Abstract
Structural analyses of bacterial ATP-binding-cassette transporters revealed that the glutamine residue in Q-loop plays roles in interacting with: 1) a metal cofactor to participate in ATP binding; 2) a putative catalytic water molecule to participate in ATP hydrolysis; 3) other residues to transmit the conformational changes between nucleotide-binding-domains and transmembrane-domains, in ATP-dependent solute transport. We have mutated the glutamines at 713 and 1375 to asparagine, methionine or leucine to determine the functional roles of these residues in Q-loops of MRP1. All these single mutants significantly decreased Mg·ATP binding and increased the Km (Mg·ATP) and Vmax values in Mg·ATP-dependent leukotriene-C4 transport. However, the Vmax values of the double mutants Q713N/Q1375N, Q713M/Q1375M and Q713L/Q1375L were lower than that of wtMRP1, implying that the double mutants cannot efficiently bind Mg·ATP. Interestingly, MRP1 has higher affinity for Mn·ATP than for Mg·ATP and the Mn·ATP-dependent leukotriene-C4 transport activities of Q713N/Q1375N and Q713M/Q1375M are significantly higher than that of wtMRP1. All these results suggest that: 1) the glutamine residues in Q-loops contribute to ATP-binding via interaction with a metal cofactor; 2) it is most unlikely that these glutamine residues would play crucial roles in ATP hydrolysis and in transmitting the conformational changes between nucleotide-binding-domains and transmembrane-domains.
1. Introduction
Over-expression of ATP-binding cassette (ABC) transporters, such as P-glycoprotein (P-gp or ABCB1) [1–3], breast cancer resistance protein (BCRP or ABCG2) [4, 5] and/or multidrug resistance protein (MRP1 or ABCC1) [6, 7], confers acquired multidrug resistance (MDR) owing to the fact that these ABC transporters catalyze ATP-dependent anticancer drug transport. Although some of the ABC transporters, such as ABCG2, contain one nucleotide binding domain (NBD) and one transmembrane domain (TMD), these transporters are thought to form either a homo- or a hetero-dimer in the plasma membrane [8–11]. Thus, all the functional ABC transporters contain two NBDs and at least two TMDs [3, 7–12]. Upon Mg·ATP binding, the two NBDs form a dimer in which the two Mg·ATP molecules are each sandwiched between the Walker A motif from one NBD and the LSGGQ ABC signature motif from another [13–19]. In addition, the residues from Walker B motif, A-loop, D-loop, H-loop and Q-loop [13, 14, 16, 17, 19–36] also contribute to Mg·ATP binding and hydrolysis. Detailed structural analyses of several NBDs of ABC transporters revealed that the amide ε-oxygen of glutamine residue in Q-loop, as shown in Figure S1, participates in Mg·ATP binding by interacting, together with other five oxygen atoms from the Walker A serine residue, the β- and γ-phosphate of the bound ATP and two water molecules, with a Mg++ cofactor to form the octahedral coordination geometry [13, 16, 21, 25, 27, 29]. The amide ε-nitrogen of the glutamine residue in Q-loop may interact with the putative hydrolytic water molecule in the active center [13], implying that this residue may play a role in ATP hydrolysis. In addition, the amide ε-nitrogen of the glutamine residue in Q-loop may also interact with an amino acid from another NBD [13] to stabilize the NBD·ATP·ATP·NBD sandwich structure. Furthermore, the residues around the glutamine residue (in the linear sequence of an NBD) in Q-loop form a Q-loop cleft that interacts with the residues in L-loop of the transmembrane subunits [14, 17], suggesting that these interactions may facilitate inter-domain communications between the transport-substrate-binding sites, such as TMDs, and ATP binding/hydrolysis sites, such as NBDs [13, 14, 22]. Thus, the interactions of the glutamine residue in Q-loop with the residues mentioned above may play very important roles in ATP binding, hydrolysis and ATP-dependent solute transport. However, whether the glutamine residue in Q-loop of human MRP1 will play so many important roles has not been experimentally proved yet. In order to determine the functional roles of the glutamine residue in the Q-loop of human MRP1, we have substituted the glutamine residue in Q-loop of MRP1 with: 1) an asparagine (Q713N and Q1375N) that remains the amide group but with one methylene shorter in asparagine than in glutamine; 2) a methionine (Q713M and Q1375M) that eliminates the amide group but contains paired electrons in the sulfur atom of the methionine residue that might potentially interact with the Mg++ cofactor; 3) a leucine (Q713L and Q1375L) that eliminates the amide group and abolishes the interactions with Mg++ cofactor and the putative hydrolytic water molecule, and used them to determine the consequence of these mutations in ATP-dependent leukotriene C4 (LTC4) transport. The data presented in this manuscript indicate that the glutamine residues in the Q-loops of MRP1 mainly play a role in Mg·ATP binding, but not in ATP hydrolysis and in inter-domain communications between the NBDs and TMDs.
2. Material and methods
2.1. Materials
Sodium orthovanadate, EGTA and ATP were purchased from Sigma. [14,15,19,20-3H]-leukotriene C4 was from PerkinElmer Life Sciences. [α-32P]-8-N3ATP and [γ-32P]-8-N3ATP were from Affinity Labeling Technologies. Fetal bovine serum was from Gemini Bio-Products. QuikChange site-directed mutagenesis kit was from Stratagene. Anti-mouse Ig conjugated with horseradish peroxidase was from Amersham Biosciences. Chemiluminescent substrates for western blotting were from Pierce.
2.2. Site-directed mutagenesis of human MRP1 cDNA
Wt N-half (NH, 1–932) and C-half (CH, 933–1531) of human MRP1 cDNA cloned into pDual expression vector [37, 38], named as pDual/NH/CH, was used as a template for the in vitro mutagenesis. The glutamine residue at position of 713 in Q-loop of NBD1 or 1375 in Q-loop of NBD2 was mutated to either asparagine (N), methionine (M) or leucine (L) by using the forward and reverse primers and the QuikChange site-directed mutagenesis kit [39]. The forward and reverse primers used to introduce these mutations are: Q713/forward, 5′-TCC GTG GCC TAT GTG CCA XXX CAG GCC TGG ATT CAG AAT-3′; Q713/reverse, 5′-ATT CTG AAT CCA GGC CTG XXX TGG CAC ATA GGC CAC GGA-3′; Q1375/forward, 5′-AAG ATC ACC ATC ATC CCC XXX GAC CCT GTT TTG TTT TCG GG-3′; Q1375/reverse, 5′-CC CGA AAA CAA AAC AGG GTC XXX GGG GAT GAT GGT GAT CTT-3′. The underlined XXX sequences are codons for mutated residues as shown in Table S1. Regions containing these mutations were sequenced to confirm that the correct clones were obtained. Recombinant viral DNA preparation and baculovirus viral particle production were carried out according to the procedures described [38].
