Abstract
The dNTP supply system genes RRM1, DCTD, TYMS, TK1 and DCK balance dNTP pools to avoid incorrect insertions of bases (i.e. DNA mismatches) and the DNA mismatch repair system genes MLH1 and MSH2 are involved in removing such mismatches. The objective of this study is to explore the possibility of interactions between these two systems, since greater mismatch production rates are expected to be more detrimental in cells that also have compromised mismatch removal rates. This conjecture was explored here specifically with respect to the development of breast cancer. More than 2400 breast cancer cases and controls are included in the Cancer Genetic Markers of Susceptibility (CGEMS) single nucleotide polymorphism (SNP) dataset. For each of these individuals, a total of 99 SNPs (69 dNTP supply SNPs and 30 mismatch repair SNPs) and 2070 SNP-SNP interactions between these two groups were evaluated for their effect on breast cancer using logistic regression to compute odds ratios (ORs) and corresponding 95% confidence intervals (CIs). Of these, 12 SNPs had found statistically significant associations with breast cancer individually (Four of them to decrease risk and eight of them to increase risk) and 697 of 2070 two-way interactions were significant associated with the risk of breast cancer. Thus, our study suggests that mismatches contribute to the formation of breast cancer.
Keywords: SNP, SNP interactions, dNTP pool imbalance, DNA mismatch repair, CGEMS, Breast Cancer
I. INTRODUCTION
According to the World Health Organization (WHO), cancer accounted for approximately 7.9 million deaths (13 percent of all deaths) in the world in 2007 [1]. Breast cancer is the most frequent malignant tumor in women, with 548,000 deaths/year reported around the world [1].
Single nucleotide polymorphisms (SNPs) account for 80% of the non-identical DNA sequence between individuals. Current evidence shows that SNPs have considerable effects on the function of proteins, regardless of whether the SNP lies within the coding sequence or not [2]. Although the effect of single SNPs may be small, collectively, and through SNP-SNP interactions, their contribution to breast [3] and other cancers [4],[5],[6] could be substantial. The number of pair-wise SNP-SNP interactions is too large to explore all of the genes of a genome wide association study, so knowledge from basic science must be used to focus studies on specific interactions.
Deoxynucleoside triphosphates (dNTPs) are the fundamental building blocks used in DNA synthesis. Genetic defects and drug treatments have been shown to result in fluctuations of the concentrations of dNTPs and as a result, increased replication error rates [7],[8]. There are also strong positive correlations between genes with mutations in mismatch repair (MMR) and the gene mutation rate [9]. After mismatch editing by specific DNA polymerase subunits, the DNA mismatch repair system is the cell's main defense mechanism for correcting mismatches. Therefore, the dNTP supply system and MMR are expected to interact as factors of mismatch mediated gene mutations.
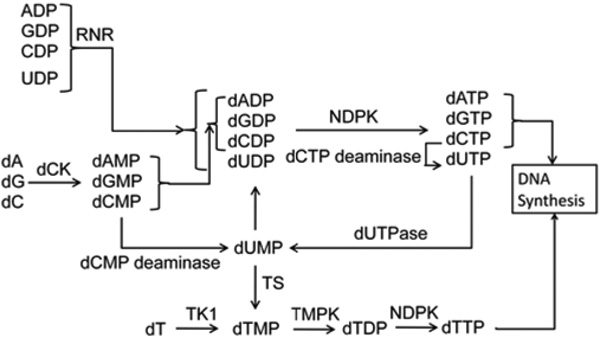
ribonucleotide reductase M1 (RRM1) is the gene that encodes the large subunit of ribonucleotide reductase (RNR), an enzyme that catalyzes the synthesis of dNTPs by converting ribonucleotide diphosphates (NDPs) into 2’-deoxyribonucleotide diphophosphates (dNDPs) which are then readily further phosphorylated into dNTPs. Other dNTP supply genes include thymidylate synthetase (TYMS), thymidine kinase 1 (TK1), dCMP deaminase (DCTD) and deoxycytidine kinase (DCK) which encode thymidylate synthetase (TS), thymidine kinase (TK), dCMP deaminase (DCTD) and deoxycytidine kinase (dCK), respectively. Collectively, these enzymes are responsible for supplying dNTPs at rates equal to those demanded by DNA synthesis. The relationship of dNTP supply enzymes is shown in Figure 1.
