantibiotic; the nisin-like, epidermin-like, Pep5-like, streptin-like and planosporocin-like lantibiotic subgroups [2,6]. The nisin-like group is named for nisin A, which is the most extensively studied bacteriocin and is currently sold in more than 50 countries as a food preservative [7]. In addition to LanA, B and C, other proteins involved in the the production of nisin A and other type 1 lantibiotics include LanP, a serine protease that cleaves the leader from the propeptide; LanT, an ABC transporter responsible for the transport of the lantibiotic precursor across the cell membrane; LanIEFG encode proteins involved in immunity and; LanK, a histidine kinase and LanR, a response regulator, that together operate as a two-component regulatory system. LanD enzymes, such as that responsible for the oxidative decarboxylation of C-terminal cysteines in epidermin [8], are less common.
Given the broad antimicrobial spectrum of many lantibiotics, the possibility of applying lantibiotics in a medicinal capacity has become the subject of much attention. This is supported by an enhanced understanding of their mechanisms of action [9] and the dearth of novel antibiotics. Of the type 1 lantibiotics, nisin, mutacin and planosporicin have been shown to be active against multi-drug resistant gram-positive pathogens [10-12], Pep5 and epidermin inhibit Staphylococcus epidermidis adhesion to catheters [13] and epidermin and gallidermin are active against Propionibacterium acnes [14]. Other lantibiotics, or their producer strains, have been used as food preservatives and as oral and gastrointestinal antimicrobials/probiotics [15-17]. As a consequence of this increased interest in lantibiotics, a concerted effort has taken place to identify new and improved forms of these peptides. Culture based screening strategies have in the past been responsible for the identification of lantibiotics produced by bacteria isolated from diverse microbial niches including the oral cavity, intestine, soil, kefir grains and milk [12,18-21]. Recently, an alternative means of identifying novel lantibiotics has emerged as a consequence of the increasing generation and availability of genomic and metagenomic sequence data. The availability of such information has recently led to the identification of the type 1 epidermin-like lantibiotic, Bsa [22] as well as type 2 lantibiotics such as haloduracin [23,24], licheniciden [25,26], as well as a range of cyanobacteria-associated lantipeptides [27]. This has prompted the development of on-line tools and repositories such as BAGEL and BACTIBASE to facilitate such screening strategies [28-31]. Notably, although an in-silico screen for lanM genes has recently resulted in the identification of 61 novel type 2 lantibiotic-like gene clusters [25], a corresponding screen for type 1 lantibiotics has not yet been described. Here we address this issue by screening for clusters containing genes homologous to the nisin A biosynthetic genes nisB (representing lanB) and nisC (representing lanC). In each case, the regions flanking the newly identified lanB/lanC genes were subjected to further in silico analysis to determine if they are potential lantibiotic/lantipeptide-associated gene clusters. This included a search of nearby open reading frames (orfs) which might encode a corresponding LanA, defined as being of relatively short length (approx 60 amino acids) and containing an uneven distribution of cysteine, threonine and serine amino acids within the propeptide region. Using this approach, 27 novel type 1 lantibiotic/lantipeptide-encoding clusters were identified. Subsequent screening using the newly identified LanA, B and C homologs as driver sequences revealed a further 22 gene clusters, resulting in a total of 49 putative novel type 1 lantibiotic clusters. Significantly, many of these clusters are present in species, genera and phyla not previously associated with lantibiotic/lantipeptide production and are predicted to encode peptides which represent completely new type 1 subclasses.
Results and Discussion
In silico screen for lanC genes
An in silico screen for LanC homologues, using the NisC sequence as a driver, resulted in the identification of 56 homologues. Of these 7 have previously been associated with lantibiotic production, 11 were orphan homologs (in that no other lantibiotic-associated genes were identified in close association) (Table 1), 9 were encoded within a cluster in which no lanA could be detected (Table 2) and one cluster contained two LanCs (but no structural peptide). The remaining 27 potential lantibiotic/lantipeptide-encoding gene clusters all contained putative lanA, B and C genes (Table 3). The genes flanking the 27 novel lanC-like genes were subjected to further bioinformatic analysis to determine the presence of other orfs that share homology with genes linked to lantibiotic production or immunity. While these 27 clusters are the primary focus of this in silico analysis, the sequences of the newly identified LanA, B and C proteins associated with these clusters were in turn used for further in silico screens. This approach uncovered an additional 22 clusters (Table 4) that were also predicted to be novel lantibiotic/lantipeptide-encoding clusters, thereby yielding a total of 49 novel type 1 clusters.
Table 1.
A selection of bacterial genomes in which isolated genes encoding LanA, LanB or LanC homologs were identified
| Clusterless Homologs | Accession No. | LanA only | LanB only | LanC only |
|---|---|---|---|---|
| Anoxybacillus flavithermus WK1 | NC_011567 | Aflv_2440 | ||
| Bacillus sp. B14905 | NZ_AAXV00000000 | BB14905_21668 | ||
| Paenibacillus larvae subsp. larvae BRL-230010 | NZ_AARF00000000 | Plarl_010100024193 | ||
| Lactobacillus crispatus MV-1A-US | NZ_ACOG00000000 | HMPREF0507_00422 | ||
| Lactobacillus crispatus JV-V01 | NZ_ACKR00000000 | HMPREF0506_0642 | ||
| Haliangium ochraceum DSM 14365 | NC_013440 | Hoch_3102 | ||
| Haliangium ochraceum DSM 14365 | NC_013440 | Hoch_4144 | ||
| Haliangium ochraceum DSM 14365 | NC_013440 | Hoch_0066 | ||
| Pedobacter sp. BAL39 | NZ_ABCM00000000 | PBAL39_02527 | ||
| Peptoniphilus lacrimalis 315-B | NZ_ADDO00000000 | HMPREF0628_0526 | ||
| Peptoniphilus lacrimalis 315-B | NZ_ADDO00000000 | HMPREF0628_0527 | ||
| Lactococcus lactis subsp. lactis KF147 | NC_013656 | |||
| Frankia alni ACN14a | NC_008278 | FRAAL2701 | ||
| Frankia sp. CcI3 | NC_007777 | Francci3_0205 | ||
| Frankia sp. CcI3 | NC_007777 | Francci3_3997 | ||
| Peptoniphilus lacrimalis 315-B | NZ_ADDO00000000 | HMPREF0628_0527 | ||
| Bifidobacterium longum subsp. infantis ATCC 15697 | NC_011593 | |||
| Streptomyces sp. AA4 | NZ_ACEV00000000 | |||
| Streptococcus pneumoniae CGSP14 | NC_010582 | |||
| Saccharopolyspora erythraea NRRL 2338 | NZ_ABFV00000000 | SACE_4959 | ||
| Kordia algicida OT-1 | NZ_ABIB00000000 | KAOT1_07113 | ||
| Streptococcus dysgalactiae GGS_124 | NC_012891 | SDEG_0295 | ||
| Microscilla marina ATCC 23134 | NZ_AAWS00000000 | M23134_07394 | ||
| Microscilla marina ATCC 23134 | NZ_AAWS00000000 | M23134_05752 | ||
| Microscilla marina ATCC 23134 | NZ_AAWS00000000 | M23134_07275 | ||
| Microscilla marina ATCC 23134 | NZ_AAWS00000000 | M23134_01545 | ||
| Microscilla marina ATCC 23134 | NZ_AAWS00000000 | M23134_07404 | ||
| Streptomyces sp. Mg1 | NZ_ABJF00000000 | SSAG_05771 | ||
| Frankia sp. EuI1c | NZ_ADDL00000000 | FraEuI1cDRAFT_6351 | ||
| Geobacillus sp. Y412MC52 | NZ_ACNM00000000 | GYMC52DRAFT_3129 | ||
| Geobacillus sp. Y412MC61 | NC_013411 | GYMC61_1158 | ||
| Streptococcus pyogenes M1 GAS | NC_002737 | SPy_1083 | ||
| Streptomyces griseus subsp. griseus NBRC 13350 | NC_010572 | SGR_6574 | ||
| Clostridium kluyveri DSM 555 | NC_009706 | CKL_3505 | ||
| Spirosoma linguale DSM 74 | NC_013730 | Slin_0903 | ||
| Spirosoma linguale DSM 74 | NC_013730 | Slin_2131 |
Table 2.
Gene clusters encoding LanB and LanC, but not LanA, homologs
| Species (Cluster No.) | Accession No. | LanB | LanC |
|---|---|---|---|
| Frankia alni ACN14a I | NC_008278 | FRAAL2701 | FRAAL2700 |
| Frankia sp. CcI3 IV | NC_007777 | Francci3_2033 | Francci3_2032 |
| Frankia EAN1pec II | NC_009921 | Franean1_2799 | Franean1_2800 |
| Frankia sp. EuI1c II | NZ_ADDL00000000 | FraEuI1cDRAFT_6786 | FraEuI1cDRAFT_6785 |
| Bacillus clausii KSM-K16 | NC_006582 | ABC3559 | ABC3558 |
| Clostridium cellulovorans 743B | NZ_ACPD00000000 | ClocelDRAFT_0447 | ClocelDRAFT_0445/_0452 |
| Bacillus cereus AH1273 | NZ_ACMT00000000 | bcere0030_58380 | bcere0030_58400 |
| Bacillus thuringiensis serovar berliner ATCC 10792 | NZ_ACNF00000000 | bthur0008_53920 | bthur0008_53930 |
| Bacillus thuringiensis IBL 200 | NZ_ACNK00000000 | bthur0013_59170 | bthur0013_59180 |
| Streptococcus pyogenes MGAS9429 | NC_008021 | MGAS9429_Spy0926 | MGAS9429_Spy0924 |
| Catenulispora acidiphila DSM 44928 | NC_013131 | Mentioned; Caci_4205 | Caci_4204 |
| Frankia sp Cc13 V | NC_007777 | Francci3_3530 | Francci3_3531 |
| Microscilla marina ATCC 23134 | NZ_AAWS00000000 | M23134_05752 | M23134_05756 |
| Staphylococcus capitis SK14 | NZ_ACFR00000000 | STACA0001_2327 | STACA0001_2326 |
| Streptomyces sp. Mg1 I | NZ_ABJF00000000 | SSAG_03540 | SSAG_03541 |
Table 3.
Bacterial genomes in which 27 uncharacterised type 1 lantibiotic clusters were identified following a NisC-led in silico screen
| Species (Cluster No.) | Accession No. |
|---|---|
| Frankia alni ACN14a (II) | NC_008278 |
| Frankia sp Cc13 (I) | NC_007777 |
| Frankia sp Cc13 (II) | NC_007777 |
| Frankia sp Cc13 (III) | NC_007777 |
| Frankia EAN1pec (I) | NC_009921 |
| Frankia EAN1pec (III) | NC_009921 |
| Frankia sp. EuI1c (I) | NZ_ADDL00000000 |
| Salinispora arenicola CNS-205* | NC_009953 |
| Stackebrandtia nassauensis DSM 44728 (I) | NC_013947 |
| Stackebrandtia nassauensis DSM 44728 (II) | NC_013947 |
| Streptomyces clavuligerus ATCC 27064 (I) | NZ_ADGD00000000 |
| Streptomyces clavuligerus ATCC 27064 (II) | NZ_ADGD00000000 |
| Streptomyces coelicolor A3(2) (I) | NC_003888 |
| Streptomyces coelicolor A3(2) (II) | NC_003888 |
| Streptomyces sp. Mg1 (II) | NZ_ABJF00000000 |
| Streptomyces griseoflavus Tu4000 (I) | NZ_ACFA00000000 |
| Streptomyces griseoflavus Tu4000 (II) | NZ_ACFA00000000 |
| Streptomyces griseoflavus Tu4000 (III) | NZ_ACFA00000000 |
| Bacillus cereus F65185 | NZ_ACMO00000000 |
| Bacillus mycoides DSM 2048 | NZ_ACMU00000000 |
| Clostridium perfringens CPE str. F4969 | NZ_ABDX00000000 |
| Enterococcus faecalis Fly1 | NZ_ACAR00000000 |
| Geobacillus kaustophilus HTA426 | NC_006510 |
| Geobacillus thermodenitrificans NG80-2 | NC_009328 |
| Geobacillus sp. G11MC16 | NZ_ABVH00000000 |
| Streptococcus thermophilus LMG 18311* | NC_006448 |
| Chitinophaga pinensis DSM 2588 I | NC_013132 |
Table 4.
