Abstract
The Protein Arginine Deiminases (PAD), which catalyze the hydrolysis of peptidyl-arginine to form peptidyl-citrulline, play important roles in a variety of cell signaling pathways including apoptosis, differentiation, and transcriptional regulation. In addition to these important cellular roles, PAD activity is dysregulated in multiple human diseases (e.g., Rheumatoid Arthritis (RA), Cancer, and Colitis), and, significantly, PAD inhibition with Cl-amidine has been shown to reduce disease severity in the Collagen Induced Arthritis model of RA. Although these enzymes play important roles in human cell signaling and disease, the mechanisms that regulate PAD activity under both physiological and pathological conditions are poorly understood. One possible mechanism for regulating PAD activity is autodeimination, which we and others have shown that PAD4 is subject to in vitro and in vivo. Herein, we demonstrate that PAD4 autodeimination does not alter the activity, substrate specificity, or calcium dependence of this isozyme. However, the results of these studies indicate a novel role for autodeimination in modulating the ability of PAD4 to interact with Histone Deacetylase 1 (HDAC1), citrullinated histone H3 (Cit H3), and Protein Arginine Methyltransferase 1 (PRMT1).
Protein arginine deiminases (PADs) are a small group of protein modifying enzymes that catalyze the hydrolysis of arginine residues to citrulline (Figure 1). This particular modification has gained much attention in recent years because of its apparent role in rheumatoid arthritis (RA) and, more recently, in cancer, multiple sclerosis, and colitis (1). Although multiple PAD isozymes (e.g., PADs 2 and 4) likely play a role in the aforementioned diseases, the fact that mutations in PAD4 confer an increased risk of developing RA in Asians, as well the fact that PAD4 is overexpressed in the disease associated tissues (2, 3), has focused most research, at the both the molecular and organismic level, on this isozyme. For example, inhibition of PAD activity with Cl-amidine, an inhibitor developed by our lab (4), has been shown to reduce disease severity in the murine Collagen Induced Arthritis model of RA (5). Cl-amidine treatment or siRNA knockdown of PAD4 has also been shown to induce either apoptosis or differentiation of a number of cancer derived cell lines, including U2OS osteosarcoma cells (apoptosis), HL60 leukemic cells (differentiation), and HT29 colon cancer cells (differentiation) (6–9). Additionally, PAD4 activity appears to play an important role in the innate immune response as the activity of this isozyme is required for the formation of Neutrophil Extracellular Traps (NETs) (10–13). Finally, PAD4 is known to be recruited to the promoters of numerous genes where it deiminates histones H3 and H4, and this modification is associated with the decreased transcription of those genes via an as yet to be defined mechanism (6, 7, 14, 15).
Figure 1.
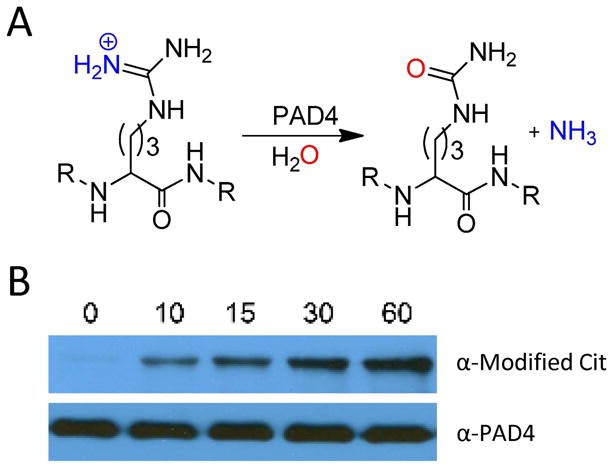
(A) PAD4 catalyzed reaction. (B) Autodeimination of PAD4. Recombinant PAD4 (0.2 μM) was incubated in the presence of 10 mM calcium at 37 °C over a range of time points (0, 10, 15, 30, and 60 min). Citrullinated PAD4 was detected by western blot using the Anti-Modified Citrulline Detection Kit (top), and anti-PAD4 was used in the detection of PAD4 to demonstrate equal protein loading (bottom).
Although our knowledge of the physiological roles of the PADs is improving, particularly with the development of inhibitors and chemical probes targeting these enzymes (4, 16–21), our understanding of the mechanisms that regulate PAD activity are less clear, even though these enzymes are known to be calcium dependent. This is particularly true when one considers the fact that high micromolar to millimolar concentrations of calcium are required to observe PAD activity in vitro. Such concentrations are far higher than the nanomolar to low micromolar concentrations of calcium found in a cell, where the PADs are clearly active (20). Thus, it seems likely that additional factors regulate PAD activity in vivo. Potential higher level regulatory mechanisms that could modulate PAD activity, as well as the amount of calcium required for in vivo activity, include both PAD interacting proteins and PTMs. The latter possibility seems particularly plausible when one considers the fact that numerous cell signaling proteins are subject to multiple PTMs (e.g., phosphorylation, acetylation, and methylation) that regulate protein-protein interactions, enzymatic activity, and cellular localization. Given the putative role of PAD4 in human disease, understanding the mechanisms that regulate its deiminating activity, under both physiological and pathological conditions, is critical for a complete understanding of PAD4 function; as such knowledge will ultimately assist in the development of inhibitors targeting this enzyme.
Quite often, protein modifying enzymes also modify themselves. As we and others (22, 23) have shown, PAD4 is subject to autodeimination (Figure 1). As this modification could potentially regulate PAD activity, substrate specificity, calcium dependence, protein-protein interactions, and protein stability, we sought to understand the regulatory effects of PAD4 autodeimination at the molecular level. Herein we report the identification of 7 sites of autodeimination that occur both in vitro and in vivo. Site directed mutagenesis was then used to probe the roles of these residues in regulating PAD4 activity, substrate specificity, calcium dependence, and protein-protein interactions. In contrast to a recent report (22), the results of these studies indicate that autodeimination has minimal effects on the activity, substrate specificity, and calcium dependence of PAD4. However, we show for the first time that autodeimination alters the ability of PAD4 to bind to HDAC1, citrullinated histone H3 (Cit H3), and Protein Arginine Methyltransferase 1 (PRMT1). In addition, we also investigated the effects of the RA-associated PAD4 haplotype, which consists of 4 exonic single nucleotide polymorphisms that result in 3 amino acid substitutions: S55G, A82V, and A112G (the fourth polymorphism is silent). The results of these studies indicate that these substitutions also do not affect the activity, substrate specificity, or calcium dependence of PAD4. On the other hand, the SNPs appear to increase the affinity of PAD4 for Cit H3, H3 and HDAC1.