Figure 1.
dNTP supply enzymes system; dA : Deoxyadenosine, dG : Deoxyguanosine, dC : Deoxycytidine, dT : Thymidine, NDPK : Nucleoside diphosphokinase and TMPK : dTMP kinase. Other acronyms are as in the text.
The MMR system is responsible for detecting and removing DNA mismatches. Detection occurs by recognizing distortions in the DNA helix and removal ensues by removal of a stretch of the more recently synthesized DNA strand which encompasses the incorrectly inserted bases [9]. The MMR genes examined here are MLH1 and MSH2.
A normal cell with imbalanced dNTP pools and without a proper defense mechanism is expected to have a high possibility of mutating to form a tumor cell. Thus, using SNP data from the Cancer Genetic Markers of Susceptibility Study (CGEMS), we explore here interactions between SNPs in enzymes that supply and balance dNTPs and SNPs in mismatch DNA repair enzymes, with respect to breast cancer risk.
II. MATERIALS AND METHODS
A. CGEMS Data
The Cancer Genetics Markers of Susceptibility (CGEMS) case-control dataset was used [10],[11]. Briefly, of 121,700 women enrolled in a longitudinal study called Nurses’ Health Study (NHS) in 1976, this data was derived from 32,826 participants who provided blood samples between 1989 and 1990 and who were followed until May 2004 [10],[11]. Of these, 1,231 patients with invasive breast cancer and 1,203 controls matched on age, use of postmenopausal hormones, ethnicity and menopausal status, had their peripheral blood lymphocytes assayed for 555,352 SNP genotypes using the Illumina HumanHap550 platform. Of these, 528,173 SNPs remained after removal of low HapMap minor allele frequencies (MAF) (<1%) [10],[11]. All participates were Caucasians and menopausal when blood was drawn. The amount of missing data in each SNP file is less than 1 percent, so the analysis should not be affected by missing values.
B. Selection of SNPs
A total of 99 CGEMS SNPs of the dNTP supply (RRM1, DCTD, TYMS, TK1 and DCK) and MMR (MLH1and MSH2) target gene sets were selected for our analysis (Table 1). To investigate interaction effects, pairs of SNPs from each of these two gene groups, totaling 2070 (= 69 × 30) two-way interactions, were then further studied.
Table 1.
the number of SNPs related with target genes:
C. Statistical analyses
Single SNP breast cancer susceptibility risks have already been individually analyzed by CGEMS using unadjusted and adjusted score tests involving 3-by-2 contingency tables of genotypes by phenotypes (diagnosed with breast cancer or not before May 2004; the adjusted test was adjusted for age and hormone replacement therapy status through stratification of the tables before formulating chi-square test p-values). The logistic regression was performed because some numbers in the interaction tables were less than 5. The Odds ratio (OR) and corresponding 95% confidence interval (95% CI) were obtained for each individual SNP with three possible genotypes to measure the association with/without breast cancer. Probability of having breast cancer in individual SNP dataset is calculated by Equation (1).
| (1) |
where P* is the probability of observation in the case group. P* at each individual SNP situation is usually not the same due to different missing SNP dataset. For example, 1231 cases and 1203 controls were observed in SNP1 dataset. P* is equal to .5058 (= 1231/ (1203+1231)) and then the regression model has a fixed intercept term: β0 at 0.023.
The logistic regression model is as follows,
where P represents the probability of having invasive breast cancer. Xi (i=1, 2, 3) is 1 if the observation carries the ith genotype and zero otherwise, β0 is a fixed number and βi (i=1, 2, 3) is a measure of the potency of the genotype.