Bacterial genomes in which 22 additional type 1 lantibiotic gene clusters were identified following an in silico screen using the LanA, B, and C homologs, corresponding to the clusters referred to in Table 3, as leader sequences
| Species (Cluster No.) | Accession No. |
|---|---|
| Thermomonospora curvata DSM 43183 | NC_013510 |
| Frankia EAN1pec (IV) | NC_009921 |
| Streptomyces viridochromogenes DSM 40736 | NZ_ACEZ00000000 |
| Streptomyces sp. SPB74 | NZ_ABJG00000000 |
| Streptomyces lividans TK24 | NZ_ACEY00000000 |
| Catenulispora acidiphila DSM 44928 | NC_013131 |
| Streptomyces sp. Mg1 (III) | NZ_ABJF00000000 |
| Nocardiopsis dassonvillei subsp. dassonvillei DSM 43111 | NZ_ABUI00000000 |
| Micromonospora aurantiaca ATCC 27029 (I) | NZ_ADBZ00000000 |
| Micromonospora aurantiaca ATCC 27029 (II) | NZ_ADBZ00000000 |
| Bacillus cereus AH1272 | NZ_ACMS00000000 |
| Staphylococcus aureus subsp. aureus D139 | NZ_ACSR00000000 |
| Staphylococcus aureus subsp. aureus H19 | NZ_ACSS00000000 |
| Actinomyces sp. oral taxon 848 | NZ_ACUY00000000 |
| Parachlamydia acanthamoebae str. Hall's coccus | NZ_ACZE00000000 |
| Corynebacterium lipophiloflavum DSM 44291 | NZ_ACHJ00000000 |
| Staphylococcus aureus A9765 | NZ_ACSN00000000 |
| Chitinophaga pinensis DSM 2588 (II) | NC_013132 |
| Spirosoma linguale DSM 74 | NC_013730 |
| Pedobacter heparinus DSM 2366 | NC_013061 |
| Kordia algicida OT-1 | NZ_ABIB00000000 |
| Microscilla marina ATCC 23134 | NZ_AAWS00000000 |
All except one of the 27 gene clusters revealed following the initial screen were located within the genomes of Firmicutes and Actinobacteria. The exception was Chitinophaga pinensis DSM 2588 of the phylum Bacteroidetes. Of the other 26, the genera most commonly associated with lantibiotic production were Bacillus, Geobacillus, Clostridium, Enterococcus, Streptococcus, Frankia and Streptomyces. In many cases the novel clusters associated with a specific genus, such as those found on the Streptomyces and Frankia genomes, showed at least some similarity to each other. It was also noted that several of the genomes in which a cluster was located also contained an additional cluster(s) (Table 3), or other genes predicted to encode additional LanA, B or C proteins (Table 2), elsewhere in the genome. The 27 clusters are described below and are grouped according to the phylum and genus of the associated strain.
Type 1 lantibiotic gene clusters in Actinobacteria
Identification of novel Frankia-associated lantibiotic gene clusters
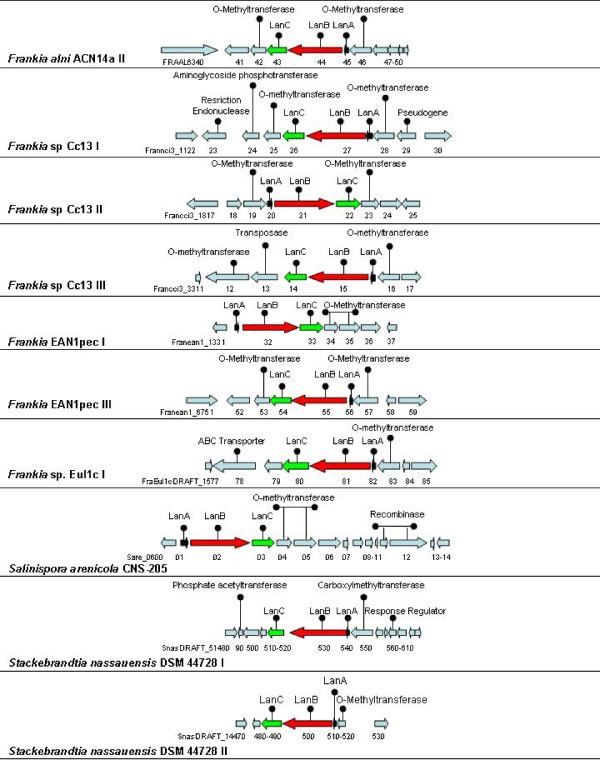
The Frankia are nitrogen-fixing, root nodule-forming filamentous Actinobacteria that live in symbiosis with actinorhizal plants. All species of Frankia are closely related [32]. To date, four Frankia genomes have been sequenced, i.e. Frankia alni ACN14a, Frankia sp. EAN1pec, Frankia sp. Cc13 and Frankia sp. EUI1c, and although no Frankia-associated bacteriocins have previously been reported, a number of predicted lantibiotic clusters can be found in each case (Figure 1) in addition to a number of apparently LanB- and LanC-encoding genes which do not have an accompanying lanA (Table 2). This latter phenomenon could be a result of the frequent rearrangements which occur in Frankia strains [32]. Of the clusters identified, many resemble clusters associated with another genus of Actinobacteria, the Streptomyces.
Figure 1.
Diagramatic representation of the non-streptomyces Actinobacteria type 1 lantibiotic operons, found in the original NisC screen, which contain genes predicted to encode a structrural peptide LanA, and the modification enzymes LanB and LanC.
Frankia alni ACN14a: The genome sequence of F. alni ACN14a [32] contains one complete cluster, F. alni ACN14a II which includes the predicted LanA prepropeptide, FRAAL6345, FRAAL6344 and FRAAL6343 (encoding a putative LanB and LanC, respectively). The LanA peptide does not resemble any of the previously characterised type 1 lantibiotic propeptides but is 79% identical to Franean1_0057 of Frankia sp. EAN1pec (see below). The LanB and LanC proteins resemble those associated with other Frankia, as well as Streptomyces, clusters. The LanC protein is also 62% identical to Tcur_4648 of Thermomonospora curvata DSM 43183 (NC_013510), which itself appears to be encoded by an orf within a novel lantibiotic gene cluster. The ACN14a II cluster is also predicted to encode two proteins which share homology with O-methyltransferases (FRAAL6342 and FRAAL6346). O-methyltransferases contribute to the production of a number of non-ribosomal antibiotics [33,34] and catalyse the methylation of hydroxyl group(s) on deoxysugar rings to protect the reactive hydroxyl group from undesired modifications and can alter the solubility and pharmacokinetic properties of the resulting molecule [35]. Although O-methyltransferases have not previously been associated with lantibiotic production, this study reveals that many Actinobacteria-associated type 1 clusters possess genes predicted to encode these enzymes.
Frankia sp. Cc13: The Frankia sp. Cc13 genome [32] contains three gene clusters of interest (Frankia sp. Cc13 I, Frankia sp. Cc13 II and Frankia sp. Cc13 III). Two O-methyltransferase-encoding genes were identified in each case and it was noted that the associated LanB and LanC proteins are similar to one another and to numerous others predicted to be produced by Frankia and Streptomyces species. In contrast, there is a lack of homology between the three lanA genes. The lanA gene from cluster I was not previously annotated and was only identified following closer inspection of the DNA sequence. The cluster II-associated LanA, Francci3_1820, most closely resembles Frankia sp. EuI1c FraEuI1cDRAFT_6351 (69% identity) while the third, and also previously unannotated LanA appears to be one of an extended group of Frankia- and Streptomyces-associated LanAs that includes Frankia sp. EuI1c FraEuI1cDRAFT_1582 (56% identity).
Frankia EAN1pec: The Frankia EAN1pec genome [32] contains 4 putative LanB-encoding genes three of which correspond to potential lantibiotic/lantipeptide-associated gene clusters (Frankia EAN1pec I, Frankia EAN1pec III and Frankia EAN1pec IV) which again resemble those of Streptomyces and other Frankia species, and contain O-methyltransferase-encoding genes. Within the first cluster, a putative LanA prepropeptide, encoded by a previously unannotated orf located between Franean1_1331 and the LanB determinant, is homologous to a number of other LanAs, including Sare_0601 of Salinispora arenicola CNS-205 (55% identity). The cluster is also noteworthy be virtue of the presence of two LanC-encoding genes, Franean1_1333 and Franean1_1336. Within Frankia EAN1pec III, the LanA peptide, encoded by Franean1_6756 is 42% identical to FraEuI1cDRAFT_6351 of Frankia sp. EuI1c while finally, a screen using F. alni ACN14a FRAAL6345 as a driver led to the identification of yet another cluster (consisting of at least Franean1_0057-0055) which closely resembles cluster II of F. alni ACN14a II.
Frankia sp. EuI1c: Frankia sp. EuI1c contains a single putative lantibiotic/lantipeptide gene cluster (Frankia sp. EuI1c I) which again contains LanB, C and O-methyltransferase genes typical of Frankia and Streptomyces clusters. The associated LanA homolog (FraEuI1cDRAFT_1582) is notable by virtue of being 46% identical to SSCG_03316, a known LanA of Streptomyces clavuligerus ATCC 27064 while a gene encoding an ABC transporter related protein (FraEuI1cDRAFT_1578) is also present.
Identification of novel Salinispora -associated lantibiotic gene clusters
Salinispora are marine Actinobacteria. There are two recognised species, S. tropicalis and S. arenicola. Representatives have been sequenced in each case and genes predicted to encode non-lantibiotic bacteriocins have been identified in both cases [36]. The existence of a putative lantibiotic/lantipeptide cluster, between Sare_0602 and Sare_0623, in the genome of S. arenicola CNS-205 was noted previously [36]. However, this cluster has not been the subject of a detailed bioinformatic characterisation. Our analysis reveals that Sare_0601 apparently encodes a LanA peptide which is 88% identical to that encoded by MicauDRAFT_5818 of Micromonospora aurantiaca ATCC 27029. The proteins encoded by Sare_0602 (LanB) and Sare_0603 (LanC) also resemble other ATCC 27029-associated proteins (encoded by MicauDRAFT_5819 (71% identity) and MicauDRAFT_5820 (75% identity)), thereby revealing an additional novel cluster in Micromonospora, a genus better known for its production of non-ribosomal antibiotics such gentamycin and netamycin [37] (Table 4).
Identification of novel Stackebrandtia-associated lantibiotic gene clusters
Stackebrandtia are aerobic, non-motile Actinobacteria which have been isolated from soil [38]. There are only 2 associated species i.e. S. albiflava and S. nassauensis and in silico analysis of S. nassauensis DSM 44728 (NC_013947) reveals the presence of two similar clusters (S. nassauensis DSM 44728 I and S. nassauensis DSM 44728 II) (Figure 1). The hypothetical LanA, encoded by Snas_5416, of the first cluster showed a singular homology of 78% identity to Snas_3601 of the second cluster. The corresponding LanBs (Snas_5417 and Snas_3602) are 62% identical while the LanCs (Snas_5418 and Snas_3603) are 68% identical.