Materials and Methods
Chemicals
Dithiothreitol (DTT), iodoacetic acid, protease inhibitor cocktail (Cat#P8465), and Benzoyl L-arginine ethyl ester (BAEE) were acquired from Sigma-Aldrich (St. Louis, MO). Wild type recombinant human PAD4 and wild type recombinant human PRMT1 were purified using previously described methods (24, 25). Histones H3 and H4 were purified as previously described (26). The synthesis and characterization of Biotin-conjugated F-amidine (BFA), F-amidine-YNE, and TEV-biotin-azide have previously been described (20).
Construction of PAD4 mutants and purification of recombinant proteins
The Quik Change Mutagenesis Kit (Stratagene) was used to introduce point mutations into the PAD4 gene. The sequences of the primers, which were obtained from IDT DNA Technologies, are listed in Table S1. The entire open reading frame of each mutant gene was sequenced to ensure that the appropriate mutation was obtained without incorporation of undesired mutations. Expression and purification of the recombinant PAD4 mutants was carried out as previously described (24, 27).
Purification of HDAC1 Fragment 1–90
A recombinant Escherichia coli expression system encoding residue 1–90 of human HDAC1 (HDAC1 1–90) was the kind gift of Dr. Francois Fuks. Briefly, the HDAC1 1–90 expression construct, which encodes the first 90 residues of human HDAC1 fused in frame to an N-terminal GST-tag, was transformed into E. coli Rosetta cells (EMB Biosciences). A single colony was used to prepare starter cultures, which were used to inoculate 1L of TB media containing ampicillin (50 μg/mL) and chloramphenicol (20 μg/mL) in a 4 L baffled flask. Cells were incubated at 37 °C with shaking (250 rpm) until an OD600 of 0.8 was reached. The culture was cooled to 16 °C and IPTG (0.3 mM final) was added to induce protein expression. After overnight incubation, the cells were harvested by centrifugation (5000 rpm, 10 min). The cell pellet was resuspended in 30 mL of lysis buffer (20 mM Tris-HCl pH 8.0, 1 mM EDTA, 1 mM DTT, 400 mM NaCl, 20% glycerol, and protease inhibitor cocktail) and incubated for 30 min at 4 °C with gentle stirring. The cell suspension was then diluted with an additional 70 mL of lysis buffer and lysed by sonication (12 cycles for 15 sec burst, duty cycle 10, 100% output with 60 sec intervals). Cellular debris was removed by centrifugation (14,000 rpm, 30 min) and the supernatant was applied to a glutathione-sepharose fast flow affinity column (GE Healthcare). The column was washed with 50 mL of low salt buffer (20 mM Tris-HCl pH 8.0, 1 mM EDTA, 1 mM DTT, 250 mM NaCl, and 10% glycerol), followed by 50 mL of high salt buffer (20 mM Tris-HCl pH 8.0, 1 mM EDTA, 1 mM DTT, 500 mM NaCl, and 10% glycerol). GST-HDAC1-1-90 was eluted from the column with 25 mL of glutathione buffer (50 mM Tris-HCl pH 8.0, 1 mM DTT, and 10 mM reduced glutathione). Protein was dialyzed against 20 mM Tris-HCl pH 7.6, 1 mM EDTA, and 2 mM DTT to remove any remaining glutathione. Dialyzed protein was applied to a S200 size exclusion column (GE Healthcare) and fractions were collected and analyzed by SDS-PAGE. GST-HDAC1-1-90 was pooled and dialyzed against long term storage buffer (20 mM Tris-HCl pH 8.0, 2 mM DTT, 500 mM NaCl, and 10% glycerol), aliquoted, and stored at −80 °C.
Kinetic characterization of mutant enzymes
All enzymatic assays were carried out as previously described (24). Briefly, assays were performed in Reaction Buffer containing 100 mM Tris-HCl pH 7.6, 50 mM NaCl, 2 mM DTT, and 10 mM CaCl2 (60 μL final volume). The reaction buffer containing the appropriate substrate (e.g., H4, H3, or BAEE) was preincubated for 10 min at 37 °C, followed by the addition of recombinant enzyme (0.2 μM final) to initiate the reaction. Citrulline production was quantified as previously described (24, 28). Experiments were performed at least in duplicate and the values generally agreed to within 20%. The kinetic values (e.g., kcat and Km) were obtained by fitting the initial velocity data to the Michaelis-Menten equation 1,
| (1) |
using Graphit software version 5.0.11 (29).
Calcium dependence
The calcium dependence of PAD4 mutants was characterized using methods that have previously been established for wild type PAD4 (24). Briefly, Reaction Buffer, containing 100 mM Tris-HCl pH 7.6, 50 mM NaCl, 2 mM DTT, 10 mM BAEE, and varying concentrations of CaCl2 (0–10 mM), was preincubated for 10 min at 37 °C, followed by the addition of recombinant enzyme (0.2 μM final) to initiate the reaction. After 15 min, the reactions were quenched and citrulline production was quantified as previously described (24). The data obtained were fit to equation 2,
| (2) |
using GraphIt version 5.0.11 (29), where v is the initial rate, Vmax is the maximum rate, and K0.5 is the concentration of calcium that yields half maximal activity.
Autodeimination of PAD4
Recombinant proteins were incubated in Reaction Buffer in the absence (no autodeimination) or presence (autodeimination) of 10 mM CaCl2 at 37 °C and analyzed over a range of time points (0 to 60 min). Following this incubation, samples were boiled in SDS-PAGE loading buffer, and subjected to electrophoresis on a 10% SDS-PAGE gel for 60 min at 200 V. Proteins were then electrotransferred onto a polyvinylidene difluoride (PVDF) membrane for 70 min at 80 V. Detection of autodeiminated PAD4 was accomplished using the Anti-Modified Citrulline Detection Kit (Upstate, Temecula, Ca).