Thus,
When all variables are zeros, the probability is .5058.
SNP-SNP interaction analysis
Each SNP has three possible genotypes and this means that interaction between two SNPs yields a total of 9 possible independently different genotype combinations, were modeled by a logistic regression as follows,
where P is the probability of invasive breast cancer, Xi is ith interaction genotype, β0 is the fixed intercept and βi represents a corresponding coefficient. A significant genotype (Xi) means that a person with this genotype will have a significant either higher or lower odds of breast cancer.
A stepwise algorithm (using glm and step in R) was performed to select the best model. The rule of the stepwise algorithm is to select the model with smallest Akaike's information Criteria (AIC). The selection of variables involves adding and removing variables until the AIC is lower than the value in a previous model. All calculations used the raw CGEMS data (http://cgems.cancer.gov/data).
III. RESULTS
ORs and their corresponding 95% CIs for all 99 individual SNPs were estimated by logistic regression. Among all SNPs investigated, 12 SNP genotypes showed significant associations with breast cancer based on crude P-values less than .05 (Table 2) and 6 of them agree with the results from CGEMS. According to CGEMS, among these 12 SNPs, genotype frequencies in the control groups do not differ from expectations under Hardy-Weinberg equilibrium (P-value > .05). A protective characteristic (OR<1) was found in 4 SNPs, and 8 were found with odds ratio greater than 1. Table 2 also shows the genotype frequencies of these 12 SNPs in the control population of the CGEMS study. SNP with main effect was called main-effect-SNPs in this study and other SNPs were called non-main-effect-SNPs.
Table 2.
Main-effect-SNPs on breast cancer. ORs and corresponding 95% confidence interval were estimated by the full model of unconditional logistic regression.
| Main-effect- SNP |
Related gene | Chromosomal Location€ |
Alteration* | Genotype | Genotype frequency (%)* |
OR | 95% CI | P-vale+ |
|---|---|---|---|---|---|---|---|---|
| OR < 1 | ||||||||
| rs1981929 | MSH2 | 2 : 47526073 | Intron Missense: |
GG | 16.24 | 0.81 | (0.66,0.98) | 0.033 |
| rs1799977 | MLH1 | 3 : 37028572 | A: Ile ->G:Val | GG | 10.25 | 0.72 | (0.55,0.94) | 0.015 |
| rs10017797 | DCTD | 4 : 184049877 | Intron | TT | 0.66 | 0.33 | (0.12,0.90) | 0.031 |
| rs2612092 | TYMS | 18 : 672399 | Intron | AG | 16.22 | 0.82 | (0.68,0.99) | 0.044 |
| OR > 1 | ||||||||
| rs13320360 | MLH1 | 3 : 37028124 | Intron | TC | 0.80 | 2.56 | (1.14,5.79) | 0.023 |
| rs6834938 | DCTD | 4 : 184047807 | 3' near gene | TT | 2.16 | 1.75 | (1.00,3.08) | 0.050 |
| rs3733399 | DCK | 4 : 72069135 | 3' UTR | CC | 3.72 | 1.61 | (1.00,2.57) | 0.048 |
| rs1474500 | RRM1 | 11 : 4080688 | Intron | TT | 0.55 | 3.19 | (1.04,9.78) | 0.043 |
| rs9948583 | TYMS | 18 : 665000 | Intron | TT | 10.18 | 1.34 | (1.05,1.71) | 0.017 |
| rs3819101 | TYMS | 18 : 667240 | Intron | TT | 9.86 | 1.28 | (1.00,1.64) | 0.047 |
| rs3786355 | TYMS | 18 : 671962 | Intron | TT | 9.92 | 1.30 | (1.01,1.66) | 0.040 |
| rs9966612 | TYMS | 18 : 639311 | 3' near gene | AA | 7.73 | 1.45 | (1.10,1.91) | 0.008 |
SNP-SNP interaction models were then selected by stepwise logistic regression. 812 combinations with significant association were ordered by crude P-value.