2.3. Cell Culture and cells expressing N-half and C-half of MRP1
Spodoptera frugiperda 21 (Sf21) cells were cultured in Grace’s insect cell medium supplemented with 5% heat-inactivated fetal bovine serum at 27 °C. Viral infection was performed according to Invitrogen’s recommendations, i.e., a multiplicity of infection (MOI) of 5 to 10 was used to infect the insect cells. The expression levels of the dually expressed N-half and C-half with varying MOI were determined by western blot analysis. The MOI producing similar amount of N-half (by comparing the wt N-half with the mutated N-half) and C-half (by comparing the wt C-half with the mutated C-half) was used to infect Sf21 cells for membrane vesicle preparations.
2.4. Identification of N-half and C-half of MRP1 protein
Western blot was performed according to the method described previously [39]. 42.4 mAb was used to identify the NBD1-containing N-half fragment of MRP1, whereas 897.2 mAb was used to detect the NBD2-containing C-half fragment [37, 39]. The secondary antibody used was anti-mouse Ig conjugated with horseradish peroxidase.
2.5. Membrane vesicle preparation
Membrane vesicles were prepared according to the procedure described previously [39]. The membrane vesicle pellet was re-suspended in a solution containing 10 mM Tris-HCl (pH 7.5), 250 mM sucrose and 1 X protease inhibitor cocktail (2 μg/ml aprotinine, 121 μg/ml benzamidine, 3.5 μg/ml E64, 1 μg/ml leupeptin and 50 μg/ml Pefabloc). Aliquots of the membrane vesicles, after passage through a Liposofast™ vesicle extruder (Avestin, Ottawa, Canada), were stored in −80 °C.
2.6. Membrane vesicle transport
ATP-dependent LTC4 transport was assayed by a rapid filtration technique [39–41]. The assays were carried out in triplicate determinations in a 30 μl solution containing 3 μg of membrane proteins, 50 mM Tris-HCl (pH 7.5), 250 mM sucrose, 10 mM MgCl2 (or 10 mM MnCl2), 200 nM LTC4 (17.54 nCi of 3H-labeled LTC4) and 4 mM AMP (used as negative control) or 4 mM ATP (with an ATP regenerating system consisting of 120 mU of creatine kinase and 10 mM creatine phosphate). For percentage determination, above components were mixed on ice, transferred to 37 ºC water bath, incubated at 37 ºC for 4 minutes, brought back to ice and diluted with 1 ml of ice-cold 1 X transport buffer (50 mM Tris-HCl, pH 7.5, 250 mM sucrose and 10 mM MgCl2). For kinetic analysis, the ATP solutions and the membrane vesicles were prepared in two different eppendorf tubes on ice, transferred to 37 ºC heating block (filled with water), warmed up to 37 ºC, mixed the two solutions, incubated at 37 ºC for 1 minute, brought back to ice and diluted with 1 ml of ice-cold 1 X transport buffer.
2.7. Photoaffinity labeling of N-half and C-half of MRP1 protein
Vanadate preparation (from sodium orthovanadate) and photoaffinity labeling of MRP1 protein were performed according to procedures described previously [39, 42, 43]. Briefly, photolabeling experiments were carried out in a 10 μl solution containing 10 μg of membrane proteins, 40 mM Tris-HCl (pH7.5), 2 mM ouabain, 10 mM MgCl2, 0.1 mM EGTA and either 10 μM [α-32P]-8-N3ATP or 10 μM [γ-32P]-8-N3ATP. The other conditions, such as incubation time, temperature and vanadate, are indicated in figure legend. After UV-irradiation (λ = 254 nm) on ice for 2 minutes, the labeled proteins were separated by polyacrylamide gel (7%) electrophoresis and electro-blotted to a nitrocellulose membrane.
2.8. Statistical analysis
The results in Figure 1C, Figure 4, Table 1 and S3 were presented as means ± SD from three independent experiments. The two-tailed P values were calculated based on the unpaired t test from GraphPad Software Quick Calcs. By conventional criteria, if P value is less than 0.05, the difference between two samples is considered to be statistically significant.
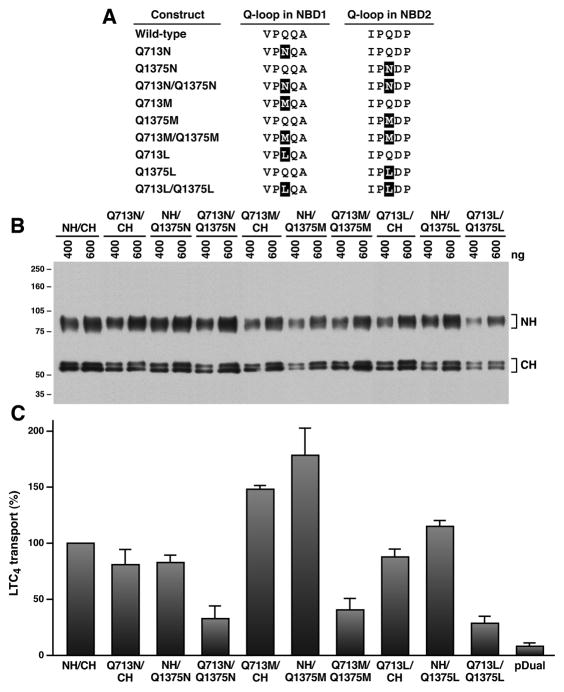
Figure 1. Substitution of Q713 in Q-loop of NBD1 or Q1375 in Q-loop of NBD2 with an amino acid that eliminates the amide group did not have a significant effect on the Mg·ATP-dependent LTC4 transport.