Partial Proteolysis experiments
The partial proteolysis of wild-type PAD4 and PAD4 mutants was carried out as previously described (24). Briefly, wild-type PAD4 and PAD4 mutants (0.2 μM) were incubated in the absence or presence of subtilisin (3.3 μg/mL) in Reaction Buffer on ice over a range of time points (0–60 min). The proteolysis reaction was quenched by the addition of phenylmethanesulfonyl fluoride (5 mM final). Proteolytic fragments were separated by SDS-PAGE, and visualized with coomassie brilliant blue staining.
Co-immunoprecipitations
Wild type PAD4 and PAD4 mutants (5 μg) were labeled with 1 μM BFA for 30 min at 37 °C in Reaction Buffer. Labeled protein (5 μg) was added to MCF-7 whole cell e xtracts (500 μg total protein) and applied to streptavidin-agarose beads (100 μL) and allowed to incubate overnight at 4 °C with end over end rocking. The beads were washed three times with PBS followed by three times with NET-2 buffer (150 mM NaCl, 0.05% NP-40, and 50 mM Tris-HCl pH 7.4). Beads were boiled in SDS buffer (2% SDS, 62.5 mM Tris-HCl pH 6.8, and 10% glycerol), and then the eluted proteins were separated on a 12% SDS-PAGE gel and subsequently transferred to a nitrocellulose membrane. Membranes were probed with a mouse monoclonal anti-p53 antibody (Abcam, ab1101), a rabbit polyclonal anti-H3 antibody (Abcam, ab1791), anti-PRMT1 (Abcam, ab7027), anti-HDAC1 (Abcam, ab19845), and an anti-citrullinated H3 antibody (Abcam, ab77164).
Stimulation of MCF7 cells
MCF-7 cells were grown and maintained to 90% confluence in DMEM containing 10% FBS at 37 °C, 5% CO2, at which point the media was exchanged with DMEM phenol red free containing 10% charcoal stripped FBS. After 48 h incubation at 37 °C and 5% CO2, estrogen (0.1 μM) was added to stimulate the cells. After a 2 h incubation with estrogen, cells were rinsed in PBS and lysed in modified RIPA buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, and 1% sodium deoxycholate) containing a complete protease inhibitor tablet (Roche).
Identification of autodeimination sites in vitro
Purified recombinant PAD4 (100 μg) was deiminated at 37 °C for 2 h in a standard reaction buffer containing 100 mM Tris-HCl pH 7.6, 50 mM NaCl and 2 mM DTT in the presence or absence of 10 mM CaCl2. For reactions performed in 18O-labelled water, the PAD4 protein was lyophilized overnight, reconstituted in 18O-labelled water, and incubated in reaction buffer under similar conditions. Autodeiminated sites were determined from a tryptic digest of the autodeiminated samples followed by mass spectrometry analysis on a Ultraflex MALDI-TOF (Bruker Daltonics, Germany). Briefly, autodeiminated samples were concentrated to 25 μL using micro spin columns (10000 MWCO, Millipore) and mixed with 50 μL of denaturing buffer (6 M Urea, 0.1 M MOPS and 1 mM EDTA, pH 7.2). Protein was reduced and derivatized with 5 μM DTT (37 °C, 2 h) and 25 μM iodoacetamide (rt, 1 h) respectively. All reactions were performed on column and the sample was mixed well by thorough pipetting before each step. In order to reduce the urea concentration, the buffer was washed off by centrifugation (5000 × g) and the column was then rinsed twice with 100 μL of water. Sequence-grade trypsin (Promega), which was reconstituted in 100 mM NH4HCO3 pH 8.0, was then added at a final enzyme:protein ratio of 1:20. Digestion was carried out overnight at 37 °C and peptides were collected in the flow through in a fresh collection tube by centrifugation at 5000 × g for 20 min. Peptides were concentrated in a speedvac to a final volume of ~30 μL, desalted using a C18 ZipTip (Millipore), and eluted in 2.5 μL of α-cyano hydroxycinnamic acid for analysis.
Identification of autodeimination sites in vivo
HL-60 cells were differentiated along the granulocyte lineage with 1 μM all-trans retinoic acid for 48 h. HL-60 granulocytes were incubated with F-amidine-YNE (100 μM final) in the presence of a calcium ionophore, A23817 (4 μM) for 1 h at 37 °C and 5% CO2. Subsequently the cells were harvested and lysed in modified RIPA buffer (25 mM Tris HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate) containing complete protease inhibitor tablet (Roche) on ice, for 10 min. The samples were denatured at 95 °C for 10 min prior to the addition of TEV-biotin-azide (10 μM) reporter tag. The ‘click’ reaction was then initiated by the addition of TCEP (2.5 mM), ligand (Tris[(1-benzyl-1H-1,2,3,-triazol-4-yl)methyl]amine) (0.119 mM final), and CuSO4 (5 mM final), as previously described (20, 30). After 1 h at rt, the protein samples were collected by centrifugation (5000 rpm, 4 °C), and the protein pellet was washed with cold methanol (2 × 100 μL). The protein pellet was resuspended in 0.2% SDS/PBS and added to 100 μL of streptavidin-agarose beads in PBS and incubated overnight at 4 °C with end over end rocking. The beads were washed by adding 500 μL of 0.2% SDS/PBS for 10 min and collected by centrifugation (1,400 × g, 3 min). The supernatant was removed and the beads were washed with PBS (3 × 5 mL). In order to elute the bound proteins, streptavidin-agarose beads were incubated in buffer A (2% SDS, 15 mM biotin, 100 mM thiourea, and 3 M urea in PBS) for 1 h at 42 °C followed by 30 min at 95 °C. The supernatant was then removed, and the beads were washed with water three times. The supernatant and wash factions were combined and concentrated to approximately 20 μL. The entire sample was loaded onto a 12% SDS-PAGE gel and proteins were visualized by coomassie blue staining. A protein band corresponding to the approximate molecular weight of PAD4 was excised from the gel and subjected to an in-gel tryptic digestion. In-gel digestion was performed as previously described (31). Peptides were concentrated in a speedvac to a final volume of ~30 μL, desalted using a C18 ZipTip (Millipore), and eluted in 2.5 μL of α-cyano hydroxycinnamic acid for analysis on an Ultraflex MALDI-TOF (Bruker Daltonics, Germany).