The interactions without main-effect-SNPs were shown in Table 3. In these interactions, individual SNP did not display an association with breast cancer risk, but interactions with other non-main-effect-SNPs did. The expected number of SNPs in RRM1 is in 10 SNP pairs with OR smaller than 1, however, six were observed here. A two proportion test was performed and the P-value of the test was 0.002919 (<0.05). That is, the observed proportion was higher than expected. In SNP–SNP interactions with OR greater than1 group, a same test for SNPs in DCTD yields a P (0.0048) also found. Among all non-main-effect-SNPs in Table 3, rs2304891 (RRM1) is synonymous SNP, half of residue SNPs fall within intron and the positions of others are not clear. In addition, all genotype distributions in related SNPs in the control population were in agreement with Hardy-Weinberg equilibrium.
Table 3.
Two way interactions without main-effect-SNPs. ORs and corresponding 95% confidence interval were estimated by the reduced model of unconditional logistic regression.
| SNP-SNP interaction | OR | 95% CI | P-Value* | |
|---|---|---|---|---|
| SNP1 | SNP2 | |||
| OR < 1 | ||||
| rs4073674(DCTD)-AC | rs7632760(MLH1)-AG | 0.74 | (0.61,0.89) | 1.24 × 10−3 |
| rs12450989(TK1)-AA | rs6544990(MSH2)-AA | 0.42 | (0.24,0.73) | 2.17 × 10−3 |
| rs1980412(RRM1)-CC | rs7372736(MLH1)-GG | 0.52 | (0.34,0.80) | 2.64 × 10−3 |
| rs1980412(RRM1)-CC | rs9852810(MLH1)-TC | 0.52 | (0.34,0.80) | 2.70 × 10−3 |
| rs4073674(DCTD)-AC | rs6789043(MLH1)-TC | 0.76 | (0.63,0.91) | 3.59 × 10−3 |
| rs1980412(RRM1)-CC | rs7611106(MLH1)-AA | 0.54 | (0.35,0.82) | 3.59 × 10−3 |
| rs2854702(TK1)-AG | rs17036614(MSH2)-AG | 0.51 | (0.32,0.80) | 3.67 × 10−3 |
| rs1980412(RRM1)-CC | rs7632760(MLH1)-GG | 0.55 | (0.37,0.83) | 3.89 × 10−3 |
| rs1980412(RRM1)-CC | rs6789043(MLH1)-CC | 0.56 | (0.37,0.83) | 3.90 × 10−3 |
| rs2304891(RRM1)-TT | rs7372736(MLH1)-GG | 0.52 | (0.33,0.81) | 4.16 × 10−3 |
| OR > 1 | ||||
| rs2464974(DCTD)-GG | rs6544990(MSH2)-AA | 1.77 | (1.25,2.50) | 1.26 × 10−3 |
| rs7698606(DCTD)-TT | rs6544990(MSH2)-AA | 1.85 | (1.26,2.72) | 1.66 × 10−3 |
| rs7698606(DCTD)-TT | rs3771281(MSH2)-CC | 1.81 | (1.24,2.63) | 2.04 × 10−3 |
| rs6831306(DCTD)-CC | rs6544990(MSH2)-AA | 2.25 | (1.33,3.80) | 2.53 × 10−3 |
| rs6831306(DCTD)-CC | rs3771281(MSH2)-CC | 2.12 | (1.30,3.48) | 2.82 × 10−3 |
| rs2464974(DCTD)-GG | rs3771281(MSH2)-CC | 1.62 | (1.17,2.25) | 3.56 × 10−3 |
| rs7698606(DCTD)-TT | rs6544992(MSH2)-CC | 1.59 | (1.16,2.18) | 3.65 × 10−3 |
| rs2298582(TYMS)-AC | rs4952887(MSH2)-TC | 1.93 | (1.24,3.03) | 3.72 × 10−3 |
| rs7698606(DCTD)-TT | rs10188090(MSH2)-GG | 1.74 | (1.20,2.53) | 3.74 × 10−3 |
| rs7698606(DCTD)-TT | rs7584256(MSH2)-CC | 1.57 | (1.16,2.14) | 3.91 × 10−3 |
Twenty SNP-SNP interactions with main-effect-SNPs were selected by smallest crud P-value and shown in Table 5. The ORs of ten interactions were less than 1 and the other ten were greater than 1. Among these interactions, 9 of 10 are combination of SNPs in TYMS and MLH1 (OR>1) where .