A. Linear amino acid sequences around the glutamine residue in Q-loop. The highlighted amino acids indicate the mutated residues. B. Expression of the N-half and C-half of MRP1 mutants in insect cells. Membrane vesicles were prepared from Sf21 cells infected with viral particles harboring pDual/NH/CH of MRP1 cDNA. The amounts (ng) of membrane proteins loaded in the gel are indicated on top of the gel. Molecular weight markers are indicated on the left. NH and CH on the right indicated the N-half and C-half of MRP1 protein. C. Relative ATP-dependent LTC4 transport activities of the wt and mutated MRP1s. ATP-dependent LTC4 transport was performed according to the methods described in Materials and methods. Since the amounts of MRP1 proteins determined in Figure 1B were different, the amounts of membrane vesicles used in these experiments were adjusted, based on the ratio of MRP1 protein in membrane vesicles listed in Table S2, to have a similar amount of MRP1 protein by adding varying amounts of membrane vesicles prepared from insect cells infected with viral particles harboring pDual expression vector. For example, 1.56 μg of wt MRP1 + 1.44 μg of pDual and 3.00 μg of Q1375M were used to determine the ATP-dependent LTC4 transport activities of wt MRP1 and Q1375M-mutated MRP1. The data, considering the transport activity catalyzed by wt MRP1 as 100%, are presented as means ± standard deviation of three independent experiments.
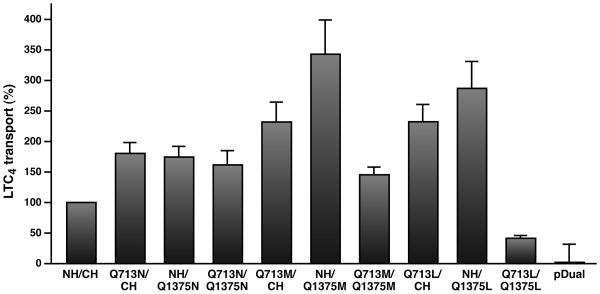
Figure 4. MnCl2 significantly enhanced the ATP-dependent LTC4 transport activities of the Q713 or Q1375 mutants.
The experiments were carried out exactly the same as in Figure 1C, except that 10 mM MnCl2, instead of 10 mM MgCl2, was used in the transport assay.
Table 1.
Vmax (LTC4) and Km (Mg·ATP) values of wild-type and mutant MRP1s.
| Construct | Vmax (pmol/mg/min)* | P value | Km (μM Mg.ATP)* | P value |
|---|---|---|---|---|
| Wt MRP1 | 104.3 ± 20.5 | 65.7 ± 4.2 | ||
| Q713N | 191.7 ± 20.9 | 0.0135 | 318.3 ± 2.4 | 0.0001 |
| Q1375N | 280.0 ± 32.7 | 0.0030 | 370.0 ± 8.2 | 0.0001 |
| Q713N/Q1375N | 63.7 ± 6.9 | 0.0566 | 916.7 ± 20.5 | 0.0001 |
| Q713M | 213.3 ± 29.5 | 0.0128 | 278.3 ± 19.3 | 0.0001 |
| Q1375M | 203.3 ± 24.9 | 0.0123 | 295.0 ± 7.1 | 0.0001 |
| Q713M/Q1375M | 77.0 ± 12.5 | 0.1249 | 1006.3 ± 12.7 | 0.0001 |
| Q713L | 120.0 ± 4.1 | 0.3489 | 293.3 ± 30.9 | 0.0005 |
| Q1375L | 390.0 ± 24.8 | 0.0002 | 900.0 ± 21.6 | 0.0001 |
| Q713L/Q1375L | 88.3 ± 6.2 | 0.3505 | 970.0 ± 80.0 | 0.0006 |
Km (Mg·ATP) and Vmax (LTC4) values (n = 3) for wild-type and Q713- or Q1375-mutated MRP1 were derived from the corresponding Michaelis-Menten curves (with variant concentration of ATP at 37 ºC for one minute). The P values for comparison of Vmax (LTC4) and Km (Mg·ATP) of wild-type versus mutants are shown.
3. Results
3.1. Substitution of Q713 in Q-loop of NBD1 or Q1375 in Q-loop of NBD2 with an amino acid that eliminates the amide group did not have a significant effect on the Mg·ATP-dependent LTC4 transport
In order to determine the functional role of the glutamine residue in Q-loop of NBD1 or NBD2, the Q713 and Q1375 were substituted with an asparagine (Fig. 1A) in the pDual construct expressing the N-half and C-half of MRP1 simultaneously [37, 38]. ATP-dependent LTC4 transport activity of each individual mutant, including Q713N and Q1375N, after adjusting the membrane vesicles to have similar amount of MRP1 protein, based on the amount of protein shown in Figure 1B and the ratio listed in Table S2, is similar to that of wt MRP1 (Fig. 1C), indicating that the amide side group of asparagine with one methylene shorter in asparagine than in glutamine plays a similar role as that of glutamine. However, the ATP-dependent LTC4 transport activity of the double mutant Q713N/Q1375N is significantly lower than that of wt MRP1 (Fig. 1C), implying that the double mutations might significantly affect the binding for Mg·ATP in both NBDs. We, then, tested whether a mutant without the amide group would significantly reduce the ATP-dependent LTC4 transport. Interestingly, substitution of the glutamine residue with a methionine, including Q713M and Q1375M, increased the transport activities, whereas substitution of the glutamine residue with a leucine, including Q713L and Q1375L, did not have a significant effect on the ATP-dependent LTC4 transport (Fig. 1C). In contrast, the double mutants, including Q713M/Q1375M and Q713L/Q1375L, significantly decreased the ATP-dependent LTC4 transport (Fig. 1C).
3.2. Substitution of the Q713 in NBD1 or Q1375 in NBD2 with an amino acid that eliminates the interaction between this residue and Mg++ cofactor significantly increased the Km value in ATP-dependent LTC4 transport
The results in Figure 1C imply that the double mutants might significantly affect the binding of Mg·ATP. If this would be the case, it meant that individual mutants, including Q713N, Q1375N, Q713M, Q1375M, Q713L and Q1375L, should also decrease the binding of Mg·ATP. We have found that decreased binding might significantly increase the Km (Mg·ATP) and Vmax values in ATP-dependent LTC4 transport [37, 38]. To test whether these mutants will also increase the Km (Mg·ATP) and Vmax values, the same amount of MRP1 protein used in Figure 1C was employed to establish the Michaelis-Menten curves. The results shown in Figure S2 clearly indicate that the Vmax and Km (Mg·ATP) values for Q1375L are significantly higher than that for wt MRP1. These experiments were repeated three times and the Vmax and Km (Mg·ATP) values derived from these Michaelis-Menten curves were listed in Table 1. The results in Table 1 indicate that the Vmax values derived from Q713N, Q1375N, Q713 M, Q1375M and Q1375L are significantly higher than that of wt MRP1, whereas the Vmax values of Q713L and the double mutants, including Q713N/Q1375N, Q713M/Q1375M and Q713L/Q1375L, are not significantly different from that of wt MRP1. Interestingly, the Km (Mg·ATP) values derived from all the mutants are significantly higher than that of wt MRP1 (Table 1). In addition, the Km (Mg·ATP) values derived from the double mutants are significantly higher than, except Q1375L, their corresponding single mutants.