Identification of novel Streptomyces-associated lantibiotic gene clusters
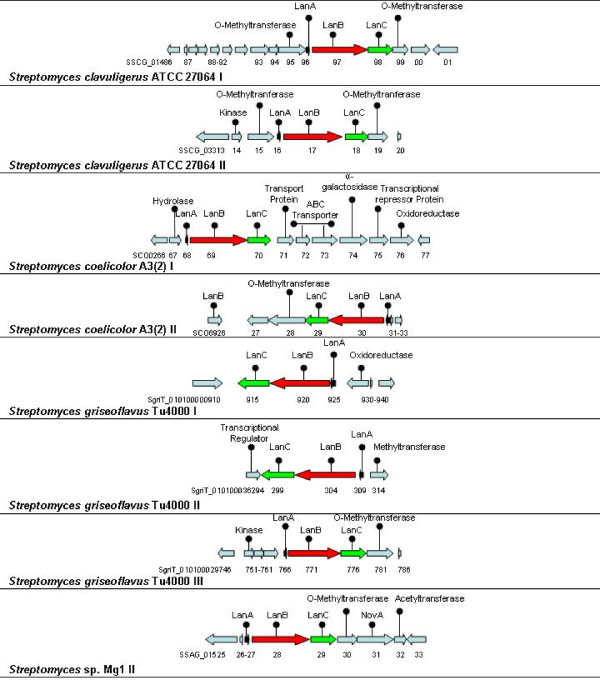
Bacteria from the genus Streptomyces, comprising over 500 species, are filamentous, high G-C bacteria found frequently in soil and rotting vegetation. They are the most numerous and ubiquitous soil bacteria [39]. Streptomyces are also responsible for the production of over two-thirds of the clinically useful antibiotics of natural origin (e.g., neomycin, chloramphenicol) [40]. Although a number of Streptomyces-associated bacteriocins, such as ancovenin [41] and cinnamycin [42], have been identified, this number is relatively small considering the size of the genus. As was apparent above, our in silico analysis has revealed that many Streptomyces possess potentially lantibiotic-encoding gene clusters which resemble those found in Frankia. Once again, the majority of these clusters contain O-methyltransferases (Figure 2).
Figure 2.
Diagramatic representation of the Streptomyces type 1 lantibiotic operons, found in the original NisC screen, which contain genes predicted to encode a structrural peptide LanA, and the modification enzymes LanB and LanC.
Streptomyces clavuligerus ATCC 27064
S. clavuligerus is an aerobic, mesophillic Streptomyces sp. While there have been no previous reports of bacteriocin production by this species, two lantibiotic clusters were found to be present on the genome of S. clavuligerus ATCC 27064. In the first of these clusters, the associated hypothetical LanA, B and C proteins (SSCG_01498-01496) are 63%, 42% and 50% identical to the corresponding proteins of Frankia sp. CcI3 II. BLAST analysis of these proteins also revealed another novel cluster in Streptomyces viridochromogenes DSM 40736 corresponding to SvirD4_23440 (LanA; 50% identity), SvirD4_23449 (LanB; 36% identity) and SvirD4_23454 (LanB; 45% identity) (Table 4). The second S. clavuligerus cluster, which contains SSCG_03316 (LanA), SSCG_03317 (LanB) and SSCG_03318 (LanC), resembled clusters present in a number of other strains such as that of Streptomyces griseus subsp. griseus NBRC 13350 [43] (73%, 56% and 64% identity, respectively). BLAST analysis of these sequences also led to the identification of yet another novel cluster in Streptomyces sp. SPB74 (SSBG_01041 [LanA] 69% identity and SSBG_01042 [LanB] 58% identity).
Streptomyces coelicolor A3(2)
Streptomyces coelicolor A3(2) (NC_003888) is the best characterised representive of its genus [44] and was the first Streptomyces strain to have its genome sequenced [45]. Although bacteriocins/bacteriocin-like peptides are known to be produced by this species (e.g. the class III morphogenic peptide SapB [4]), such peptides have not previously been associated with this strain. Here BLAST analysis revealed the presence of two lantibiotic/lantipeptide clusters (S. coelicolor A3(2) I and S. coelicolor A3(2) II). The first of these clusters, containing SCO0268 (LanA), SCO0269 (LanB) and SCO0270 (LanC), very closely resembles Streptomyces griseoflavus Tu4000 cluster II (see below). Subsequent BLAST searches with the A3(2) cluster I-associated proteins led in turn to the discovery of an almost identical cluster in Streptomyces lividans TK24 which contains SSPG_07329 (LanA; 100% identity), SSPG_07328 (LanB; 99% identity) and SSPG_07327 (LanC; 100% identity) (Table 4). The second cluster, Streptomyces coelicolor A3(2) II, is predicted to encode two LanA peptides, (SCO6932 [43aa] and SCO6931 [59aa]) which are 97% identical to each other, as well as LanB (SCO6930) and LanC (SCO6929) proteins with homology (39-46% identity) with corresponding proteins associated with Frankia sp. CcI3. Such analysis also revealed another cluster of interest in the actinomycete, Catenulispora acidiphila DSM 44928 (NC_013131; [Table 4]).
Streptomyces griseoflavus Tu4000
Three lantibiotic/lantipeptide clusters were identified on the genome of this anaerobic, terrestrial Streptomyces. Although the LanA encoded within the first cluster (SgriT_010100000925) does not significantly resemble any other protein, the associated LanB (SgriT_010100000920) and LanC (SgriT_010100000915) proteins are homologous to the corresponding proteins of S. coelicolor A3(2) cluster I and Tu4000 cluster II. BLAST searches using the cluster I proteins as drivers also resulted in the identification of several additional clusters in Nocardiopsis dassonvillei DSM 43111 (NdasDRAFT_3161 [LanB] 30% identity), Streptomyces sp. Mg1 (SSAG_05771 [LanB] 37% identity) and two clusters on the genome of M. aurantiaca ATCC 27029 (NZ_ADBZ00000000; MicauDRAFT_5820 and MicauDRAFT_3008 [both LanBs] 35% identity). In addition to the components of the second cluster referred to above, an associated LanA (SgriT_010100036309) was also noted. In addition to the Tu4000 I and A3(2) I clusters, this cluster is also highly identical to that of S. lividans TK24 (SSPG_07329 [LanA] 97% identity; SSPG_07328 [LanB] 87% identity and SSPG_07327 [LanC] 89% identity). The LanA associated with the final cluster (SgriT_010100029766) again bears no homology with any other known peptides whereas the LanB (SgriT_010100029771) and LanC (SgriT_010100029776) corresponded to those of Frankia sp. EAN1pec II (39% and 44% identity, respectively).
Type 1 lantibiotic gene clusters in Firmicutes
Identification of novel Bacillus-associated lantibiotic gene clusters
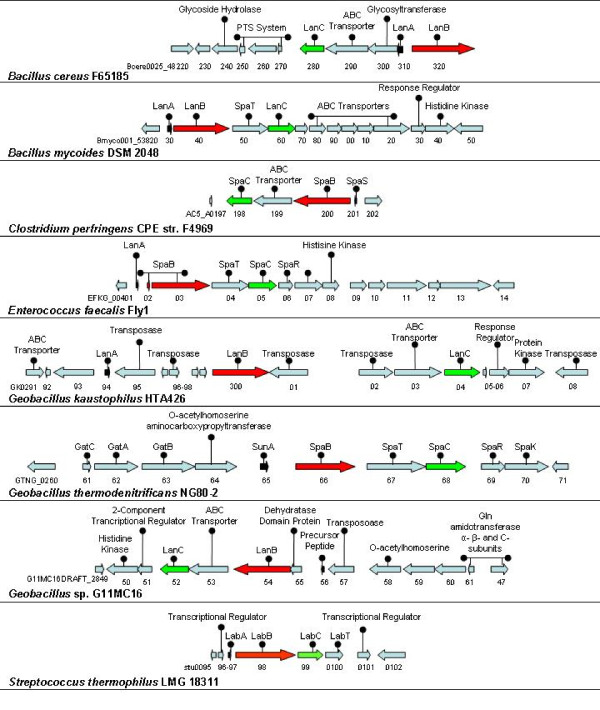
Bacillus is a large and diverse genus of rod-shaped, sporulating, obligate aerobes which contains both free living and pathogenic species. A number of type 1 lantibiotics have previously been characterized in this genus (e.g. subtilin [46] and ericin [47]). The NisC-driven screen highlighted the presence of a type 1 lantibiotic cluster in the genomes of two Bacillus strains i.e. Bacillus cereus F65185 and Bacillus mycoides DSM 2048 (Figure 3). Bioinformatic analysis of these clusters revealed two further clusters in B. cereus ATCC 14579 and B. cereus AH1272.
Figure 3.
Diagramatic representation of the Firmicutes type 1 lantibiotic operons, found in the original NisC screen, which contain genes predicted to encode a structrural peptide LanA, and the modification enzymes LanB and LanC.
Bacillus cereus F65185
B. cereus F65185 is a mesophilic bacterium sourced from a human wound containing one lantibiotic/lantipeptide cluster which is unusual in that 3 orfs separate the putative LanB and C genes and the two have a divergent orientation. The predicted LanA (bcere0025_48310) does not resemble any other known lantibiotic prepropeptides. The LanB homolog (bcere0025_48320) resembles a putative LanB associated with Clostridium cellulovorans 743B (ClocelDRAFT_0452, 30% identity) while the predicted LanC (bcere0025_48280) most closely resembles two further 743B proteins (ClocelDRAFT_0452, 34% identity and ClocelDRAFT_0446, 28% identity). However the 743B strain lacks an associated LanA. Further BLAST analysis with the F65185-associated LanB highlighted the presence of a related protein within thiocillin-encoding gene cluster in B. cereus ATCC 14579 [48].
Bacillus mycoides DSM 2048
B. mycoides is a non-motile, non-pathogenic, saprophytic Bacillus, strains of which have been investigated with a view to their application as biological pesticides. Although representatives of this species have been associated with bacteriocin production [49], there are no published reports of lantibiotic-producing B. mycoides. The DSM 2048 genome contains a lantibiotic/lantipeptide cluster that contains putative lanA, B, C and other lantibiotic-associated genes which is very similar to other novel clusters in B. cereus AH1272 and B. cereus AH1273. Bmyco0001_53830 is the predicted prepropeptide and is 100% identical to the products of the B. cereus AH1272-associated bcere0029_28240 and bcere0029_28250. It is also 58% identical to prepropeptides associated with several Staphylococcus-associated Bsa lantibiotics, such as BsaA2RF122 of S. aureus RF122 [22]. It is thus apparent that the LanA is a member of the epidermin-like peptides. In addition to homologues in B. cereus AH1273 and AH1272, use of the DSM 2048-associated LanB and LanC sequences as drivers also surprisingly highlighted a gene cluster present in Actinomyces sp. oral taxon 848 (Table 4). In addition to homology with respect to LanB and LanC proteins (HMPREF0972_00932 and HMPREF0972_00933, respectively), the proposed LanA (HMPREF0972_00931; 86aa) is 43% identical to bmyco0001_53830 and bcere0029_28240 and bcere0029_28250 of Bacillus mycoides DSM 2048 and Bacillus cereus AH1272, respectively.
Identification of novel Clostridium-associated lantibiotic gene clusters
The Clostridia are gram postitive anaerobic, endospore-forming Firmicutes of which there are approximately 100 species. These include important pathogens such as Clostridium difficile, Clostridium perfingens and Clostridium tetani. Several Clostridium-associated bacteriocins have been characterised from this genus [50-52], but no type 1 lantibiotic producers have been identified to date. Here, bioinformatic analysis revealed one Clostridium-associated lantibiotic/lantipeptide cluster, located on the genome of C. perfringens CPE str. F4969 (Figure 3).
C. perfringens CPE str. F4969
C. perfringens is one of the leading causes of food-borne illness in the developed world, usually as a result of the improper sterilization of canned foods in which endospores have germinated. It can also be responsible for wound and surgical infections [53,54]. The predicted LanA (AC5_A0201) of strain F4969 is most closely related to the LanAs of B. mycoides DSM 2048 and B. cereus AH1272 (51% identity) while the proteins encoded by AC5_A0200 (LanB) and AC5_A0198 (LanC) most closely resemble proteins associated with Geobacillus sp. G11MC16 (G11MC16DRAFT_2954 and G11MC16DRAFT_2952; 34% and 35% identity respectively). Surprisingly, BLAST analysis also highlighted the presence of a related LanB homolog encoded within a lantibiotic/lantipeptide-like gene cluster in the genome of Parachlamydia acanthamoebae (phlylum Chlamydiae) str. Hall's coccus (pah_c028o031; 25% identity). Adjacent genes of note within the P. acanthamoebae cluster include pah_c028o029 (LanA) and pah_c028o030 (LanC).