IV. DISCUSSION
This study of more than 1200 cases and 1200 controls tested the hypothesis that dNTP supply enzymes and DNA MMR enzymes are associated with breast cancer risk. To achieve this goal, 69 and 30 SNPs were selected from these two systems, respectively. Their effects for breast cancer risk were measured via a logistic regression.
Twelve of 99 SNPs were found significant associations (P-value < .05) with breast cancer; ORs of four SNPs were less than 1 and of eight were greater than 1. Others did not have statistically significant effects on the breast cancer.
rs1799977 locates to MLH1 and the A-to-G polymorphism changes the amino acid from Isoleucine to Valine . Our results shown that there is a smaller odds of breast cancer for people with the GG genotype. This result coincides with previous studies in lung cancer, prostate cancer and breast cancer, [12], [13], [14]. However, the population in all of these studies is Caucasians. No similar association between rs1799977 and breast cancer risk has been found in Korean women population [15].
The associations between an individual SNP and breast cancer risk may not be found, but this individual SNP could be the one in the significant SNP-SNP interactions which related with breast cancer. The main objective in this study was to identify interactions between two groups of SNPs that individually display no influence on breast cancer risk. 697 of 2070 (69 × 30) interactions were significant (crude P-value < .05) associated with breast cancer. The observed number of SNPs in RRM1 and DCTD are greater than expected. All 6 SNP-SNP interactions are composed of RRM1 and MLH1 and 9 SNP-SNP interactions are composed of DCTD and MSH2. DCTD encodes dCMP deaminase. The function of this enzyme is to catalyze the deamination of dCMP to dUMP. dUMP is used to form dTMP which is further phosphate to form thymidine triphosphate (dTTP), one of four dNTP building blocks used in DNA replication and repair.
People who had these significant genotypes in this study were suggested with lower/higher odds of breast cancer than people without them in this study. That is, the defects in dNTP supply and MMR interact to modulate the risk of breast cancer.
In this study, rs3819101 (TYMS), rs3786355 (TYMS) and rs9948583 (TYMS) display similar ORs not only in individual SNP analyses but also in SNP-SNP interaction logistic regression analyses. Also, these three chromosomal locations are very close.
In the future work, other races and populations could be considered to discover the association between breast cancer and SNPs in genes which are related with DNA mismatches, such as dNTP pool enzymes, DNA polymerase and MMR enzymes. Moreover, a dummy variable could be used to code single SNP/ SNP pairs in logistic regression analysis. Further demographic information, such as age, education and income, might be added in the model to investigate the potential breast cancer risk.
Table 4.
Two way interactions with main-effect-SNPs (main-effect-SNPs are bold). ORs and corresponding 95% confidence interval were estimated by the reduced model of unconditional logistic regression.