3.3. Substitution of the Q713 in NBD1 with an amino acid that diminishes the interaction between this residue and Mg++ cofactor significantly decreased Mg·ATP binding at NBD1
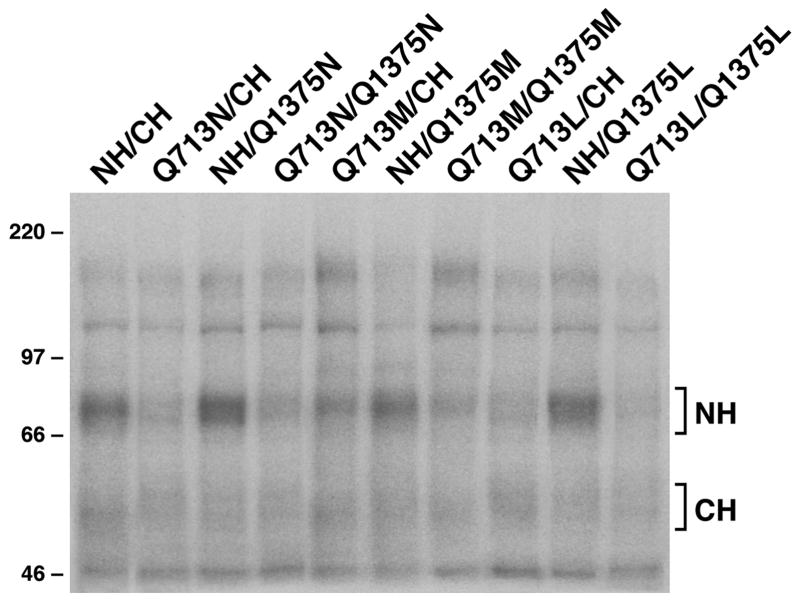
The above results imply that all these mutants, regardless of whether they are single mutants or double mutants, may significantly reduce the affinity for Mg·ATP. In order to test this hypothesis, [γ-32P]-8-N3ATP was used to label the NBD1 (within N-half) and NBD2 (within C-half) of MRP1 on ice by taking the advantage of expressing these mutants in N-half and C-half so that we can check the effects of these mutants on ATP binding or ADP-trapping directly without partial trypsin digestion. Since [γ-32P]-8-N3ATP was used to label the NBD1 and NBD2 of MRP1 on ice without vanadate, the labeling reflects the Mg·ATP binding, but not ATP hydrolysis and vanadate-dependent trapping. As shown in Figure 2, the labeling at NBD2 (within CH) is too weak to distinguish whether the mutations affect the Mg·ATP binding or not. However, the labeling at NBD1 (within NH) is high enough to distinguish whether the mutations affect the Mg·ATP binding or not. As shown in Figure 2, the mutations at NBD2, including Q1375N, Q1375M and Q1375L, did not have a significant effect on Mg·ATP binding at NBD1, whereas the mutations at NBD1, including Q713N, Q713N/Q1375N, Q713M, Q713M/Q1375M, Q713L and Q713L/Q1375L, significantly reduced the Mg·ATP binding at NBD1.
Figure 2. Substitution of Q713 with an amino acid that eliminates the interaction of the amide group (ε-oxygen) of glutamine residue in Q-loop with Mg++ cofactor significantly reduced Mg·ATP binding at NBD1.
Photolabeling experiments were carried out in the presence of 10 μM [γ-32P]-8-N3ATP on ice for 10 minutes. Molecular weight markers (kDa) are indicated on the left. NH and CH on the right indicate the 32P-labeled N-half and C-half of MRP1 protein.
3.4. Substitution of Q1375 with an amino acid that eliminates the interaction of the amide side group (ε-oxygen) of glutamine residue in Q-loop with Mg++ cofactor significantly decreased the vanadate-dependent Mg·ADP trapping at NBD2
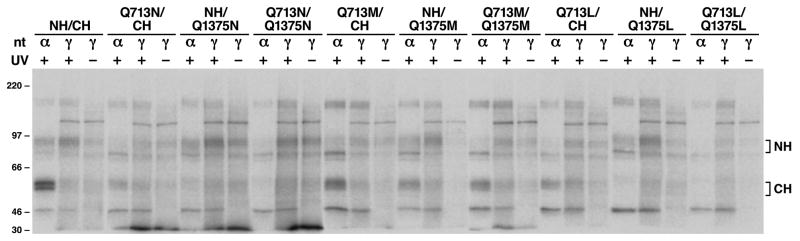
Although the labeling at NBD2 was too weak to distinguish whether the mutations would affect the Mg·ATP binding or not, the above results imply that the mutations at NBD2 should also affect Mg·ATP binding at NBD2. We have found that vanadate-dependent ATP-hydrolysis-product ADP trapping mainly occured at NBD2 [39, 42]. In order to test whether the mutations at NBD2 would also affect Mg·ATP binding and Mg·ADP trapping at NBD2, [γ-32P]-8-N3ATP and [α-32P]-8-N3ATP were used to label the N-half and C-half of MRP1 at 37 ºC in the presence of vanadate. As shown in Figure 3, [γ-32P]-8-N3ATP, without UV treatment, labeled N-half and C-half of MRP1, reflecting the phosphorylation events occurred in MRP1 protein [39]. However, upon treatment with UV, the labeling with [γ-32P]-8-N3ATP at the NBD1 (within NH) is significantly higher than that without UV treatment, indicating that the bound [γ-32P]-8-N3ATP at NBD1 is not efficiently hydrolyzed. The labeling at NBD2 (within CH) with [γ-32P]-8-N3ATP, regardless of whether they were treated with UV or not, was weak, suggesting that the bound ATP was hydrolyzed. In contrast, the labeling at NBD2 with [α-32P]-8-N3ATP, including wt MRP1, Q713N, Q713M, Q1375M, Q713M/Q1375M and Q713L, is stronger than the corresponding labeling with [γ-32P]-8-N3ATP, indicating that the bound Mg·ATP was hydrolyzed and trapped there by vanadate. However, the labeling at the mutated NBD2, including Q1375N, Q713N/Q1375N, Q1375M, Q713M/Q1375M, Q1375L and Q713L/Q1375L, with [α-32P]-8-N3ATP was significantly lower than the corresponding labeling at wt MRP1, implying that much less [α-32P]-8-N3ATP bound to the mutated NBD2 than to wt MRP1 or the ATP-hydrolysis-product ADP had not been firmly trapped there by vanadate.