Identification of novel Enterococcus-associated lantibiotic gene clusters
The enterococci are gram positive lactic acid bacteria which are common commensal organisms in the intestines of humans but can also be pathogens. Many Enterococcus-associated bacteriocins (enterocins) have been identified [55]. Only one Enterococcus-associated lantibiotic, the type 2 peptide cytolysin, has been identified to date [56]. Here we describe genes which potentially encode the first type 1 Enterococcus-associated lantibiotic (Figure 3).
Enterococcus faecalis Fly1: E. faecalis
Fly1 (NZ_ACAR00000000) is a non-motile, facultative anaerobe. Within its genome we identified a previously unannotated LanA determinant, through analysis of raw sequence data. The corresponding peptide is homologous to C. perfringens CPE str. F4969 (AC5_A0201; 68% identity), as well as a number of epidermin-like LanAs in other bacilli. The putative LanB protein is split across two orfs, EFKG_00402 (80 amino acids) and EFKG_0403 (942 amino acids), with both components most closely resembling the N-terminus of the dehydratase of Streptococcus pyogenes MGAS10270, MGAS10270_Spy0922. It is unclear whether the apparent frameshift in the Fly1 lanB is genuine or the result of a sequencing error. The LanC-like EFKG_00405, was most closely related to the corresponding protein in G. thermodenitrificans NG80-2 (SpaC GTNG_0268; 35% identity).
Identification of novel Geobacillus-associated lantibiotic gene clusters
Geobacilli are thermophillic (45-70°C), aerobic, spore-forming Firmicutes. They have been isolated from various terrestrial and marine environments, in geothermal, temperate and permanently cold habitats. Reclassified in 2001 [57], these bacteria are of industrial interest as sources of thermostable enzymes. Bacteriocins have been identified in Geobacillus stearothermophilus [58] and Geobacillus thermoleovorans [59], and while screening for LanM-producing gene clusters has highlighted the potential existence of a number of type 2 lantibiotics [25], associated type 1 lantibiotics have not previously been described. Here, three putative type 1 lantibiotic/lantipeptide-encoding clusters within the genomes of Geobacillus kaustophilus HTA426, Geobacillus thermodenitrificans NG80-2 and Geobacillus sp. G11MC16 (Figure 3) are described.
Geobacillus kaustophilus HTA426
G. kaustophilus grows optimally in aquatic environments at 60°C with an upper temperature limit of 74°C. From a lantibiotic persepective, genome xsequencing of HTA426 revealed a hypothetical protein annotated as a 'lantibiotic precursor' GK0294. Our analysis revealed that this putative LanA is 91% identical to another prepropeptide encoded by the closely located GK0286 gene. It is also 100% identical to orphan 'lantibiotic precursor' homologs (GYMC52DRAFT_3129 and GYMC61_1158) in Geobacillus sp. Y412MC52 and Geobacillus sp. Y412MC61, respectively. More distantly related LanAs (79% identity) are also associated with the genomes of Geobacillus thermodenitrificans NG80-2 (GTNG_0265) and Geobacillus sp. G11MC16 (G11MC16DRAFT_2956). The homology between the Geobacillus LanAs is highest within the leader regions, but, as is the case with nisin-, epidermin- and streptin-like lantibiotics, a conserved serine and CTPGC motif in the N-terminus of the propeptide is present, which is believed to be involved in the binding of these lantibiotics to lipid II in the cell wall in gram positive bacteria [60]. BLAST analysis of the GK0286-encoded LanA highlighted the presence of another potential lantibiotic/lantipeptide cluster in Corynebacterium lipophiloflavum DSM 44291 (57% identity with HMPREF0298_1795). Within the HTA426 cluster, the proteins predicted to be encoded by GK0300/301 (an apparently frameshifted lanB) and GK0304 are homologous to those associated with many other geobacilli. It was also noted that this cluster is less condensed than typical lantibiotic gene clusters in that there are insertions of 7, 5 and 3 genes (predicted to encode many transposases and small, hypothetical proteins) between the lantibiotic associated genes.
Geobacillus thermodenitrificans NG80-2 and Geobacillus sp. G11MC16:
G. thermodenitrificans are facultative soil bacteria with denitrification qualities. Representatives of this species grow between 45°C and 73°C (optimum 65°C). NG80-2 was isolated from a deep-subsurface oil reservoir in Dagang oilfield, Northern China [61] and on the basis of in-silico analysis is potentially the producer of both a type 1 (see below) and type 2 lantibiotic [25]. Our analysis reveals that the type I lantibiotic/lantipeptide operons in G. thermodenitrificans NG80-2 and Geobacillus sp. G11MC16 are very highly conserved. The two LanAs are 100% identical and the homology between these, and indeed the associated B and Cs, and the corresponding G. kaustophilus HTA426 proteins is discussed above. It was noted that the lanB of Geobacillus sp. G11MC16 is apparently frameshifted (G11MC16DRAFT_2955 (176aa) and G11MC16DRAFT_2954 (848aa)) but that this is not the case in G. thermodenitrificans NG80-2 (GTNG_0266).
Identification of novel Staphylococcus-associated lantibiotic gene clusters
The staphylococci are non-sporeforming, non-motile Firmicutes. The genus Staphylococcus contains 33 species, most of which are harmless and reside normally on the skin and mucous membranes of humans and other organisms. However, staphylococci can also cause a wide variety of diseases either through toxin production or penetration and are a common cause of food poisoning and nosocomial infections. Several strains of Staphylococcus epidermidis have been shown to be producers of type 1 lantibiotics, including epidermin [62], Pep5 [63], epicidin 280 [64] and epilancin K7 [65], gallidermin was isolated from S. gallinarum [14] while Staphylococcin Au26 [66] and Bsa [22] were isolated from S. aureus. BLAST analysis has revealed that several other S. aureus strains possess gene clusters similar to those associated with Bsa and BsaRF122 [22]. These clusters were identified in S. aureus A9765, D139 and H19. In A9765, SAPG_01762 and SAPG_01760 correspond to the BsaA1 and BsaA2 peptides of S. aureus MW2 (97% and 100% identity, respectively). The precursor peptides of the D139 (SATG_00575 and SATG_00574; 76% identical to each other) and H19 (SAUG_01228 and SAUG_01229; 76% identical to each other) strains are 100% identical. The peptides encoded by SATG_00575 and SAUG_01229 are 93% identical to BsaA1RF122 of S. aureus RF122 (93% identity) while those corresponding to.SATG_00574 and SAUG_01228 are 100% identical to BsaA2RF122.
Identification of novel Streptococcus-associated lantibiotic gene clusters
These facultative anaerobes of the phylum Firmicutes are spherical in shape and grow in long chains. Many species are part of the normal commensal flora of the mouth, skin, intestine and upper respiratory tract of humans but the genus also includes numerous human pathogens such as Streptococcus pneumoniae, pyogenes and agalactiae. The streptococci are known to producers of type 1 lantibiotics [67,68], such as streptin [69], some mutacins [21,70-72], nisin U and nisin U2 [73], as well as several non-lantibiotic bacteriocins. Here we discuss two clusters, identified in strains of S. pyogenes and S. thermophilus LMG 18311.
Streptococcus pyogenes MGAS10270
S. pyogenes (or Group A Streptococcus, GAS) is the cause of many important human diseases ranging from mild superficial skin infections to life-threatening systemic diseases. Bacteriocin production by these strains may give them a competitive advantage against the natural skin microbiota. It has previously been established that many S. pyogenes strains, as well as strains of Streptococcus salivarius, produce the type 2 lantibiotic salivaricin A or closely related variants [16]. The type 1 streptins (1 and 2) and type 2 streptococcin A-FF22 are also S. pyogenes associated [69,74]. Here our analysis focuses on a type 1 cluster within the genome of S. pyogenes MGAS10270 [75]. This includes MGAS10270_Spy0919, which is 100% identical to the propeptide sequence of streptin. While this lantibiotic is thus not novel, subsequent BLAST searches were revealing in that they highlighted the presence of a LanA with 97% identity in S. pyogenes MGAS10750 (MGAS10750_Spy0955) which is contained within a cluster which also encodes a LanB (MGAS10750_Spy0958) and LanC (MGAS10750_Spy0957).
Streptococcus thermophilus LMG 18311
S. thermophilus is a thermophillic, non-pathogenic Streptococcus. It is of major importance to the fermented dairy food industry. A number of non-lantibiotic bacteriocins (thermophilins) from this species have been characterized, including thermophilin 347 [76], thermophilin A [77] and thermophilin ST-1 [78]. Strain LMG 18311 was sequenced in 2004 and at the time it was noted that bacteriocin production was one of the characteristics that distinguishes it from strain CNRZ1066 [79]. While the existence of a putative lantibiotic/lantipeptide gene cluster in LMG 18311 has been reported [79,80], this cluster (Figure 3) has not been the focus of a detailed in silico analysis. The associated LanA, encoded by stu0097, is homologous with that predicted to be encoded by SPCG_0144 of S. pneumoniae CGSP14 (88% identical) which, on the basis of previous in silico analysis, is also within a lantibiotic gene cluster [81]. The LanB protein (Stu0098) is 73% identical to SPCG_0145 of S. pneumoniae CGSP14 and 97% identical to a truncated LanB associated with S. thermophilus CNRZ1066 [79].
Type 1 lantibiotic gene clusters in Bacteroidetes
The Bacteroidetes are a highly diverse phylum found in soil, seawater and the skin and intestines of animals. The Bacteroidales class, which includes the genus Bacteroides, are the best-studied of the phylum. Bacteroides comprises the most substantial portion of the human gastrointestinal tract [82] some of which are opportunistic pathogens [83].
Identification of novel Chitinophaga-associated lantibiotic gene clusters
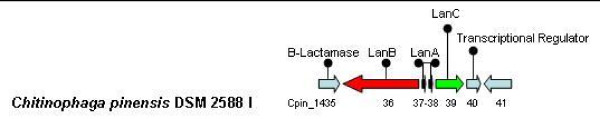
Chitinophaga are rod-shaped mesophiles of the phylum Bacteroidetes which are are noted for their ability to degrade chitin [84]. There have been no reports to date of bacteriocin production by any of the 10 Chitinophaga species. Chitinophaga pinensis DSM 2588 (NC_013132) is unusual in that it appears to be a Bacteroidetes possessing genes encoding a type 1 lantibiotic (Figure 4). There are two predicted LanA peptides, corresponding to Cpin_1438 and Cpin_1437, which are 50% identical as a consequence of similar N-terminii. Adjacent orfs of note include Cpin_1435 and Cpin_1440, predicted to encode a β-lactamase and a 2-component transcriptional regulator of the LuxR family, respectively. BLAST analysis of the associated LanB and LanC proteins (Cpin_1436 and Cpin_1439 respectively) revealed another putative LanB (Cpin_3392; 36% identity) and LanC (Cpin_3397; 23% identity) encoded within the same genome. Within this second C. pinensis-associated cluster, Cpin_3393 possess a number of features which suggest that it may be a LanA-encoding gene. Interestingly, BLAST analysis of the Cpin_3397-encoded LanC also led to the identification of a number of additional homologs apparently encoded within the genomes of strains not previously associated with lantibiotic production. The genome of another Bacteroidetes, Spirosoma linguale DSM 74, is notable in that it contains 4 putative LanB-encoding genes and 5 putative LanC-encoding genes. Of these only one LanB protein (Slin_4704; 31% identity) and one LanC protein (Slin_4705; 26% identity), are encoded within what appears to be a novel lantibiotic-associated gene cluster. This cluster contains 3 potentially LanA-encoding genes, Slin_4706-4708. Slin_4706 and Slin_4707, which are identical and share 58% identity with Slin_4708. The genome of another Bacteroidetes species, Pedobacter heparinus DSM 2366 (NC_013061), contains a cluster encoding two LanBs (Phep_0556 and Phep_0557; 37% and 36% identity, respectively), a LanC (Phep_0555; 33% identity) and a potential LanA (Phep_0553; no significant BLAST hits). A cluster within Kordia algicida OT-1 contains LanB (KAOT1_15523; 36% identity), LanC (KAOT1_15518; 30% identity) and LanA determinants (KAOT1_15533; no significant BLAST hits) while Microscilla marina ATCC 23134 potentially has five associated LanB homologs, but of these, the LanB corresponding to M23134_03921 (28% identity) was the only one to be located in close proximity to one of multiple LanC proteins (M23134_03925; 22% identity). The putative LanA, M23134_03926, does not resemble any other known proteins.