Results and Discussion
Autodeimination of PAD4 in vitro
Many enzymes that generate PTMs also modify themselves as a mode of regulating their enzymatic activity. Given this fact, we were interested in determining whether PAD4 autodeiminates. For these studies, wild type recombinant PAD4 was incubated in the absence (t = 0 min) or presence of calcium (t = 10, 15, 30, and 60 min) for various lengths of time, and the presence of citrullinated residues was detected by western blotting using an anti-modified citrulline antibody. The results of these studies indicated that PAD4 is autodeiminated in a time dependent manner with near maximal deimination occurring at the 60 min time point (Figure 1B). These results are consistent with those obtained by Andrade et. al (22).
Effects of autodeimination on PAD4 activity
To investigate the effects of autodeimination on PAD4 activity, the steady-state kinetic parameters were determined for a variety of small molecule and protein substrates (i.e., histones H3 and H4, as well as BAEE), using both recombinant PAD4 and autodeiminated PAD4 (adPAD4); adPAD4 was generated by incubating the enzyme in the presence of 10 mM calcium for 1 h. To control for the effects of incubation time on PAD4 activity, we also incubated PAD4 in the absence of calcium for 1 h to generate control PAD4 (conPAD4). As shown in Table 1, the kinetic parameters obtained for BAEE, H3, and H4 with adPAD4 are generally comparable to those obtained with PAD4 and conPAD4; the effects on kcat/Km are ≤ 2.3-fold. The one exception to this trend is the result obtained with H3. Here, incubation in the absence of calcium led to a 4.7-fold reduction in kcat/Km. However, when the enzyme was incubated for 1 h in the presence of calcium, little to no change in kcat/Km, relative to PAD4, was observed. The reasons for this small, but nonetheless significant, loss in activity for H3 are unclear, but overall the results are inconsistent with the notion that autodeimination negatively impacts PAD4 activity.
Table 1.
Steady-state kinetic parameters of autodeiminated PAD4
| 1 h autodeimination | 2 h autodeimination | |||||
|---|---|---|---|---|---|---|
| kcat (s−1) | Km (mM) | kcat/Km (s−1 M−1) | kcat (s−1) | Km (mM) | kcat/Km (s−1 M−1) | |
| H4 | ||||||
| WT PAD4 | 2.1 ± 0.6 | 0.21 ± 0.10 | 10000 | 2.1 ± 0.6 | 0.21 ± 0.10 | 10000 |
| adPAD4 | 0.6 ± 0.1 | 0.10 ± 0.04 | 6400 | ND | ND | 3000 |
| conPAD4 | 1.0 ± 0.4 | 0.23 ± 0.13 | 4300 | ND | ND | 3000 |
|
| ||||||
| H3 | ||||||
| WT PAD4 | ND | ND | 3700 | ND | ND | 3700 |
| adPAD4 | ND | ND | 3500 | ND | ND | 3100 |
| conPAD4 | ND | ND | 790 | ND | ND | 780 |
|
| ||||||
| BAEE | ||||||
| WT PAD4 | 2.8 ± 0.6 | 0.58 ± 0.12 | 4800 | 2.8 ± 0.6 | 0.58 ± 0.12 | 4800 |
| adPAD4 | 2.7 ± 0.1 | 0.91 ± 0.14 | 3000 | 1.0 ± 0.1 | 3.8 ± 0.6 | 250 |
| conPAD4 | 2.6 ± 0.1 | 0.69 ± 0.07 | 3800 | 0.9 ± 0.1 | 2.5 ± 0.89 | 340 |
Recombinant wild-type (WT) PAD4 (0.2 μM) was incubated in the absence (conPAD4) or presence (adPAD4) of 10 mM calcium for 1 or 2 h at 37 °C. The kinetic parameters were then determined by incubating enzyme with histone H4, H3, and BAEE over a range of concentrations at 37 °C for 6 min. WT PAD4 = non-incubated protein; adPAD4 = autodeiminated PAD4, conPAD4 = non-autodeiminated incubated control; and ND = Not Determined
Given that Andrade et. al have previously suggested that the autodeimination of PAD4 inhibits its deiminating activity (22), we were understandably concerned with our results. The major differences between our two studies are: i) the incubation time (2 h versus the 1 h used here); and ii) the choice of substrate (Andrade et. al used HL60 cell extracts as PAD4 substrates and detected changes in protein citrullination using the anti-modified citrulline antibody). To determine whether incubation time could explain the discrepancy, the kinetic parameters were determined for adPAD4 and conPAD4 after a 2 h incubation in the presence and absence of calcium. The results of these studies (Table 1) indicate that there was little to no difference in the kinetic parameters obtained for adPAD4 and conPAD4 that were incubated for 2 h. The one exception is again the case of H3, where the kcat/Km for H3 with conPAD4 is ~4-fold lower than that obtained with adPAD4. Again, while the reason for this discrepancy is not known, the effect is relatively small. It is interesting to note, however, that significant activity is lost after incubating the enzyme for 2 h as compared to WT PAD4. For example, a pronounced decrease in kcat/Km was observed with both H4 (~3.3 fold) and BAEE (~14–19 fold) after the 2 h incubation (compare the adPAD4 and conPAD4 data to the non-incubated control) (Table 1). The fact that both adPAD4 and conPAD4 are less active after the 2 h incubation suggests that the loss of activity is of a non-specific nature. This seems likely given the fact that the loss of activity is less pronounced at the 1 h time point. Taken together, these results indicate that autodeimination of PAD4 has little to no effect on the steady-state kinetic parameters of PAD4, at least in vitro.