Figure 3. Substitution of Q713 or Q1375 with an amino acid that eliminates the interaction of the amide group (ε-nitrogen) of the glutamine residue in Q-loop with the putative hydrolytic water molecule in the active center did not abolish ATP hydrolysis at NBD2.
Photolabeling experiments were carried out in the presence of 0.8 mM vanadate and either 10 μM [α-32P]-8-N3ATP (α) or 10 μM [γ-32P]-8-N3ATP (γ) at 37 °C for 2 minutes. The reaction mixture was either loaded to the gel directly (−) or subjected to UV-irradiation (+) and then loaded to the gel. Molecular weight markers (kDa) are indicated on the left. NH and CH on the right indicate the 32P-labeled N-half and C-half of MRP1 protein.
3.5. Mn·ATP significantly enhanced the ATP-dependent LTC4 transport activities of the Q-mutants
The above results were interpreted as that the glutamine residues in Q-loops of MRP1 participated in, via interaction with metal cofactor, Mg·ATP binding, but did not play a crucial role in ATP hydrolysis and in inter-domain communications between the NBDs and the TMDs. If that would be the case, the double mutant Q713L/Q1375L might require higher concentration of Mg++ to perform the ATP-dependent LTC4 transport. To test this hypothesis, varying concentration of MgCl2 was used to do the ATP-dependent LTC4 transport. As shown in Figure S3, wt MRP1 transported detectable amount of LTC4 into the membrane vesicles at 0.5 mM MgCl2, whereas Q713L/Q1375L required 5 mM MgCl2 to transport detectable amount of LTC4 into the membrane vesicles. Substitution of the glutamine residue (with an amide side group) at 713 with asparagine (with an amide side group) significantly reduced the Mg·ATP binding at NBD1 (Fig. 2), suggesting that the length of the side chain might also play an important role in Mg·ATP binding. We wondered whether a bigger metal cofactor, for example, Mn++ (with a radius of 0.80 Å) or Ca++ (0.99 Å) is bigger than Mg++ (0.66 Å), would increase the nucleotide binding of Q713N. Interestingly, the Mn·ATP-dependent LTC4 transport activities of Q713N/Q1375N and Q713M/Q1375M were significantly higher than that of wt MRP1 (Fig. S4A), whereas the Ca·ATP-dependent LTC4 transport activity of wt MRP1 was significantly higher than that of the double mutants (Fig. S4B). Due to these results, Mn·ATP and Mg·ATP were used to establish Michaelis-Menten curves of wt MRP1. Interestingly, the Km value for Mn·ATP (32.5 μM) is significantly less than that of the value for Mg·ATP (67.0 μM) and the Vmax value for Mn·ATP-dependent LTC4 transport is also significantly less than that for Mg·ATP (Table S3), implying that MRP1 may have higher affinity for Mn·ATP than for Mg·ATP.
Mn·ATP was used to determine the LTC4 transport activities of wt MRP1 and Q-mutants. Interestingly, the Mn·ATP-dependent LTC4 transport activity of Q713N or Q1375N is ~ 191% or 175% of wt MRP1 (Fig. 4), whereas the Mg·ATP-dependent LTC4 transport activity of the corresponding mutant is ~ 81% or 83% of the wt MRP1 (Fig. 1C). Strikingly, the Mn·ATP-dependent LTC4 transport activity of the double mutant Q713N/Q1375N is ~ 162% of wt MRP1 (Fig. 4), whereas the Mg·ATP-dependent LTC4 transport activity of this mutant is ~ 33% of the wt MRP1 (Fig. 1C). Similarly, the percentages (comparing to wt MRP1 as shown in Figure 4) of the Mn·ATP-dependent LTC4 transport activities of other mutants, including Q713M, Q1375M, Q713M/Q1375M, Q713L and Q1375L, are also significantly higher than their corresponding percentages (comparing to wt MRP1 as shown in Figure 1C) of the Mg·ATP-dependent LTC4 transport activities, suggesting that these mutants might have higher affinity for Mn·ATP than for Mg·ATP. If that would be the case, at lower concentration of divalent cation, wt MRP1 should have higher Mn·ATP-dependent LTC4 transport activity than that of Mg·ATP. Indeed, at 2 mM divalent cation, the Mn·ATP-dependent LTC4 transport activity of wt MRP1 is significantly higher than that of Mg·ATP (Fig. S5), suggesting that the glutamine residues in Q-loops of MRP1 contribute to ATP binding via interaction with metal cofactor.
4. Discussion
Structural analyses of several NBDs of ABC transporters suggested that the conserved glutamine residue in Q-loop may interact: 1) with Mg++ cofactor to participate in Mg·ATP binding; 2) with the putative hydrolytic water molecule in the active center to participate in ATP hydrolysis; 3) with an amino acid from another NBD to stabilize the NBD·ATP·ATP·NBD sandwich structure. In addition, the residues in Q-loop form a cleft that interacts with the residues in L-loop of the transmembrane subunits to play a role in transmitting the conformational changes induced by ATP binding/hydrolysis from NBDs to TMDs. However, the data derived from our studies indicate that the glutamine residues in the Q-loops of MRP1 mainly play a role in Mg·ATP binding, but not in ATP hydrolysis and in inter-domain communications between the NBDs and the TMDs. This conclusion is based on the facts that substitution of the Q713 in NBD1 or Q1375 in NBD2 with an amino acid that eliminates the amide group: 1) significantly reduced the Mg·ATP binding at the mutated NBD1 (Fig. 2) or vanadate-dependent ADP trapping at the mutated NBD2 (Fig. 3); 2) did not abolish the Mg·ATP- (Fig. 1C) or Mn·ATP-dependent (Fig. 4) LTC4 transport; 3) significantly increased the Km and Vmax values in ATP-dependent LTC4 transport (Table 1).
Of note, although substitution of the glutamine residues in Q-loops of MRP1 significantly decreased the affinity for ATP, these mutations significantly increased the ATP-dependent LTC4 transport activities (Fig. 1C, 4 and Table 1). These results were interpreted as that a decreased affinity for Mg·ATP and/or Mg·ATP hydrolysis product Mg·ADP in these mutants might facilitate the release of the bound nucleotide from the mutated NBDs and bring the molecule back to its original conformation so that this protein can start a new cycle of ATP-dependent solute transport [37, 38, 44].