Figure 4.
Diagramatic representation of the Chitinophaga pinensis (Bacteroidetes) type 1 lantibiotic operons, found in the original NisC screen, which contain genes predicted to encode a structrural peptide LanA, and the modification enzymes LanB and LanC.
Phylogenetics of LanABC
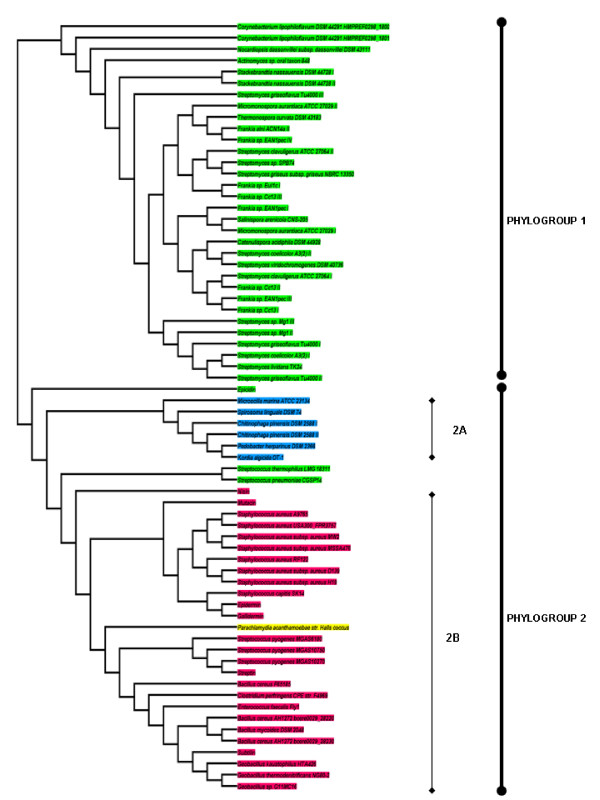
The conserved nature of LanB and LanC proteins facilitated a phylogenetic analysis of their relatedness. The resultant cladogram of LanB enzymes (all those identified in both screens, as well as a number of LanBs from previously analysed clusters) highlights the existence of two distinct phylogroups (Figure 5). The first phylogroup contains Actinobacteria-associated LanBs, all of which are from strains not previously known to be producers of lantibiotics/lantipeptides. The second contains a variety of lanthionine synthetases associated with known lantibiotics (nisin, subtilin, epidermin etc), some uncovered by previous in silico analysis (e.g. S. pneumoniae CGSP14 [81]), novel clusters from genera with which lantibiotic production has previously been attributed as well as genera not previously associated with lantibiotic production. Within this second phylogroup one finds two subgroups; one consisting of Bacteroidetes-associated LanBs (2A) and a second consisting of Firmicutes-associated LanB's (2B) as well as that from P. ancanthamoeba. Among the Firmicutes-associated LanBs further subclustering is evident. One common branch contains three offshoots; (i) Bacillus/Geobacillus/Enterococcus/Clostridium, (ii) S. pyogenes and (iii) P. acanthamoebae str. Hall's coccus LanBs. The Staphylococcus LanBs and that of mutacin 1140 (those associated with epidermin-like peptides) also form a distinct subgroup as do those encoded with the genomes of S. thermophilus LMG 18311 and S. pneumoniae CGSP14. Curiously the epicidin (S. epidermidis) LanB does not group with any other LanB.
Figure 5.
Cladogram of all the LanB enzymes from clusters encountered during the screen. Also included are dehydratases from some well-known lantibiotics. Green = Actinobacteria; Pink = Firmicutes; Blue = Bacteroidetes; Yellow = Chlamydiae
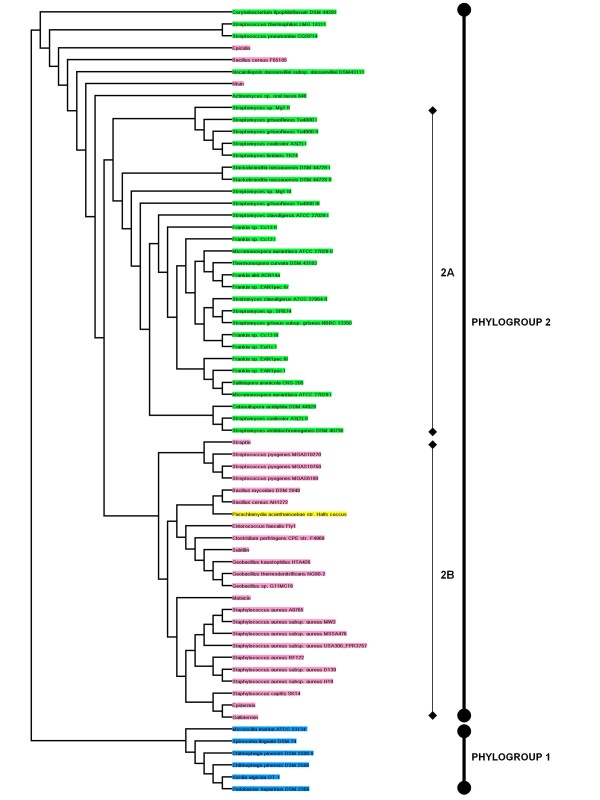
The cladogram of the corresponding lanthionine synthetases (LanCs) is quite similar to that of the dehydratases (Figure 6). All can be positioned into one of two phylogroups (phylogroups 1 and 2). Phylogroup 1 contains six Bacteroidetes-associated LanC's. In contrast phylogroup 2 is large and can be further divided into group 2A, which are Actinobacteria-associated, and the Firmicutes-associated group 2B. Further subgrouping follows the patterns identified from analysis of the LanB cladogram. 8 exceptional LanCs that avoid subgrouping include those associated with C. lipophiloflavum DSM 44291, S. thermophilus LMG 18311/S. pneumoniae CGSP14, epicidin, B. cereus F65185, N. dassonvillei subsp. dassonvillei DSM 43111, nisin-producing lactococci and Actinomyces sp. oral taxon 848.
Figure 6.
Cladogram of all the LanC enzymes from clusters encountered during the screen, as well as some from well-known lantibiotics. Green = Actinobacteria; Pink = Firmicutes; Blue = Bacteroidetes; Yellow = Chlamydiae
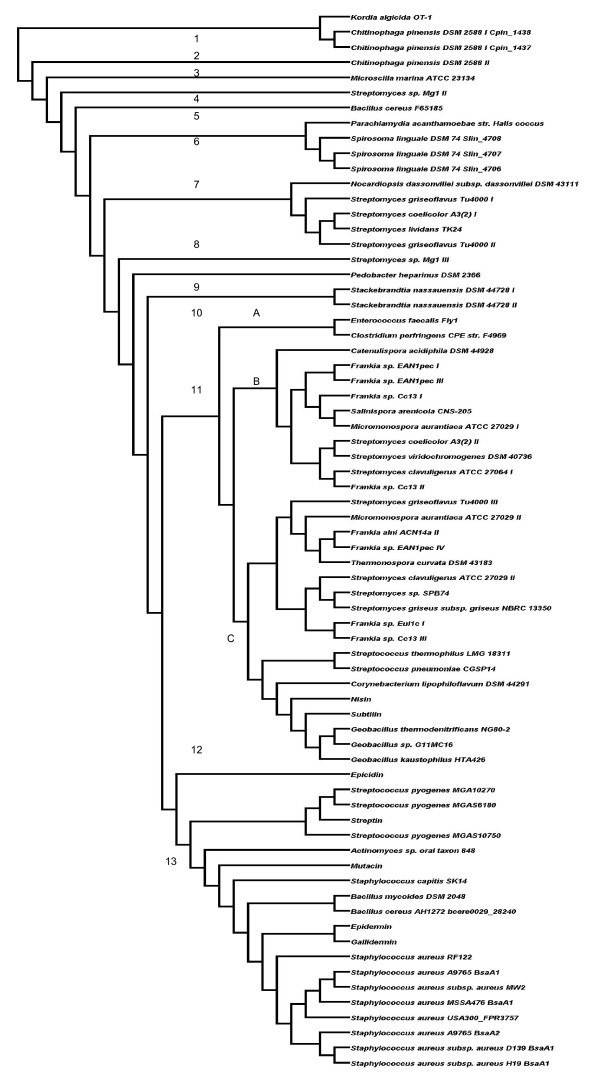
A cladogram of the less highly conserved LanAs revealed 13 major branches, several of which contain only one corresponding LanA (Figure 7). Notably the various phylogroups do not group in a phylum specific manner to the same extent as was evident in LanB and LanC cladograms. The largest phylogroups, i.e. phylogroups 11 and 13, are those containing the nisin-like and epidermin-like peptides, respectively. While phylogroup 13 is, with the exception of the Actinomyces sp. oral taxon 848-associated LanA, composed of Firmicutes-associated LanAs, phylogroup 11 contains LanAs from both Firmicutes and Actinobacteria. This phylogroup contains three subgroups, with subgroup 11A containing LanAs from E. faecalis Fly1 and C. perfringens F4969, 11B contains Actinobacteria-associated LanAs and 11C contains both Actionobacteria- and Firmicutes-associated LanAs. Of the other phylogroups, phylogroups 6 and 7 are largest and contain Chlamydiae/Bacteroidetes- and Actinobacteria-associated LanAs, respectively.
Figure 7.
A cladogram of all the LanA prepeptides identified in this study, as well as a number of previously identified LanAs.
Alignment of LanABC
The availability of a significant number of LanA, B and C protein sequences enabled further in silico analysis to identify conserved motifs and residues. Alignment of 66 LanB proteins highlighted a number conserved motifs which are summarised in Table 5. A YxxR motif (corresponding to residues 80-83 of NisB) is conserved in 89% of the LanB enzymes, while a GxG motif (363-365) is present in 92% of LanBs, with the LanB of C. lipophiloflavum being exceptional by virtue of lacking both glycine residues. A GRF motif (463-465) is fully conserved in 86% of LanBs with the Streptomyces sp. Mg1 III LanB being the only protein to lack this motif. An RxTPFG motif (87-94) is present in 77% of LanBs but is completely absent from the LanBs of Geobacillus sp. G11MC16, Streptomyces sp. Mg1 III, M. aurantiaca and C. lipophiloflavum. A FxxxYG motif (342-347) is present in 82% of LanBs and, although present in only 50% of LanBs, a PxxxRxxNV (501-509) motif is at least partially conserved in many such proteins i.e. 94% contain the proline, 71% contain the NV residues and the least conserved is the arginine with 71% conservation. Elsewhere, there is a RFL motif (585-587) conserved in 51% of LanBs, a RYG motif (826-828) conserved in 85% of LanBs and a HxxxNR motif (961-966) in 70% of the dehydratases. n addition to these, there are multiple highly conserved residues such as aspartates at residues 121, 299, 648 and 843, prolines at 612 and 639 and a leucine, tryptophan and phenylalanine at 97, 616 and 840.
Table 5.