Given that Andrade et. al had used whole cell extracts as the source of proteins for their activity measurements, we hypothesized that a protein present in those extracts, or some other unknown component, could potentially be acting to inhibit PAD4 activity. To examine this possibility, we prepared whole cell extracts from unstimulated and estrogen stimulated MCF7 cells – MCF7 cells are a breast adenocarcinoma derived cell line that is known to express PAD4 – and examined the effects of these extracts on the kinetic parameters of adPAD4 and conPAD4. The results of these studies (Figure S1 and Table S2) indicate that neither extract altered the activity of PAD4 as the kcat/Km values obtained with adPAD4 and conPAD4 are virtually identical. Thus, there appears to be no endogenous factor in MCF-7 cell extracts that can inhibit PAD4 activity. Although it is unclear why we see such dramatically different results, it is worth noting that Andrade et. al used very low amounts of β-mercaptoethanol in their incubation buffers and the observed loss of activity may be due to oxidation of the active site Cys, as we and others have previously shown that enzyme activity is highly dependent on the presence of a reducing agent (24, 28).
Effect of autodeimination on the calcium dependence of PAD4
Given that PAD4 requires millimolar amounts of calcium for activity in vitro (24), but only nanomolar to low millimolar levels of calcium are found in vivo, we were curious to see whether autodeimination could alter the calcium-dependence of this enzyme. For these studies, adPAD4 was generated by incubating the enzyme in Reaction Buffer plus 10 mM calcium for either 1 h or 2 h. The excess calcium was then removed by dialysis, and then PAD4 activity was measured as a function of increasing calcium. Since the activity versus calcium concentration plots are sigmoidal, the data were fit to the Hill equation to provide values for the Hill coefficient (n) and K0.5 for calcium activation (Figure S2). The results of these studies indicated that when PAD4 was autodeiminated for 1 h no significant effect was present on either the K0.5 or the hill coefficient (K0.5 = 407 ± 106 μM, n = 2.3 ± 0.37) compared to non-incubated control (K0.5 = 500 ± 91 μM, n = 1.5 ± 0.37). Similar results were obtained when PAD4 was autodeiminated for 2 h (K0.5 = 1200 ± 170, n = 2.0 ± 0.37). Although the K0.5 is increased by 2.4-fold with this treatment, the effect is relatively small and inconsistent with the hypothesis that autodeimination could decrease the concentration of calcium required for PAD4 activity. Taken together, the data suggests that autodeimination has little to no effect on the calcium dependence of PAD4.
Identification of the sites of autodeimination
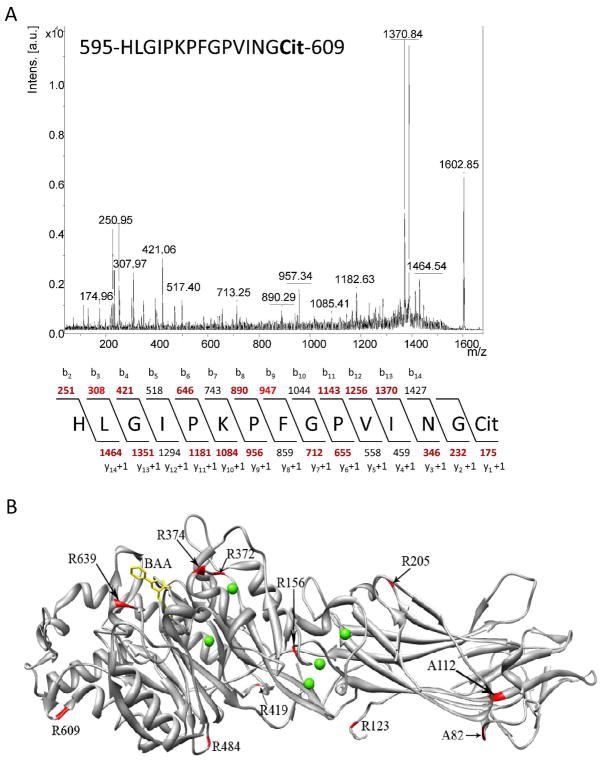
MS analyses were used to identify the sites of PAD4 autodeimination, both in vitro and in vivo. To identify the in vitro sites of deimination, recombinant PAD4 was either reconstituted in 18O-labelled water and autodeiminated for 2 h or autodeiminated directly for 2 h. Subsequently, adPAD4 was subjected to in solution tryptic digestion and MS and MS/MS analyses. Note that these experiments were performed in 18O-labelled water so as to enhance our ability to identify the sites of autodeimination because deimination in normal water results in only a 0.98 Da mass increase whereas in 18O-labelled water an ~3 Da increase is observed. The results of these studies identified six in vitro sites of autodeimination, i.e. R123, R156, R205, R419, R484, and R639 (Table S3). Note that sequence coverage was 64%. To identify the sites of autodeimination that occur in vivo, we isolated endogenous PAD4 from HL-60 granulocytes utilizing a recently described PAD4-targeted ABPP, i.e., F-amidine-YNE (20). For these studies, F-amidine-YNE (100 μM final), in the presence of a calcium ionophore (i.e., A23817; 4 μM final), was added to HL60 granulocytes for 1 h. Subsequently the cells were harvested, lysed, and then denatured by boiling at 95 °C for 10 min. The TEV-biotin-azide reporter tag was then added in the presence of TCEP, ligand, and copper sulphate to afford the coupling of the reporter tag to F-amidine-YNE via the copper catalyzed azide-alkyne [3 + 2] cycloaddition reaction. Biotinylated proteins were then isolated on streptavidin agarose, eluted, and separated by SDS-PAGE. A band corresponding to the size of PAD4 was excised from the gel and subjected to in gel tryptic digestion. The results of these studies identified two in vivo sites (i.e., R123 and R609) of autodeimination (see Figure 2A for representative MS data). As depicted in Figure 2B, the majority of the autodeiminated arginine residues clustered in the C-terminal catalytic domain (i.e., R419, R484, R609, and R639), while the remaining three arginines are located in the N-terminus of the protein (i.e., R123, R156, and R205). Note that all of these residues are not directly accessible to the active site; thus, the deimination of these residues must occur in trans. Also note that while Andrade et. al identified 10 sites of autodeimination (i.e., R205, R212, R218, R372, R374, R383, R394, R495, R536, and R544), only one of these sites, i.e. R205, is shared between these two analyses. The reason for this discrepancy is unknown, but may be related to the different methods of detection (LC-MS and MS/MS versus MALDI-TOF/TOF) or the fact that we do not observe an effect of autodeimination on PAD4 activity.