Functional roles of the glutamine residue in Q-loop have also been studied in other ABC transporters [45–47]. Substitution of the Q82 in Q-loop of MalK, a bacterial maltose transporter, with either a lysine (Q82K) or a glutamic acid (Q82E) reduced the ATPase activities to 61% or 84% and maltose transport activities to 29% or 24% [46], indicating that introducing a charged group at this position did not abolish the ATP binding/hydrolysis and ATP-dependent maltose transport. Substitution of the Q88 in Q-loop of DrrA with a glutamic acid (Q88E) reduced the level of ATP binding [45]. However, this mutation retained the doxorubicin stimulated ATP binding [45], indicating that this mutation did not abolish the inter-domain signal communication. In contrast, however, substitution of the glutamine residue Q471 in Q-loop of NBD1 or Q1114 in Q-loop of NBD2 in mouse P-gp with either a glutamic acid or an alanine did not significantly affect ATP binding, but decreased the ATPase activities to ~ 10% [47], implying that the interaction of the amide group of the glutamine residue in Q-loop with the putative catalytic water molecule plays a crucial role for ATP hydrolysis. Furthermore, the P-gp substrates, such as verapamil or vinblastine, stimulated the ATPase activities of wt P-gp and the Q-mutants. However, the substrate-stimulated ATPase activities of the Q-mutants remained less than 10% of the wt P-gp [47], implying that the glutamine residue in Q-loop of mouse P-gp may interact with the putative hydrolytic water molecule in the active center to participate in ATP hydrolysis. Based on these results, the authors concluded that “the primary role of this glutamine residue is in inter-domain signal communication.”
The results presented in this manuscript do not support the above conclusion because the Mn·ATP-dependent LTC4 transport activities of all Q-mutants, except Q713L/Q1375L, are significantly higher than that of wt MRP1 (Fig. 4). This might reflect the structural and functional differences between the human MRP1 and the mouse P-gp. In other words, the glutamine residues in Q-loops of P-gp might interact with the residues from intracellular loops of TMDs whereas the glutamine residues in Q-loops of human MRP1 might not. To test this possibility, we have established the nucleotide-free inward-facing (Fig. S6B) or the nucleotide-bound outward-facing (Fig. S6C) MRP1 models and searched for the amino acids located within 4 Å of Q713 or Q1375. Interestingly, we have not found any residue from TMD1 or TMD2 that is located within 4 Å of Q713 or Q1375 (Tables S4, S5 and S6), regardless of whether the nucleotide-free inward-facing or the nucleotide-bound outward-facing model was used. In contrast, many residues from intracellular loop 7 in TMD2, such as K1181 or R1173, are located within 4 Å of the residues, such as E724, in Q-loop of NBD1 (Table S5) and many residues from intracellular loop 5 in TMD1, such as E521, are located within 4 Å of the residues, such as S1381 (Table S6), in Q-loop of NBD2. Of note, the relative locations of the residues may be changed in these two models. For example, R1173 was located within 4 Å of E724 in Q-loop of NBD1 in the nucleotide-free inward-facing model, whereas R1173 is not located within 4 Å of E724, instead, K1181 is located within 4 Å of E724 in the nucleotide-bound outward-facing model, perhaps reflecting the conformational changes upon ATP binding. In fact, the distance changes between a residue from the Q-loop (rather than the glutamine residue itself) and a residue from an intracellular loop of TMDs have been found in the nucleotide-free and nucleotide-bound BmrA [48], a Bacillus ABC transporter. Thus, the interactions between the residues listed in Tables S5 and S6, rather than the conserved glutamine residues in Q-loops, may play a crucial role in inter-domain signal communication in human MRP1.
Supplementary Material
Acknowledgments
We thank Irene Beauvais for processing of the manuscript and Marv Ruona for preparation of the graphics. This work was partly supported by the grant CA89078 (Xiu-bao Chang) from the National Cancer Institute, National Institutes of Health.
Abbreviations
- Sf21
Spodoptera frugiperda 21
- ABC
ATP binding cassette
- P-gp
P-glycoprotein
- BCRP
breast cancer resistance protein
- MRP1
multidrug resistance protein 1
- NBD
nucleotide binding domain
- TMD
transmembrane domain
- LTC4
leukotriene C4
- 8-N3ATP
8-azidoadenosine 5′-triphosphate
- PBS
phosphate-buffered saline
- EGTA
ethylene glycol-bis(β-aminoethyl ether) N,N,N,N-tetraacetic acid
- SDS
sodium dodecyl sulfate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Juliano RL, Ling V. A surface glycoprotein modulating drug permeability in Chinese hamster ovary cell mutants. Biochim Biophys Acta. 1976;455:152–162. doi: 10.1016/0005-2736(76)90160-7. [DOI] [PubMed] [Google Scholar]
- 2.Gros P, Croop J, Housman D. Mammalian multidrug resistance gene: complete cDNA sequence indicates strong homology to bacterial transport proteins. Cell. 1986;47:371–380. doi: 10.1016/0092-8674(86)90594-5. [DOI] [PubMed] [Google Scholar]
- 3.Chen CJ, Chin JE, Ueda K, Clark DP, Pastan I, Gottesman MM, Roninson IB. Internal duplication and homology with bacterial transport proteins in the mdr1 (P-glycoprotein) gene from multidrug-resistant human cells. Cell. 1986;47:381–389. doi: 10.1016/0092-8674(86)90595-7. [DOI] [PubMed] [Google Scholar]
- 4.Allikmets R, Schriml LM, Hutchinson A, Romano-Spica V, Dean M. A human placenta-specific ATP-binding cassette gene (ABCP) on chromosome 4q22 that is involved in multidrug resistance. Cancer Res. 1998;58:5337–5339. [PubMed] [Google Scholar]