Highly conserved residues shared by 66 cluster-associated (including those from the 49 novel clusters referred to in this paper) lanthionine dehydratases (LanB).
| From Alignment | NisB Position | % Conservation |
|---|---|---|
| Motifs | ||
| YxxR | 80-83 | 89% |
| RxTPFG | 87-94 | 77% |
| FxxxYG | 342-347 | 82% |
| GxG | 363-365 | 92% |
| GRF | 463-465 | 86% |
| PxxxRxxNV | 501-509 | 50% |
| RFL | 585-587 | 51% |
| RYG | 826-828 | 85% |
| HxxxNR | 961-966 | 70% |
| Single Residues | ||
| R | 14 | 86% |
| D | 121; 299; 648; 843 | 86%; 94%; 94%; 85% |
| N | 145 | 86% |
| L | 217 | 97% |
| P | 612; 639 | 100%; 95% |
| E | 975 | 89% |
| W | 616 | 98% |
| F | 840 | 95% |
| V | 352 | 83% |
Residues are numbered according to their position in NisB
Alignment of the LanC protein also revealed several conserved regions (summarised in Table 6). Of these, CHG and WCYG motifs were particularly notable. The CHG motif (corresponding to residues 330-333 of NisC) was found to be conserved in 98% of the LanCs. The cysteine330 and histidine331 residues, which act as ligands to the zinc in the active site of NisC, have been shown to be necessary for enzyme activity [85]. The WCYG motif (283-286) was present in 95% of the aligned enzymes. Within the WCYG motif, tryptophan283 (W) and cysteine284 (C) have been shown to be vital to subtilin and nisin biosynthesis (residue numbers refer to location in NisC) [85,86]. It has previously been shown that although alanine subsititution of tyrosine285 (Y) results in enzyme inactivation, a phenylalanine change is tolerated indicating that the presence of an aromatic ring at this position is of key importance [85]. In the same study, a preceding arginine residue (Arg280), present in 86% of these enzymes, was found not to be essential for enzyme activity. 92% of LanCs also contained a closely located Gly289 residue. The histidine212 of another highly conserved motif, GxAHGxxG (209-216; conserved in 83% of LanCs), together with a conserved aspartic acid141 (91% of LanCs) are thought to be involved in the electrophilic activation of the carbonyl group of dehydroalanine/dehydrobutyrine or in the protonation of the enolate (thiol substrate) [85]. The HG of this latter motif was conserved in 98% of the enzymes (the exception being S. aureus subsp. aureus D139). In addition to these, other motifs of note included LxxG (39-42; conserved in 83% of LanCs), GxxxGxxGxxLxL (377-389; 73%) and YDxxxGxxG (140-148; 67%). Highly conserved single residues include Gly90 (94%) and Tryp258 and Tryp401 (83% and 92% respectively).
Table 6.
Highly conserved residues shared by 66 cluster-associated (including those from 49 novel clusters) lanthionine cyclases. Residues are numbered according to their position in NisC
| Conserved Residues | NisC Position | % Conservation |
|---|---|---|
| Motifs | ||
| LxxG | 39-42 | 83% |
| YDxxxGxxG | 140-148 | 67% |
| GxAHGxxG | 209-216 | 83% |
| WCYG | 283-286 | 95% |
| CHG | 330-332 | 98% |
| GxxxGxxGxxLxL | 377-389 | 73% |
| Single Residues | ||
| G | 90 | 94% |
| W | 258; 401 | 83%; 92% |
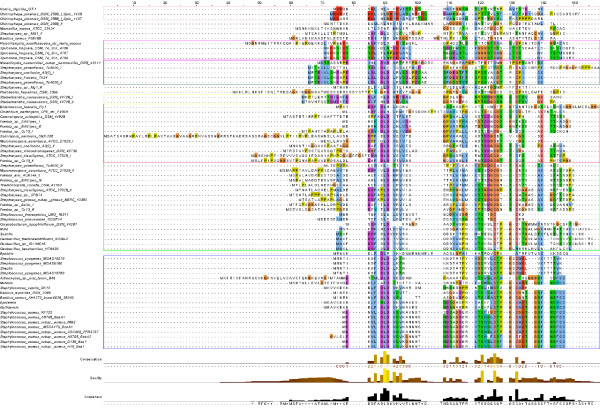
Although LanA peptides are less conserved than their modification enzymes, some motifs were evident (Figure 8). A DLD motif present in the leader region of the almost all phylogroup 13 LanAs is also found in many other LanAs. Indeed, the leucine of this motif is conserved across 93% of the 70 aligned peptides. The only peptides lacking this residue are those from M. marina, B. cereus F65185, N. dassonvillei subsp. dassonvillei and P. heparinus as well as mutacin 1140. The leader regions from many actinobacteria, and especially those from phylogroup 11, are also distinctive as a consequence of the frequent presence of proline residues.
Figure 8.
Alignment of the LanA prepeptides identified in this study, as well as a number of previously identified LanAs.
Within the propeptide, the most highly conserved residues are cysteines corresponding to positions 30 and 34 of the Nisin A prepropeptide [60] which significantly are within the lipid II-binding region of the peptide. These cysteines are each found in 86% of the type 1 LanAs. The Actinobacteria in phylogroups 10 and 11 also share a conserved or partially conserved DGCG motif in the propeptide region. A less highly conserved AC motif which is closer to the C terminus is also evident. In addition to motifs which are conserved across motifs, a large number of motifs which are conserved within phylogroups are evident.
Conclusions
The in silico strategy adopted here resulted in the initial identification of 56 proteins which share 20-30% identity with NisC. Further investigation of novel LanC proteins in turn led to the identification of even more homologs, revealing novel lantibiotic/lantipeptide associated clusters and establishing the existence of subgroups of LanA, B and C proteins. Theoretically, additional homologs could be identified through a continuation of this method but the rate at which new homologs would be identified would begin to level off. The approach taken led to the identification of 49 novel clusters which, prior to this study, had not been the subject of a detailed bioinformatic analysis. While the in silico identification of gene clusters in a strain will not always be confirmed by detection of an associated lantibiotic/lantipeptide, past experience [22-25] suggests that there is likely to be a strong correlation. It is thus anticipated that the peptides produced by these gene clusters will represent a valuable resource, as will be the associated biosynthetic proteins.
This study reveals new details regarding type 1 lantibiotics and their associated clusters. Type 1 lantibiotics have been predominantly associated with the Firmicutes, with the Actinobacteria-produced planosporicin and microbisporicin being notable exceptions. It is thus interesting to find type 1 clusters distributed among the genomes of bacteria representing four different phyla, the Actinobacteria, Firmicutes, Bacteroidetes and Chlamydiae, which have been isolated from a diverse range of habitats including soil, skin, intestines and the deep-sea. Indeed, based on these investigations, it would appear that such clusters are as common among Actinobacteria as they are among Firmicutes, with Streptomyces and Frankia sp. being particularly rich sources. The Actinobacteria clusters are, in general, quite similar, typically encoding a LanA, B, C and a methyltransferase. The role of the methyltransferase is not clear but may serve to protect specific serine and threonine residues from LanB-mediated dehydration. The presence of five clusters within the genomes of five Bacteroidetes, a phylum in which bacteriocin production is purportedly quite rare, is particularly noteworthy. However, the sequencing of additional representatives of this species may well reveal this to be a common feature. The P. acanthamoebae cluster is unusual by virtue of its presence in a representative of the Chlamydiae. Phylogenetic analysis indicates that the LanB and LanC proteins from this strain are closely related to those of several Firmicutes and thus the cluster may originally have been acquired from such a source.
The availability of a much larger collection of LanA, B and C sequences for further in silico analysis is also extremely useful for a number of other reasons. In addition to providing greater certainty with respect to the proposed conservation of particular motifs, it also reveals the existence of a greater number of subgroups of sequences than was previously apparent. This is particularly important with respect to LanAs as alignment of these peptides has previously been employed as a means of subgrouping type 1 lantibiotics [2,6]. Ultimately, the most significant outcome has been the number of new type 1 lantibiotic gene clusters. When one considers that less than 25 type 1 lantibiotics had been identified prior to this study, this represents a major expansion. While the genome sequenced strains themselves can be accessed with a view to purifying the associated peptides and/or utilising the biosynthetic machinery, the information gathered will also encourage researchers to include Actinobacteria and Bacteroiodetes when carrying out wet lab-based screens for novel lantibiotic producers. A combination of this approach and analysis of newly generated bacterial genome sequence data will ensure that many more lantibiotics and lantipetides will soon be discovered which are associated with unusual microorganisms and a wide variety of environments.
Methods
Screening of genomic databases
Using the nisin modification enzyme NisC (GenBank accession number CAA79470) as a driver sequence, all fully sequenced genomic sequences (approx. 1178 at time of study; Dec 2009) were mined for homologs using Genomic-BLAST (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi). BLASTs were carried out with default parameters; criteria for homolog detection were a threshold of 1e-7 and greater than 20% identity.
Bioinformatic anlaysis of lanC-containing gene clusters
In cases where novel lanC-like genes encoding enzymes were identified, the arrangement of adjacent genes was visualised using the genome viewer on NCBI, and individual orfs were subjected to BLAST analysis to identify those potentially involved in lantibiotic production or immunity. The predicted LanA, LanB and LanC proteins from these operons were each in turn used for further in silico screens to determine their similarities to corresponding proteins associated with known lantibiotics and to identify additional novel clusters. In instances where a LanC- and LanB-, but not a LanA-, encoding gene were annotated, intergenic regions were inspected following translation by the Seqbuilder program of the DNASTAR Lasergene 8 software package to investigate the presence of potentially unidentified lanA genes. The tblastn program was then used to search all sequenced DNA for related peptides.
Phylogenetic analysis
Protein alignments were generated by MUSCLE [87]. Sequence alignment were viewed and edited for publication with Jalview alignment editor [88]. These alignments were used to establish phylogenetic trees in Phylip [89] which were subsequently visualised using the Dendroscope package [90].
Authors' contributions
PDC conceived the study and designed the project. OOS participated in the design and coordination of the project. AJM performed the screening of the databases and the bioinformatic and phylogenetic analysis. AJM and PDC wrote the manuscript. OOS, PRR and CH contributed in the preparation of the manuscript. All authors have read and approved the final manuscript.
Contributor Information
Alan J Marsh, Email: alan.marsh@teagasc.ie.
Orla O'Sullivan, Email: orla.osullivan@teagasc.ie.
R Paul Ross, Email: paul.ross@teagasc.ie.
Paul D Cotter, Email: paul.cotter@teagasc.ie.
Colin Hill, Email: c.hill@ucc.ie.
Acknowledgements
This work was supported by the Science Foundation of Ireland funded Centre for Science, Engineering and Technology (SFI-CSET): the Alimentary Pharmabiotic Centre (APC).