Figure 2.
(A) Representative mass spectrometry data for autodeimination sites found in PAD4 (B) Location of PAD4 AD sites and SNPs. The crystal structure was generated using the UCSF Chimera software package (37) and PDB file 1WDA (32). The location of the deiminated arginines and the SNPs are shown in red on the ribbon structure. Bound calcium ions (ball-shaped) are shown in green and BAA (benzoyl-L-arginine amide) is shown in yellow.
Site directed mutagenesis
Site directed mutagenesis was next used to investigate the contribution of these autodeiminated residues to the in vitro activity and calcium dependence of PAD4. In order to mimic the effects of arginine deimination, R123, R156, R205, R419, R484, R609, and R639, as well as R372 and R374 (previously described sites of autodeimination (22)), were mutated to glutamine and the steady state kinetic parameters determined for BAEE and histone H4. Although the side chain of glutamine is ~2 methylene units shorter than the side chain of citrulline, we considered that the carboxamide moiety present on glutamine would provide a reasonable mimic of the ureido group in citrulline as it possesses similar hydrogen bond acceptors and donors. These residues were also mutated to lysine to serve as non-deiminatable controls. As shown in Table 3, only minor effects on the kinetic parameters are observed when R123, R156, R205, R484, R609, R639, and R374 are mutated to either a glutamine or a lysine. These results are consistent with our results demonstrating that autodeimination has only minor effects on PAD4 activity. The two exceptions are the data obtained for the R372 mutants and the R419Q mutant when BAEE is used as the substrate (see below). The effect of these arginine substitutions on the calcium dependence of PAD4 was also analyzed. For the R156, R374, R484, and R609 mutants, only very minor effects on the Hill coefficient and K0.5 for calcium were observed. The exceptions are the data obtained for R123, R205, R419, and R639. The effects of mutating these residues, as well as R372, on the calcium dependence and activity of PAD4 are described below. Note that partial proteolysis experiments were performed on all of the PAD4 mutants described. The fact that these mutants behaved identically to the wild type enzyme in these assays indicates that the observed changes in activity are not due to gross structural perturbations. Also note that while we generally define minor effects as being ≤2-fold, we recognize that such small effects in vitro could potentially have profound effects in vivo. Nevertheless, to limit speculation we have generally focused our discussion on only those changes that are ≥2-fold.
Table 3.
Calcium dependence of PAD4 mutants
| N | K0.5 (μM) | |
|---|---|---|
| wild-type | 1.5 ± 0.19 | 500 ± 91 |
| R123K | 2.2 ± 0.25 | 106 ± 29 |
| R123Q | 4.5 ± 0.55 | 310 ± 66 |
| R156K | 1.4 ± 0.36 | 410 ± 150 |
| R156Q | 2.2 ± 0.24 | 550 ± 83 |
| R205K | 3.8 ± 0.61 | 105 ± 46 |
| R205Q | 2.9 ± 0.65 | 109 ± 61 |
| R372K | ND | ND |
| R372Q | ND | ND |
| R374K | 2.8 ± 0.51 | 230 ± 83 |
| R374Q | 1.8 ± 0.14 | 420 ± 51 |
| R419K | 2.4 ± 0.26 | 66 ± 23 |
| R419Q | 2.1 ± 0.56 | 300 ± 140 |
| R484K | 2.6 ± 0.22 | 320 ± 47 |
| R484Q | 2.6 ± 0.37 | 380 ± 89 |
| R609K | 1.9 ± 0.39 | 480 ± 140 |
| R609Q | 1.7 ± 0.25 | 530 ± 106 |
| R639K | 2.4 ± 0.77 | 85 ± 75 |
| R639Q | 3.2 ± 0.81 | 100 ± 67 |
Values were obtained by incubating protein (0.2 μM) and BAEE (10 mM) over a range of calcium concentrations (0–10 mM) at 37 °C for 10 min. ND = not determined
R123
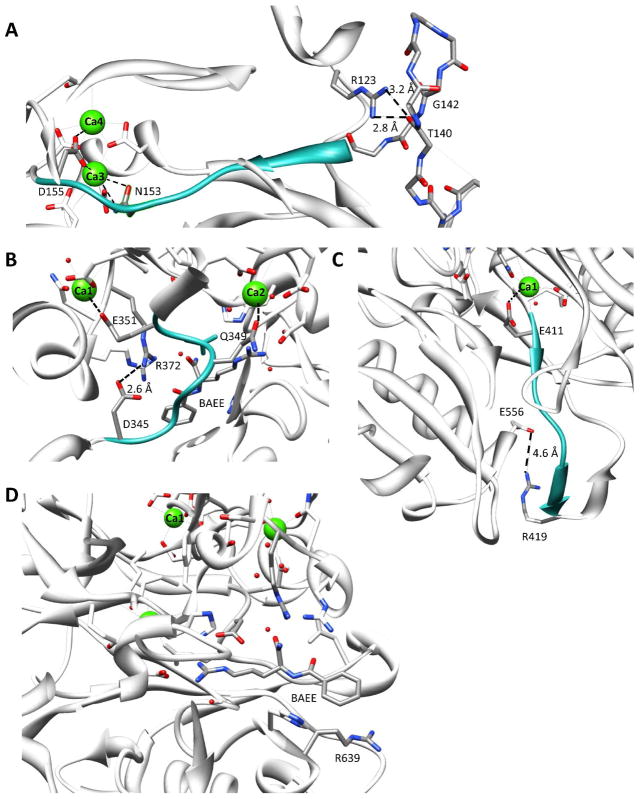
R123 is present on the tip of a loop in subdomain 1 (Figure 3A) and likely stabilizes a second loop encompassing residues 138-146 via hydrogen bond interactions with the backbone carbonyls of T140 (r = 2.8 Å) and G142 (r = 3.2 Å). As deimination of this residue would be expected to disrupt these interactions, one might expect that this would lead to the destabilization of this loop. Given that this loop directly connects to the region of the enzyme that binds to calciums 1, 2, 3, one might further expect that its deimination would alter the calcium dependence of the enzyme. Consistent with this notion is the fact that the K0.5 for calcium obtained for the R123Q mutant (K0.5 = 310 ± 66 μM) is significantly higher than that obtained with the R123K mutant (K0.5 = 106 ± 29 μM), which mimics a non-deiminatable arginine. While the K0.5 of the R123K mutant is significantly lower than that observed for the wild type enzyme (~5-fold), it remains higher than physiologically relevant. Thus, deimination of this residue cannot fully explain the in vitro requirement for such high levels of calcium.