- 5.Doyle LA, Yang W, Abruzzo LV, Krogmann T, Gao Y, Rishi AK, Ross DD. A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc Natl Acad Sci U S A. 1998;95:15665–15670. doi: 10.1073/pnas.95.26.15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mirski SE, Gerlach JH, Cole SP. Multidrug resistance in a human small cell lung cancer cell line selected in adriamycin. Cancer Res. 1987;47:2594–2598. [PubMed] [Google Scholar]
- 7.Cole SP, Bhardwaj G, Gerlach JH, Mackie JE, Grant CE, Almquist KC, Stewart AJ, Kurz EU, Duncan AM, Deeley RG. Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line [see comments] Science. 1992;258:1650–1654. doi: 10.1126/science.1360704. [DOI] [PubMed] [Google Scholar]
- 8.Ozvegy C, Varadi A, Sarkadi B. Characterization of drug transport, ATP hydrolysis, and nucleotide trapping by the human ABCG2 multidrug transporter. Modulation of substrate specificity by a point mutation. J Biol Chem. 2002;277:47980–47990. doi: 10.1074/jbc.M207857200. [DOI] [PubMed] [Google Scholar]
- 9.Kage K, Tsukahara S, Sugiyama T, Asada S, Ishikawa E, Tsuruo T, Sugimoto Y. Dominant-negative inhibition of breast cancer resistance protein as drug efflux pump through the inhibition of S-S dependent homodimerization. Int J Cancer. 2002;97:626–630. doi: 10.1002/ijc.10100. [DOI] [PubMed] [Google Scholar]
- 10.Litman T, Jensen U, Hansen A, Covitz KM, Zhan Z, Fetsch P, Abati A, Hansen PR, Horn T, Skovsgaard T, Bates SE. Use of peptide antibodies to probe for the mitoxantrone resistance-associated protein MXR/BCRP/ABCP/ABCG2. Biochim Biophys Acta. 2002;1565:6–16. doi: 10.1016/s0005-2736(02)00492-3. [DOI] [PubMed] [Google Scholar]
- 11.Han B, Zhang JT. Multidrug resistance in cancer chemotherapy and xenobiotic protection mediated by the half ATP-binding cassette transporter ABCG2. Curr Med Chem Anti-Canc Agents. 2004;4:31–42. doi: 10.2174/1568011043482205. [DOI] [PubMed] [Google Scholar]
- 12.Georges E, Tsuruo T, Ling V. Topology of P-glycoprotein as determined by epitope mapping of MRK-16 monoclonal antibody. J Biol Chem. 1993;268:1792–1798. [PubMed] [Google Scholar]
- 13.Smith PC, Karpowich N, Millen L, Moody JE, Rosen J, Thomas PJ, Hunt JF. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Molecular Cell. 2002;10:139–149. doi: 10.1016/s1097-2765(02)00576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J, Lu G, Lin J, Davidson AL, Quiocho FA. A tweezers-like motion of the ATP-binding cassette dimer in an ABC transport cycle. Mol Cell. 2003;12:651–661. doi: 10.1016/j.molcel.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 15.Moody JE, Millen L, Binns D, Hunt JF, Thomas PJ. Cooperative, ATP-dependent association of the nucleotide binding cassettes during the catalytic cycle of ATP-binding cassette transporters. J Biol Chem. 2002;277:21111–21114. doi: 10.1074/jbc.C200228200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verdon G, Albers SV, Dijkstra BW, Driessen AJ, Thunnissen AM. Crystal structures of the ATPase subunit of the glucose ABC transporter from Sulfolobus solfataricus: nucleotide-free and nucleotide-bound conformations. J Mol Biol. 2003;330:343–358. doi: 10.1016/s0022-2836(03)00575-8. [DOI] [PubMed] [Google Scholar]
- 17.Locher KP, Lee AT, Rees DC. The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science. 2002;296:1091–1098. doi: 10.1126/science.1071142. [DOI] [PubMed] [Google Scholar]
- 18.Hollenstein K, Frei DC, Locher KP. Structure of an ABC transporter in complex with its binding protein. Nature. 2007;446:213–216. doi: 10.1038/nature05626. [DOI] [PubMed] [Google Scholar]
- 19.Oldham ML, Khare D, Quiocho FA, Davidson AL, Chen J. Crystal structure of a catalytic intermediate of the maltose transporter. Nature. 2007;450:515–521. doi: 10.1038/nature06264. [DOI] [PubMed] [Google Scholar]
- 20.Gaudet R, Wiley DC. Structure of the ABC ATPase domain of human TAP1, the transporter associated with antigen processing. Embo J. 2001;20:4964–4972. doi: 10.1093/emboj/20.17.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu G, Westbrooks JM, Davidson AL, Chen J. ATP hydrolysis is required to reset the ATP-binding cassette dimer into the resting-state conformation. Proc Natl Acad Sci U S A. 2005;102:17969–17974. doi: 10.1073/pnas.0506039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer JA. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 23.Yuan YR, Blecker S, Martsinkevich O, Millen L, Thomas PJ, Hunt JF. The crystal structure of the MJ0796 ATP-binding cassette. Implications for the structural consequences of ATP hydrolysis in the active site of an ABC transporter. J Biol Chem. 2001;276:32313–32321. doi: 10.1074/jbc.M100758200. [DOI] [PubMed] [Google Scholar]
- 24.Hung LW, Wang IX, Nikaido K, Liu PQ, Ames GF, Kim SH. Crystal structure of the ATP-binding subunit of an ABC transporter. Nature. 1998;396:703–707. doi: 10.1038/25393. [DOI] [PubMed] [Google Scholar]
- 25.Procko E, Ferrin-O’Connell I, Ng SL, Gaudet R. Distinct structural and functional properties of the ATPase sites in an asymmetric ABC transporter. Mol Cell. 2006;24:51–62. doi: 10.1016/j.molcel.2006.07.034. [DOI] [PubMed] [Google Scholar]
- 26.de Wet H, McIntosh DB, Conseil G, Baubichon-Cortay H, Krell T, Jault JM, Daskiewicz JB, Barron D, Di Pietro A. Sequence requirements of the ATP-binding site within the C-terminal nucleotide-binding domain of mouse P-glycoprotein: structure-activity relationships for flavonoid binding. Biochemistry. 2001;40:10382–10391. doi: 10.1021/bi010657c. [DOI] [PubMed] [Google Scholar]