| SNP-SNP interaction | OR | 95% CI | P-Value* | |
|---|---|---|---|---|
| SNP1 | SNP2 | |||
| OR < 1 | ||||
| rs3819101(TYMS)-CC | rs1981929(MSH2)-GG | 0.56 | (0.42,0.75) | 1.21 × 10−4 |
| rs3786355(TYMS)-CC | rs1981929(MSH2)-GG | 0.56 | (0.42,0.76) | 1.38 × 10−4 |
| rs11873007(TYMS)-CC | rs1981929(MSH2)-GG | 0.57 | (0.43,0.77) | 2.28 × 10−4 |
| rs9948583(TYMS)-CC | rs1981929(MSH2)-GG | 0.58 | (0.43,0.78) | 3.64 × 10−4 |
| rs2292235(TK1)-AA | rs1799977(MLH1)-GG | 0.23 | (0.10,0.52) | 4.23 × 10−4 |
| rs13147196(DCTD)-AA | rs1981929(MSH2)-GG | 0.62 | (0.46,0.82) | 9.66 × 10−4 |
| rs4861536(DCTD)-GG | rs1799977(MLH1)-GG | 0.59 | (0.43,0.81) | 1.00 × 10−3 |
| rs13147196(DCTD)-AA | rs1799977(MLH1)-GG | 0.53 | (0.36,0.78) | 1.23 × 10−3 |
| rs12499918(DCTD)-AA | rs1981929(MSH2)-GG | 0.61 | (0.45,0.83) | 1.33 × 10−3 |
| rs1065769(TK1)-GG | rs1981929(MSH2)-GG | 0.62 | (0.46,0.83) | 1.63 × 10−3 |
| OR > 1 | ||||
| rs9966612(TYMS)-AA | rs7611106(MLH1)-GG | 2.42 | (1.38,4.25) | 2.09 × 10−3 |
| rs9966612(TYMS)-AA | rs2241031(MLH1)-CC | 2.45 | (1.37,4.38) | 2.40 × 10−3 |
| rs9966612(TYMS)-AA | rs9852810(MLH1)-GG | 2.37 | (1.35,4.18) | 2.75 × 10−3 |
| rs9966612(TYMS)-AA | rs7372736(MLH1)-AA | 2.36 | (1.34,4.16) | 2.83 × 10−3 |
| rs9948583(TYMS)-TT | rs1799977(MLH1)-AA | 1.71 | (1.20,2.44) | 3.07 × 10−3 |
| rs3786355(TYMS)-TT | rs1799977(MLH1)-AA | 1.71 | (1.19,2.46) | 3.72 × 10−3 |
| rs9966612(TYMS)-AA | and rs1421(MSH2)-AA | 1.60 | (1.16,2.19) | 3.75 × 10−3 |
| rs2304891(RRM1)-CC | rs1981929(MSH2)-AA | 1.40 | (1.11,1.78) | 4.47 × 10−3 |
| rs3819101(TYMS)-TT | rs1799977(MLH1)-AA | 1.68 | (1.18,2.41) | 4.48 × 10−3 |
| rs11873007(TYMS)-TT | rs1799977(MLH1)-AA | 1.68 | (1.17,2.40) | 5.00 × 10−3 |
P-value based on logistic Regression
Acknowledgment
This work was supported by the National Cancer Institute (K25CA104791)
REFERENCES
- 1.WHO website. < http://www.who.int/mediacentre/factsheets/fs297/en/index.html>.