Figure 3.
Structural basis for the effects of autodeimination on the activity and calcium dependence of PAD4. (A) R123: This image shows a close up of the region surrounding R123 and highlights its proximity to calciums 3 and 4 and its interaction with the backbone carbonyl of T140. (B) R372: This image highlights the interactions between R372 and D345 and how disruption of this interaction might alter the position of D350, a key catalytic residue. (C) R419: Proximity of R419 to E556. This panel shows a close up of the R419·E556 interaction. Disruption of this interaction would be expected to disrupt the stability of the β-strands linked to E411, a ligand for calcium 1. (D) R639: This panel highlights the position of R639 in the structure of the PAD4·calcium·BAA complex. Note that calcium ions are green balls and BAA is shown in elemental colors. These figures were generated using the UCSF Chimera software package (37) and PDB file 1WDA.
R205
Mutation of R205 to either a lysine or a glutamine decreases the kcat/Km for histone H4 and BAEE by 4.8-fold and 2.7-fold, respectively. Additionally, these mutations reduce the K0.5 for calcium by ~5-fold. Although the molecular basis for these effects is not known, it may be due to effects on dimerization as R205 is present on a short loop connecting β-strand 12 in subdomain 2 to helix α4, and appears to form hydrogen bonds with the backbone carbonyl and amide groups of S437 the second subunit in the PAD4 dimer. As such, the inability to form the correct hydrogen bonding patterns, which would be expected with both mutants, would be expected to at least partially destabilize the PAD4 dimer.
R372
Although we did not observe autodeimination of R372, we were interested in determining the importance of this residue because Andrade et. al had suggested that this residue is deiminated and that its mutation to a lysine abolished PAD4 activity. Consistent with previous results (22, 32), the conversion of R372 to either a lysine or glutamine resulted in a >830-fold decrease in kcat/Km when either BAEE or histone H4 was tested as the substrate (Table 2). Given the magnitude of these effects, which essentially dropped the activity to baseline, it was not possible to examine the effect of these mutations on the calcium dependence of PAD4. Based on the structure of PAD4 there are at least two possible reasons for the loss in activity. First, R372 forms a water mediated hydrogen bond to the backbone carbonyl C-terminal to the site of deimination (Figure 3B), and loss of this interaction would be expected to increase the Km for the substrate. Second, when bound to calcium, the side chain guanidinium of R372 forms an electrostatic interaction with D345 that likely helps position D350, an essential active site residue (27), so that it can bind and orient the substrate guanidinium for nucleophilic attack by the active site cysteine, i.e. C645. This interaction likely occurs in a calcium dependent manner because N373, which is one residue C-terminal to R372, acts a ligand for calcium 2. Given the magnitude of the effect of mutating this residue on PAD4 activity, which is on the order of mutating D350, the latter explanation seems most likely.
Table 2.
Steady-state kinetic parameters of PAD4 mutants
| H4 | BAEE | |||||
|---|---|---|---|---|---|---|
| kcat (s−1) | Km (mM) | kcat/Km (s−1M−1) | kcat (s−1) | Km (mM) | kcat/Km (s−1M−1) | |
| wild-type | 2.1 ± 0.64 | 0.21 ± 0.10 | 10000 | 2.8 ± 0.58 | 0.58 ± 0.12 | 4800 |
| R123K | 1.7 ± 0.52 | 0.24 ± 0.11 | 7100 | 3.7 ± 0.06 | 0.86 ± 0.05 | 4300 |
| R123Q | 0.90 ± 0.15 | 0.10 ± 0.03 | 9000 | 2.3 ± 0.12 | 1.4 ± 0.24 | 1600 |
| R156K | 1.4 ± 0.27 | 0.18 ± 0.06 | 7800 | 0.76 ± 0.03 | 0.50 ± 0.08 | 1500 |
| R156Q | 0.79 ± 0.17 | 0.11 ± 0.05 | 7200 | 3.6 ± 0.27 | 1.6 ± 0.37 | 2200 |
| R205K | ND | ND | 3700 | 3.8 ± 0.21 | 1.5 ± 0.27 | 2500 |
| R205Q | ND | ND | 2100 | 1.2 ± 0.04 | 0.48 ± 0.07 | 2500 |
| R372K | ND | ND | ≤ 3.1 | ND | ND | ≤ 0.6 × 10−6 |
| R372Q | ND | ND | ≤ 12.0 | ND | ND | ≤ 1.0 ×10−6 |
| R374K | 0.49 ± 0.08 | 0.088 ± 0.03 | 5600 | 3.0 ± 0.18 | 1.7 ± 0.31 | 1800 |
| R374Q | 0.77 ± 0.05 | 0.15 ± 0.02 | 5100 | 2.3 ± 0.16 | 1.2 ± 0.28 | 1900 |
| R419K | 1.3 ± 0.32 | 0.14 ± 0.06 | 9300 | 3.1 ± 0.23 | 0.72 ± 0.21 | 4300 |
| R419Q | 1.2 ± 0.32 | 0.22 ± 0.09 | 5500 | 0.55 ± 0.02 | 0.87 ± 0.14 | 630 |
| R484K | 1.6 ± 0.35 | 0.26 ± 0.08 | 6200 | 1.1 ± 0.05 | 0.45 ± 0.10 | 2400 |
| R484Q | 1.6 ± 0.40 | 0.30 ± 0.10 | 5300 | 1.2 ± 0.09 | 0.45 ± 0.16 | 2700 |
| R609K | ND | ND | 4900 | 2.6 ± 0.26 | 1.5 ± 0.49 | 1700 |
| R609Q | 1.5 ± 0.1 | 0.18 ± 0.02 | 8300 | 1.9 ± 0.07 | 0.89 ± 0.11 | 2100 |
| R639K | 1.3 ± 0.19 | 0.15 ± 0.04 | 8700 | 1.9 ± 0.14 | 0.69 ± 0.20 | 2800 |
| R639Q | 0.78 ± 0.10 | 0.094 ± 0.02 | 8300 | 1.6 ± 0.09 | 1.2 ± 0.23 | 1300 |
Kinetic parameters determined by incubating enzyme (0.2 μM) with substrates: H4 and BAEE over a range of concentrations at 37 °C for 6 min. ND = Not Determined
R419
With respect to the activity of the R419Q mutant, the kcat/Km obtained for BAEE is decreased 7.6-fold relative to the wild type enzyme, whereas the R419K mutant retains near wild-type activity (Table 3). As the decrease in kcat/Km is driven mostly by a ~5-fold decrease in kcat, these results suggest that the loss in activity may be due to an inability to properly adopt the active conformation of the enzyme. However, the fact that less than a 2-fold effect on kcat/Km is observed when histone H4 is used as a substrate suggests that long range interactions between the enzyme and the substrate can compensate for the loss of the positively charged side chain of R419.