References
- Cotter PD, Hill C, Ross RP. Bacteriocins: Developing innate immunity for food. Nat Rev Microbiol. 2005;3(10):777–788. doi: 10.1038/nrmicro1273. [DOI] [PubMed] [Google Scholar]
- Rea M, Ross RP, Cotter PD, Hill C. In Prokaryotic antimicrobial peptides. Springer Press; 2010. Classification of bacteriocins from gram positive bacteria. in press . [Google Scholar]
- Willey JM, van der Donk WA. Lantibiotics: Peptides of diverse structure and function. Annual Review of Microbiology. 2007;61:477–501. doi: 10.1146/annurev.micro.61.080706.093501. [DOI] [PubMed] [Google Scholar]
- Kodani S, Hudson ME, Durrant MC, Buttner MJ, Nodwell JR, Willey JM. The SapB morphogen is a lantibiotic-like peptide derived from the product of the developmental gene ramS in Streptomyces coelicolor. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(31):11448–11453. doi: 10.1073/pnas.0404220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Li B, Claesen J, Shi Y, Bibb M, Van der Donk W. Discovery of unique lanthionine synthetases reveals new mechanistic and evolutionary insights. PloS Biology. 2010;8(3) doi: 10.1371/journal.pbio.1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper C, Cotter P, Ross R, Hill C. Discovery of medically significant lantibiotics. Current Drug Discovery Technologies. 2009;6(1):1–18. doi: 10.2174/157016309787581075. [DOI] [PubMed] [Google Scholar]
- Delves-Broughton J. Nisin as a food preservative. Food Australia. 2005;57(12):525–527. [Google Scholar]
- Kupke T, Stevanovic S, Sahl HG, Gotz F. Purification and Characterization of Epid, a Flavoprotein Involved in the Biosynthesis of the Lantibiotic Epidermin. Journal of Bacteriology. 1992;174(16):5354–5361. doi: 10.1128/jb.174.16.5354-5361.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierbaum G, Sahl HG. Lantibiotics: Mode of Action, Biosynthesis and Bioengineering. Current Pharmaceutical Biotechnology. 2009;10(1):2–18. doi: 10.2174/138920109787048616. [DOI] [PubMed] [Google Scholar]
- Severina E, Severin A, Tomasz A. Antibacterial efficacy of nisin against multidrug-resistant Grampositive pathogens. J Antimicrob Chemother. 1998;41(3):341–347. doi: 10.1093/jac/41.3.341. [DOI] [PubMed] [Google Scholar]
- Parrot M, Charest M, Lavoie MC. Production of Mutacin-Like Substances by Streptococcus-Mutans. Canadian Journal of Microbiology. 1989;35(3):366–372. doi: 10.1139/m89-056. [DOI] [PubMed] [Google Scholar]
- Castiglione F, Cavaletti L, Losi D, Lazzarini A, Carrano L, Feroggio M, Ciciliato I, Corti E, Candiani G, Marinelli F. et al. A novel lantibiotic acting on bacterial cell wall synthesis produced by the uncommon actinomycete Planomonospora sp. Biochemistry. 2007;46(20):5884–5895. doi: 10.1021/bi700131x. [DOI] [PubMed] [Google Scholar]
- Fontana MBC, de Bastos MDF, Brandelli A. Bacteriocins Pep5 and epidermin inhibit Staphylococcus epidermidis adhesion to catheters. Current Microbiology. 2006;52(5):350–353. doi: 10.1007/s00284-005-0152-5. [DOI] [PubMed] [Google Scholar]
- Kellner R, Jung G, Horner T, Zahner H, Schnell N, Entian KD, Gotz F. Gallidermin - a New Lanthionine-Containing Polypeptide Antibiotic. European Journal of Biochemistry. 1988;177(1):53–59. doi: 10.1111/j.1432-1033.1988.tb14344.x. [DOI] [PubMed] [Google Scholar]
- Cleveland J, Montville TJ, Nes IF, Chikindas ML. Bacteriocins: safe, natural antimicrobials for food preservation. International Journal of Food Microbiology. 2001;71(1):1–20. doi: 10.1016/S0168-1605(01)00560-8. [DOI] [PubMed] [Google Scholar]
- Wescombe PA, Upton M, Dierksen KP, Ragland NL, Sivabalan S, Wirawan RE, Inglis MA, Moore CJ, Walker GV, Chilcott CN. et al. Production of the lantibiotic salivaricin A and its variants by oral streptococci and use of a specific induction assay to detect their presence in human saliva. Appl Environ Microbiol. 2006;72(2):1459–1466. doi: 10.1128/AEM.72.2.1459-1466.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wescombe PA, Heng NCK, Burton JP, Chilcott CN, Tagg JR. Streptococcal bacteriocins and the case for Streptococcus salivarius as model oral probiotics. Future Microbiology. 2009;4(7):819–835. doi: 10.2217/fmb.09.61. [DOI] [PubMed] [Google Scholar]
- Dabard J, Bridonneau C, Phillipe C, Anglade P, Molle D, Nardi M, Ladire M, Girardin H, Marcille F, Gomez A. et al. Ruminococcin A, a new lantibiotic produced by a Ruminococcus gnavus strain isolated from human feces. Appl Environ Microbiol. 2001;67(9):4111–4118. doi: 10.1128/AEM.67.9.4111-4118.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MP, Rea MC, Hill C, Ross RP. An application in cheddar cheese manufacture for a strain of Lactococcus lactis producing a novel broad-spectrum bacteriocin, lacticin 3147. Appl Environ Microbiol. 1996;62(2):612–619. doi: 10.1128/aem.62.2.612-619.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beasley SS, Saris PEJ. Nisin-producing Lactococcus lactis strains isolated from human milk. Appl Environ Microbiol. 2004;70(8):5051–5053. doi: 10.1128/AEM.70.8.5051-5053.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman JD, Novak J, Sagura E, Gutierrez JA, Brooks TA, Crowley PJ, Hess M, Azizi A, Leung KP, Cvitkovitch D. et al. Genetic and biochemical analysis of mutacin 1140, a lantibiotic from Streptococcus mutans. Infection and Immunity. 1998;66(6):2743–2749. doi: 10.1128/iai.66.6.2743-2749.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly KM, Upton M, Sandiford SK, Draper LA, Wescombe PA, Jack RW, O'Connor PM, Rossney A, Gotz F, Hill C. et al. Production of the Bsa Lantibiotic by Community-Acquired Staphylococcus aureus Strains. Journal of Bacteriology. 2010;192(4):1131–1142. doi: 10.1128/JB.01375-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClerren AL, Cooper LE, Quan C, Thomas PM, Kelleher NL, van der Donk WA. Discovery and in vitro biosynthesis of haloduracin, a two-component lantibiotic. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(46):17243–17248. doi: 10.1073/pnas.0606088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton EM, Cotter PD, Hill C, Ross RP. Identification of a novel two-peptide lantibiotic, Haloduracin, produced by the alkaliphile Bacillus halodurans C-125. FEMS Microbiol Lett. 2007;267(1):64–71. doi: 10.1111/j.1574-6968.2006.00539.x. [DOI] [PubMed] [Google Scholar]
- Begley M, Cotter PD, Hill C, Ross RP. Identification of a Novel Two-Peptide Lantibiotic, Lichenicidin, following Rational Genome Mining for LanM Proteins. Appl Environ Microbiol. 2009;75(17):5451–5460. doi: 10.1128/AEM.00730-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dischinger J, Josten M, Szekat C, Sahl HG, Bierbaum G. Production of the Novel Two-Peptide Lantibiotic Lichenicidin by Bacillus licheniformis DSM 13. Plos One. 2009;4(8) doi: 10.1371/journal.pone.0006788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Sher D, Kelly L, Shi Y, Huang K, Knerr P, Joewono I, Rusch D, Chisholm S, van der Donk W. Catalytic promiscuity in the biosynthesis of cyclic peptide secondary metabolites in planktonic marine cyanobacteria. Proceedings of the National Academy of Sciences of the United States of America. 2010. [DOI] [PMC free article] [PubMed]
- de Jong A, van Hijum SAFT, Bijlsma JJE, Kok J, Kuipers OP. BAGEL: a web-based bacteriocin genome mining tool. Nucleic Acids Research. 2006;34:W273–W279. doi: 10.1093/nar/gkl237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong A, van Heel A, Kok J, Kuipers O. BAGEL2: mining for bacteriocins in genomic data. Nucleic Acids Research. 2010;38:W647–W651. doi: 10.1093/nar/gkq365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammami R, Zouhir A, Ben Hamida J, Fliss I. BACTIBASE: a new web-accessible database for bacteriocin characterization. Bmc Microbiology. 2007;7 doi: 10.1186/1471-2180-7-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammami R, Zouhir A, Le Lay C, Ben Hamida J, Fliss I. BACTIBASE second release: a database and tool platform for bacteriocin characterization. Bmc Microbiology. 2010;10 doi: 10.1186/1471-2180-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normand P, Lapierre P, Tisa LS, Gogarten JP, Alloisio N, Bagnarol E, Bassi CA, Berry AM, Bickhart DM, Choisne N. et al. Genome characteristics of facultatively symbiotic Frankia sp strains reflect host range and host plant biogeography. Genome Research. 2007;17(1):7–15. doi: 10.1101/gr.5798407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SY, Anzai Y, Kinoshita K, Kato F, Sherman DH. Functional Analysis of MycE and MycF, Two O-Methyltransferases Involved in the Biosynthesis of Mycinamicin Macrolide Antibiotics. Chembiochem. 2009;10(8):1297–1301. doi: 10.1002/cbic.200900088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada S, Anzai Y, Li SY, Kinoshita K, Sherman DH, Kato F. Gene targeting for O-methyltransferase genes, mycE and mycF, on the chromosome of Micromonospora griseorubida producing mycinamicin with a disruption cassette containing the bacteriophage C31 attB attachment site. FEMS Microbiol Lett. 2010;304(2):148–156. doi: 10.1111/j.1574-6968.2010.01899.x. [DOI] [PubMed] [Google Scholar]
- Zubieta C, He XZ, Dixon RA, Noel JP. Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases. Nature Structural Biology. 2001;8(3):271–279. doi: 10.1038/85029. [DOI] [PubMed] [Google Scholar]
- Penn K, Jenkins C, Nett M, Udwary DW, Gontang EA, McGlinchey RP, Foster B, Lapidus A, Podell S, Allen EE. et al. Genomic islands link secondary metabolism to functional adaptation in marine Actinobacteria. Isme Journal. 2009;3(10):1193–1203. doi: 10.1038/ismej.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdy J. Bioactive microbial metabolites - A personal view. Journal of Antibiotics. 2005;58(1):1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- Labeda DP, Kroppenstedt RM. Stackebrandtia nassauensis gen. nov., sp nov and emended description of the family Glycomycetaceae. International Journal of Systematic and Evolutionary Microbiology. 2005;55:1687–1691. doi: 10.1099/ijs.0.63496-0. [DOI] [PubMed] [Google Scholar]
- Hodgson DA. Primary metabolism and its control in streptomycetes: A most unusual group of bacteria. Advances in Microbial Physiology, Vol 42. 2000;42:47–238. doi: 10.1016/s0065-2911(00)42003-5. full_text. [DOI] [PubMed] [Google Scholar]
- Watve MG, Tickoo R, Jog MM, Bhole BD. How many antibiotics are produced by the genus Streptomyces? Archives of Microbiology. 2001;176(5):386–390. doi: 10.1007/s002030100345. [DOI] [PubMed] [Google Scholar]
- Kido Y, Hamakado T, Yoshida T, Anno M, Motoki Y, Wakamiya T, Shiba T. Isolation and Characterization of Ancovenin, a New Inhibitor of Angiotensin-I Converting Enzyme, Produced by Actinomycetes. Journal of Antibiotics. 1983;36(10):1295–1299. doi: 10.7164/antibiotics.36.1295. [DOI] [PubMed] [Google Scholar]
- Widdick DA, Dodd HM, Barraille P, White J, Stein TH, Chater KF, Gasson MJ, Bibb MJ. Cloning and engineering of the cinnamycin biosynthetic gene cluster from Streptomyces cinnamoneus cinnamoneus DSM 40005. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(7):4316–4321. doi: 10.1073/pnas.0230516100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi Y, Ishikawa J, Hara H, Suzuki H, Ikenoya M, Ikeda H, Yamashita A, Hattori M, Horinouchi S. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. Journal of Bacteriology. 2008;190(11):4050–4060. doi: 10.1128/JB.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood DA. Forty years of genetics with Streptomyces: from in vivo through in vitro to in silico. Microbiology-Sgm. 1999;145:2183–2202. doi: 10.1099/00221287-145-9-2183. [DOI] [PubMed] [Google Scholar]
- Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D. et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417(6885):141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- Nishio C, Komura S, Kurahashi K. Peptide Antibiotic Subtilin Is Synthesized Via Precursor Proteins. Biochemical and Biophysical Research Communications. 1983;116(2):751–758. doi: 10.1016/0006-291X(83)90588-0. [DOI] [PubMed] [Google Scholar]
- Stein T, Borchert S, Conrad B, Feesche J, Hofemeister B, Hofemeister J, Entian KD. Two different lantibiotic-like peptides originate from the ericin gene cluster of Bacillus subtilis A1/3. Journal of Bacteriology. 2002;184(6):1703–1711. doi: 10.1128/JB.184.6.1703-1711.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown LCW, Acker MG, Clardy J, Walsh CT, Fischbach MA. Thirteen posttranslational modifications convert a 14-residue peptide into the antibiotic thiocillin. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(8):2549–2553. doi: 10.1073/pnas.0900008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N, Gautam N. Antibacterial activity and characterization of bacteriocin of Bacillus mycoides isolated from whey. Indian Journal of Biotechnology. 2008;7(1):117–121. [Google Scholar]
- Dineen SS, Bradshaw M, Johnson EA. Cloning, nucleotide sequence, and expression of the gene encoding the bacteriocin boticin B from Clostridium botulinum strain 213B. Appl Environ Microbiol. 2000;66(12):5480–5483. doi: 10.1128/AEM.66.12.5480-5483.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li AW, Verpoorte JA, Lewis RG, Mahony DE. Characterization of Bacteriocin 28 Produced by Clostridium-Perfringens. Canadian Journal of Microbiology. 1982;28(7):860–873. doi: 10.1139/m82-128. [DOI] [PubMed] [Google Scholar]
- Nieves BM, Castillo FJ. Properties of a Bacteriocin Produced by Clostridium-Bifermentans 18137. Acta Cientifica Venezolana. 1982;33(1):38–43. [PubMed] [Google Scholar]
- Narayan KG. Food Borne Infection with Clostridium-Perfringens Type-A. International Journal of Zoonoses. 1982;9(1):12–32. [PubMed] [Google Scholar]
- Pearson RD, Valenti WM, Steigbigel RT. Clostridium-Perfringens Wound-Infection Associated with Elastic Bandages. Jama-Journal of the American Medical Association. 1980;244(10):1128–1130. doi: 10.1001/jama.244.10.1128. [DOI] [PubMed] [Google Scholar]
- Franz CMAP, van Belkum MJ, Holzapfel WH, Abriouel H, Galvez A. Diversity of enterococcal bacteriocins and their grouping in a new classification scheme. Fems Microbiology Reviews. 2007;31(3):293–310. doi: 10.1111/j.1574-6976.2007.00064.x. [DOI] [PubMed] [Google Scholar]
- Booth MC, Bogie CP, Sahl HG, Siezen RJ, Hatter KL, Gilmore MS. Structural analysis and proteolytic activation of Enterococcus faecalis cytolysin, a novel lantibiotic. Molecular Microbiology. 1996;21(6):1175–1184. doi: 10.1046/j.1365-2958.1996.831449.x. [DOI] [PubMed] [Google Scholar]
- Nazina TN, Tourova TP, Poltaraus AB, Novikova EV, Grigoryan AA, Ivanova AE, Lysenko AM, Petrunyaka VV, Osipov GA, Belyaev SS. et al. Taxonomic study of aerobic thermophilic bacilli: descriptions of Geobacillus subterraneus gen. nov., sp nov and Geobacillus uzenensis sp nov from petroleum reservoirs and transfer of Bacillus stearothermophilus Bacillus thermocatenulatus, Bacillus thermoleovorans, Bacillus kaustophilus, Bacillus thermoglucosidasius and Bacillus thermodenitrificans to Geobacillus as the new combinations G-stearothermophilus, G-thermocatenulatus, G-thermoleovorans, G-kaustophilus, G-thermoglucosidasius and G-thermodenitrificans. International Journal of Systematic and Evolutionary Microbiology. 2001;51:433–446. doi: 10.1099/00207713-51-2-433. [DOI] [PubMed] [Google Scholar]
- Pokusaeva K, Kuisiene N, Jasinskyte D, Rutiene K, Saleikiene J, Chitavichius D. Novel bacteriocins produced by Geobacillus stearothermophilus. Central European Journal of Biology. 2009;4(2):196–203. doi: 10.2478/s11535-009-0009-1. [DOI] [Google Scholar]
- Novotny JF, Perry JJ. Characterization of Bacteriocins from 2 Strains of Bacillus-Thermoleovorans, a Thermophilic Hydrocarbon-Utilizing Species. Appl Environ Microbiol. 1992;58(8):2393–2396. doi: 10.1128/aem.58.8.2393-2396.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu STD, Breukink E, Tischenko E, Lutters MAG, de Kruijff B, Kaptein R, Bonvin AMJJ, van Nuland NAJ. The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nature Structural & Molecular Biology. 2004;11(10):963–967. doi: 10.1038/nsmb830. [DOI] [PubMed] [Google Scholar]
- Feng L, Wang W, Cheng JS, Ren Y, Zhao G, Gao CX, Tang Y, Liu XQ, Han WQ, Peng X. et al. Genome and proteome of long-chain alkane degrading Geobacillus thermodenitrificans NG80-2 isolated from a deep-subsurface oil reservoir. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(13):5602–5607. doi: 10.1073/pnas.0609650104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allgaier H, Jung G, Werner RG, Schneider U, Zahner H. Elucidation of the Structure of Epidermin, a Ribosomally Synthesized, Tetracyclic Heterodetic Polypeptide Antibiotic. Angewandte Chemie-International Edition in English. 1985;24(12):1051–1053. doi: 10.1002/anie.198510511. [DOI] [Google Scholar]
- Kaletta C, Entian KD, Kellner R, Jung G, Reis M, Sahl HG. Pep5, a New Lantibiotic - Structural Gene Isolation and Prepeptide Sequence. Archives of Microbiology. 1989;152(1):16–19. doi: 10.1007/BF00447005. [DOI] [PubMed] [Google Scholar]
- Heidrich C, Pag U, Josten M, Metzger J, Jack RW, Bierbaum G, Jung G, Sahl HG. Isolation, characterization, and heterologous expression of the novel lantibiotic epicidin 280 and analysis of its biosynthetic gene cluster. Appl Environ Microbiol. 1998;64(9):3140–3146. doi: 10.1128/aem.64.9.3140-3146.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandekamp M, Vandenhooven HW, Konings RNH, Bierbaum G, Sahl HG, Kuipers OP, Siezen RJ, Devos WM, Hilbers CW, Vandeven FJM. Elucidation of the Primary Structure of the Lantibiotic Epilancin K7 from Staphylococcus-Epidermidis K7 - Cloning and Characterization of the Epilancin-K7-Encoding Gene and Nmr Analysis of Mature Epilancin K7. European Journal of Biochemistry. 1995;230(2):587–600. doi: 10.1111/j.1432-1033.1995.tb20600.x. [DOI] [PubMed] [Google Scholar]
- Scott JC, Sahl HG, Carne A, Tagg JR. Lantibiotic-Mediated Anti-Lactobacillus Activity of a Vaginal Staphylococcus-Aureus Isolate. FEMS Microbiol Lett. 1992;93(1):97–102. doi: 10.1111/j.1574-6968.1992.tb05047.x. [DOI] [PubMed] [Google Scholar]
- Geis A, Singh J, Teuber M. Potential of Lactic Streptococci to Produce Bacteriocin. Appl Environ Microbiol. 1983;45(1):205–211. doi: 10.1128/aem.45.1.205-211.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nes IF, Diep DB, Holo H. Bacteriocin diversity in Streptococcus and Enterococcus. Journal of Bacteriology. 2007;189(4):1189–1198. doi: 10.1128/JB.01254-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wescombe PA, Tagg JR. Purification and characterization of streptin, a type A1 lantibiotic produced by Streptococcus pyogenes. Appl Environ Microbiol. 2003;69(5):2737–2747. doi: 10.1128/AEM.69.5.2737-2747.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MotaMeira M, Lacroix C, LaPointe G, Lavoie MC. Purification and structure of mutacin B-Ny266: A new lantibiotic produced by Streptococcus mutans. Febs Letters. 1997;410(2-3):275–279. doi: 10.1016/S0014-5793(97)00425-0. [DOI] [PubMed] [Google Scholar]
- Qi FX, Chen P, Caufield PW. Purification of mutacin III from group III Streptococcus mutans UA787 and genetic analyses of mutacin III biosynthesis genes. Appl Environ Microbiol. 1999;65(9):3880–3887. doi: 10.1128/aem.65.9.3880-3887.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi FX, Chen P, Caufield PW. Purification and biochemical characterization of mutacin I from the group I strain of Streptococcus mutans, CH43, and genetic analysis of mutacin I biosynthesis genes. Appl Environ Microbiol. 2000;66(8):3221–3229. doi: 10.1128/AEM.66.8.3221-3229.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirawan RE, Kleese NA, Jack RW, Tagg JR. Molecular and genetic characterization of a novel nisin variant produced by Streptococcus uberis. Appl Environ Microbiol. 2006;72(2):1148–1156. doi: 10.1128/AEM.72.2.1148-1156.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes WL, Ferretti JJ, Tagg JR. Cloning of the Gene Encoding Streptococcin a-Ff22, a Novel Lantibiotic Produced by Streptococcus-Pyogenes, and Determination of Its Nucleotide-Sequence. Appl Environ Microbiol. 1993;59(6):1969–1971. doi: 10.1128/aem.59.6.1969-1971.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres SB, Richter EW, Nagiec MJ, Sumby P, Porcella SF, Deleo FR, Musser JM. Molecular genetic anatomy of inter- and intraserotype variation in the human bacterial pathogen group A Streptococcus. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(18):7059–7064. doi: 10.1073/pnas.0510279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villani F, Pepe O, Mauriello G, Salzano G, Moschetti G, Coppola S. Antilisterial Activity of Thermophilin-347, a Bacteriocin Produced by Streptococcus-Thermophilus. International Journal of Food Microbiology. 1995;25(2):179–190. doi: 10.1016/0168-1605(94)00153-W. [DOI] [PubMed] [Google Scholar]
- Ward DJ, Somkuti GA. Characterization of a Bacteriocin Produced by Streptococcus-Thermophilus St134. Applied Microbiology and Biotechnology. 1995;43(2):330–335. doi: 10.1007/BF00172834. [DOI] [PubMed] [Google Scholar]
- Aktypis A, Kalantzopoulos G. Purification and characterization of thermophilin ST-1, a novel bacteriocin produced by Streptococcus thermophilus ACA-DC 0001. Lait. 2003;83(5):365–378. doi: 10.1051/lait:2003024. [DOI] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Renault P, Sorokin A, Ehrlich SD, Kulakauskas S, Lapidus A, Goltsman E, Mazur M, Pusch GD. et al. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nature Biotechnology. 2004;22(12):1554–1558. doi: 10.1038/nbt1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MJ, Siezen RJ, Nauta A. In Silico Prediction of Horizontal Gene Transfer Events in Lactobacillus bulgaricus and Streptococcus thermophilus Reveals Protocooperation in Yogurt Manufacturing. Appl Environ Microbiol. 2009;75(12):4120–4129. doi: 10.1128/AEM.02898-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, Tang P, Hsu MH, Cui P, Hu S, Yu J, Chiu CH. Genome evolution driven by host adaptations results in a more virulent and antimicrobial-resistant Streptococcus pneumoniae serotype 14. Bmc Genomics. 2009. p. 10. [DOI] [PMC free article] [PubMed]
- Qin JJ, Li RQ, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–U70. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobles ER. Bacteroides infections. Annals of Surgery. 1973;177(5) [PMC free article] [PubMed] [Google Scholar]
- Sangkhobol V, Skerman VBD. Chitinophaga, A New Genus of Chitinolytic Myxobacteria. International Journal of Systematic Bacteriology. 1981;31(3):285–293. doi: 10.1099/00207713-31-3-285. [DOI] [Google Scholar]
- Li B, van der Donk WA. Identification of essential catalytic residues of the cyclase NisC involved in the biosynthesis of nisin. Journal of Biological Chemistry. 2007;282(29):21169–21175. doi: 10.1074/jbc.M701802200. [DOI] [PubMed] [Google Scholar]
- Helfrich M, Entian KD, Stein T. Structure-function relationships of the lanthionine cyclase SpaC involved in biosynthesis of the Bacillus subtilis peptide antibiotic subtilin. Biochemistry. 2007;46(11):3224–3233. doi: 10.1021/bi062124f. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:1–19. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP - Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R. Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007. p. 8. [DOI] [PMC free article] [PubMed]