- 27.Ramaen O, Leulliot N, Sizun C, Ulryck N, Pamlard O, Lallemand JY, Tilbeurgh H, Jacquet E. Structure of the human multidrug resistance protein 1 nucleotide binding domain 1 bound to Mg2+/ATP reveals a non-productive catalytic site. J Mol Biol. 2006;359:940–949. doi: 10.1016/j.jmb.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 28.Zaitseva J, Jenewein S, Jumpertz T, Holland IB, Schmitt L. H662 is the linchpin of ATP hydrolysis in the nucleotide-binding domain of the ABC transporter HlyB. Embo J. 2005;24:1901–1910. doi: 10.1038/sj.emboj.7600657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis HA, Buchanan SG, Burley SK, Conners K, Dickey M, Dorwart M, Fowler R, Gao X, Guggino WB, Hendrickson WA, Hunt JF, Kearins MC, Lorimer D, Maloney PC, Post KW, Rajashankar KR, Rutter ME, Sauder JM, Shriver S, Thibodeau PH, Thomas PJ, Zhang M, Zhao X, Emtage S. Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator. Embo J. 2004;23:282–293. doi: 10.1038/sj.emboj.7600040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karpowich N, Martsinkevich O, Millen L, Yuan YR, Dai PL, MacVey K, Thomas PJ, Hunt JF. Crystal structures of the MJ1267 ATP binding cassette reveal an induced-fit effect at the ATPase active site of an ABC transporter. Structure (Camb) 2001;9:571–586. doi: 10.1016/s0969-2126(01)00617-7. [DOI] [PubMed] [Google Scholar]
- 31.Diederichs K, Diez J, Greller G, Muller C, Breed J, Schnell C, Vonrhein C, Boos W, Welte W. Crystal structure of MalK, the ATPase subunit of the trehalose/maltose ABC transporter of the archaeon Thermococcus litoralis. Embo J. 2000;19:5951–5961. doi: 10.1093/emboj/19.22.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zaitseva J, Jenewein S, Oswald C, Jumpertz T, Holland IB, Schmitt L. A molecular understanding of the catalytic cycle of the nucleotide-binding domain of the ABC transporter HlyB. Biochem Soc Trans. 2005;33:990–995. doi: 10.1042/BST20050990. [DOI] [PubMed] [Google Scholar]
- 33.Zaitseva J, Oswald C, Jumpertz T, Jenewein S, Wiedenmann A, Holland IB, Schmitt L. A structural analysis of asymmetry required for catalytic activity of an ABC-ATPase domain dimer. Embo J. 2006;25:3432–3443. doi: 10.1038/sj.emboj.7601208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dawson RJ, Hollenstein K, Locher KP. Uptake or extrusion: crystal structures of full ABC transporters suggest a common mechanism. Mol Microbiol. 2007;65:250–257. doi: 10.1111/j.1365-2958.2007.05792.x. [DOI] [PubMed] [Google Scholar]
- 35.Hollenstein K, Dawson RJ, Locher KP. Structure and mechanism of ABC transporter proteins. Curr Opin Struct Biol. 2007;17:412–418. doi: 10.1016/j.sbi.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 36.Pinkett HW, Lee AT, Lum P, Locher KP, Rees DC. An inward-facing conformation of a putative metal-chelate-type ABC transporter. Science. 2007;315:373–377. doi: 10.1126/science.1133488. [DOI] [PubMed] [Google Scholar]
- 37.Zhao Q, Chang XB. Mutation of the aromatic amino acid interacting with adenine moiety of ATP to a polar residue alters the properties of multidrug resistance protein 1. J Biol Chem. 2004;279:48505–48512. doi: 10.1074/jbc.M408343200. [DOI] [PubMed] [Google Scholar]
- 38.Yang R, McBride A, Hou YX, Goldberg A, Chang XB. Nucleotide dissociation from NBD1 promotes solute transport by MRP1. Biochim Biophys Acta. 2005;1668:248–261. doi: 10.1016/j.bbamem.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 39.Hou Y, Cui L, Riordan JR, Chang XB. Allosteric interactions between the two non-equivalent nucleotide binding domains of multidrug resistance protein MRP1. J Biol Chem. 2000;275:20280–20287. doi: 10.1074/jbc.M001109200. [DOI] [PubMed] [Google Scholar]
- 40.Leier I, Jedlitschky G, Buchholz U, Keppler D. Characterization of the ATP-dependent leukotriene C4 export carrier in mastocytoma cells. Eur J Biochem. 1994;220:599–606. doi: 10.1111/j.1432-1033.1994.tb18661.x. [DOI] [PubMed] [Google Scholar]
- 41.Loe DW, Almquist KC, Deeley RG, Cole SP. Multidrug resistance protein (MRP)-mediated transport of leukotriene C4 and chemotherapeutic agents in membrane vesicles. Demonstration of glutathione-dependent vincristine transport. J Biol Chem. 1996;271:9675–9682. doi: 10.1074/jbc.271.16.9675. [DOI] [PubMed] [Google Scholar]
- 42.Hou YX, Cui L, Riordan JR, Chang XB. ATP binding to the first nucleotide-binding domain of multidrug resistance protein MRP1 increases binding and hydrolysis of ATP and trapping of ADP at the second domain. J Biol Chem. 2002;277:5110–5119. doi: 10.1074/jbc.M107133200. [DOI] [PubMed] [Google Scholar]
- 43.Hou YX, Riordan JR, Chang XB. ATP binding, not hydrolysis, at the first nucleotide-binding domain of multidrug resistance-associated protein MRP1 enhances ADP.Vi trapping at the second domain. J Biol Chem. 2003;278:3599–3605. doi: 10.1074/jbc.M210480200. [DOI] [PubMed] [Google Scholar]
- 44.Chang XB. A molecular understanding of ATP-dependent solute transport by multidrug resistance-associated protein MRP1. Cancer Metastasis Rev. 2007:15–37. doi: 10.1007/s10555-007-9041-7. [DOI] [PubMed] [Google Scholar]
- 45.Rao DK, Kaur P. The Q-loop of DrrA is involved in producing the closed conformation of the nucleotide binding domains and in transduction of conformational changes between DrrA and DrrB. Biochemistry. 2008;47:3038–3050. doi: 10.1021/bi701699a. [DOI] [PubMed] [Google Scholar]
- 46.Walter C, Wilken S, Schneider E. Characterization of site-directed mutations in conserved domains of MalK, a bacterial member of the ATP-binding cassette (ABC) family [corrected] FEBS Lett. 1992;303:41–44. doi: 10.1016/0014-5793(92)80473-t. [DOI] [PubMed] [Google Scholar]
- 47.Urbatsch IL, Gimi K, Wilke-Mounts S, Senior AE. Investigation of the role of glutamine-471 and glutamine-1114 in the two catalytic sites of P-glycoprotein. Biochemistry. 2000;39:11921–11927. doi: 10.1021/bi001220s. [DOI] [PubMed] [Google Scholar]
- 48.Dalmas O, Orelle C, Foucher AE, Geourjon C, Crouzy S, Di Pietro A, Jault JM. The Q-loop disengages from the first intracellular loop during the catalytic cycle of the multidrug ABC transporter BmrA. J Biol Chem. 2005;280:36857–36864. doi: 10.1074/jbc.M503266200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.