- 2.Chakravarti A. It's raining SNPs, hallelujah? Nat Genet. 1998 Jul;vol. 19:216–217. doi: 10.1038/885. [DOI] [PubMed] [Google Scholar]
- 3.Onay VU, Briollais L, Knight JA, Shi E, Wang Y, Wells S, Li H, Rajendram I, Andrulis IL, Ozcelik H. SNP-SNP interactions in breast cancer susceptibility. BMC Cancer. 2006;vol. 6:114. doi: 10.1186/1471-2407-6-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kidokoro K, Ino K, Hirose K, Kajiyama H, Hosono S, Suzuki T, Kawase T, Hiraki A, Hamajima N, Tanaka H, Tajima K, Kikkawa F, Matsuo K. Association between CYP19A1 polymorphisms and sex hormones in postmenopausal Japanese women. J Hum Genet. 2009 Jan 16; doi: 10.1038/jhg.2008.11. [DOI] [PubMed] [Google Scholar]
- 5.Chang BL, Cramer SD, Wiklund F, Isaacs SD, Stevens VL, Sun J, Smith S, Pruett K, Romero LM, Wiley KE, Kim ST, Zhu Y, Zhang Z, Hsu FC, Turner AR, Adolfsson J, Liu W, Kim JW, Duggan D, Carpten J, Zheng SL, Rodriguez C, Isaacs WB, Gronberg H, Xu J. Fine mapping association study and functional analysis implicate a SNP in MSMB at 10q11 as a causal variant for prostate cancer risk. Hum Mol Genet. 2009 Jan 19; doi: 10.1093/hmg/ddp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.T K, M J, M P, M V, M Z, Z K, T S, A Z. Polymorphism −23HPhI in the promoter of insulin gene and pancreatic cancer: A pilot study. Neoplasma. 2009;vol. 56:26–32. doi: 10.4149/neo_2009_01_26. [DOI] [PubMed] [Google Scholar]
- 7.Bebenek K, Roberts JD, Kunkel TA. The effects of dNTP pool imbalances on frameshift fidelity during DNA replication. J Biol Chem. 1992 Feb 25;vol. 267:3589–3596. [PubMed] [Google Scholar]
- 8.Echols H, Goodman MF. Fidelity mechanisms in DNA replication. Annu Rev Biochem. 1991;vol. 60:477–511. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- 9.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;vol. 74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 10.National Cancer Institute Cancer Genetics Markers of Susceptibility (CGEMS) [Google Scholar]
- 11.K. P. Hunter DJ, Jacobs KB, Cox DG, Yeager M, Hankinson SE, Wacholder S, Wang Z, Welch R, Hutchinson A, Wang J, Yu K, Chatterjee N, Orr N, Willett WC, Colditz GA, Ziegler RG, Berg CD, Buys SS, McCarty CA, Feigelson HS, Calle EE, Thun MJ, Hayes RB, Tucker M, Gerhard DS, Fraumeni JF, Jr, Hoover RN, Thomas G, Chanock SJ. A Genome-Wide Association Study Identifies Alleles in FGFR2 Associated with Risk of Sporadic Postmenopausal Breast Cancer. Nat Genet. 2007;vol. 39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landi S, Gemignani F, Canzian F, Gaborieau V, Barale R, Landi D, Szeszenia-Dabrowska N, Zaridze D, Lissowska J, Rudnai P, Fabianova E, Mates D, Foretova L, Janout V, Bencko V, Gioia-Patricola L, Hall J, Boffetta P, Hung RJ, Brennan P. DNA repair and cell cycle control genes and the risk of young-onset lung cancer. Cancer Res. 2006 Nov 15;vol. 66:11062–11069. doi: 10.1158/0008-5472.CAN-06-1039. [DOI] [PubMed] [Google Scholar]
- 13.Burmester JK, Suarez BK, Lin JH, Jin CH, Miller RD, Zhang KQ, Salzman SA, Reding DJ, Catalona WJ. Analysis of candidate genes for prostate cancer. Hum Hered. 2004;vol. 57:172–178. doi: 10.1159/000081443. [DOI] [PubMed] [Google Scholar]
- 14.Smith TR, Levine EA, Freimanis RI, Akman SA, Allen GO, Hoang KN, Liu-Mares W, Hu JJ. Polygenic model of DNA repair genetic polymorphisms in human breast cancer risk. Carcinogenesis. 2008 Nov;vol. 29:2132–2138. doi: 10.1093/carcin/bgn193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee KM, Choi JY, Kang C, Kang CP, Park SK, Cho H, Cho DY, Yoo KY, Noh DY, Ahn SH, Park CG, Wei Q, Kang D. Genetic polymorphisms of selected DNA repair genes, estrogen and progesterone receptor status, and breast cancer risk. Clin Cancer Res. 2005 Jun 15;vol. 11:4620–4626. doi: 10.1158/1078-0432.CCR-04-2534. [DOI] [PubMed] [Google Scholar]