With respect to the calcium dependence of the R419 mutants, the K0.5 of the R419K mutant (66 ± 23 μM) is decreased by 7.6-fold relative to the wild type enzyme (500 ± 91 μM) (Table 3), whereas the values obtained for the glutamine mutant are similar to those obtained for wild type PAD4 (300 ± 140 μM). As the glutamine mutant mimics a constitutively deiminated form of the enzyme, these data are consistent with the notion that the deimination of R419 could increase the K0.5 for calcium. To provide a molecular basis for this effect, we examined the structure of PAD4 bound to calcium, and found that this residue, which is present on the β-turn that connects β-strand 26 and β-strand 27, points towards the C-terminus of helix α12 and likely stabilizes the structure of the helix via electrostatic interactions with the helix dipole. Additional stabilization likely comes via an electrostatic interaction with E556, which is present at the C-terminus of this helix (Figure 3C). These two interactions also likely stabilize the structure of β-strand 26. Given that R419 is only eight residues C-terminal to E411 (E411 is present on β-strand 25, binds to calcium 1, and appears to be essential for activity), the deimination of this residue could destabilize this region of PAD4, and potentially increase the amount of calcium required to trigger the conformational change that converts PAD4 from an inactive to active conformation. If true, such a mechanism would explain the results obtained with the R419K mutant, a non-deiminatable PAD4 mutant, and that lysine can act as a functional mimic of the arginine residue at this position.
R639
Mutation of R639 to either a lysine or a glutamine also decreased the K0.5 for calcium by ~5-fold. This residue, which lines the top of the substrate binding cleft on helix α16, points out into solution and is far from the sites of calcium binding (Figure 3D). Thus, it is unclear how the mutation of this residue leads to a decreased requirement for calcium. It is, however, worth noting that the fact that this residue lines the substrate binding cleft suggests that the deimination of this residue could impact PAD4 substrate recognition, although that does not appear to be the case with either BAEE or histone H4.
Autodeimination of PAD4 reduces protein-protein interactions
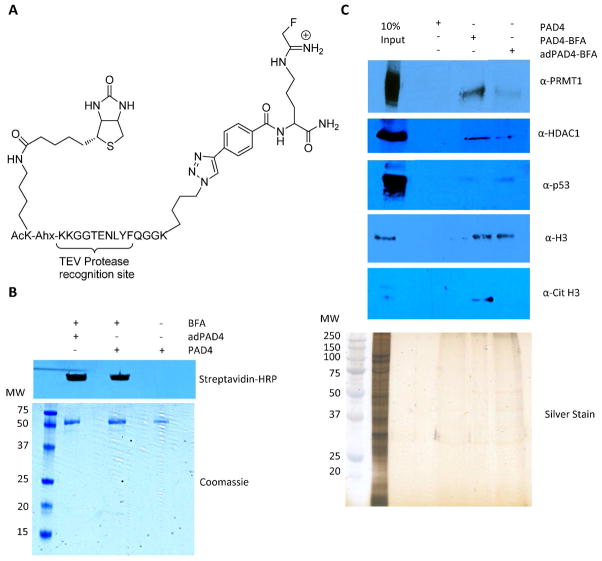
Given our results showing that autodeimination has only minor effects on the catalytic activity and calcium dependence of PAD4, we hypothesized that this PTM may indirectly affect PAD4 activity by altering its ability to interact with its binding partners. This seems reasonable when one considers that the deimination of arginines residues results in a net loss of positive charge, and such a change can have profound effects on local electrostatic interactions and hydrogen bonding with proximal residues or with potential binding proteins. To examine this possibility, we investigated the ability of adPAD4 to coimmunoprecipitate with known PAD4 binding proteins from stimulated MCF-7 WCE. Wild type PAD4 was autodeiminated at 37 °C for 1 h and labeled with BFA, a recently described PAD4-targeted Activity-based Protein Profiling (ABPP) reagent (Figure 4A) (20). Wild type non-deiminated PAD4 was labeled with BFA in an identical fashion (Figure 4B). The labeled protein was then used to “fish” out known binding proteins from the extracts. The PAD4 binding proteins evaluated were histone H3, Cit H3, p53, HDAC1 and PRMT1. Note that while PRMT1 has not previously been shown to be a PAD4 binding protein, coimmunoprecipitation experiments were used to confirm this finding (Figure S3). The results of these experiments indicated that the binding affinity of adPAD4 for p53 and H3 were similar to non-deiminated PAD4 (Figure 5). In contrast, the affinity of adPAD4 for HDAC1, PRMT1, and Cit H3 was reduced by 3.7-, 4.5-, and >10-fold, respectively (Figure 5). These results suggest that the autodeimination of PAD4 in vivo could affect its functional activity by regulating its ability to interact with binding partners.
Figure 4.
(A) Structure of BFA (B) Labeling of conPAD4 and adPAD4 with BFA. (C) Enrichment of PAD4 binding proteins using conPAD4-BFA and adPAD4-BFA. conPAD4-BFA and adPAD4-BFA were incubated with MCF-7 WCE to isolate known PAD4 binding proteins, including H3, Cit H3, HDAC1 and PRMT1. The proteins were enriched on streptavidin agarose and analyzed by western blot.
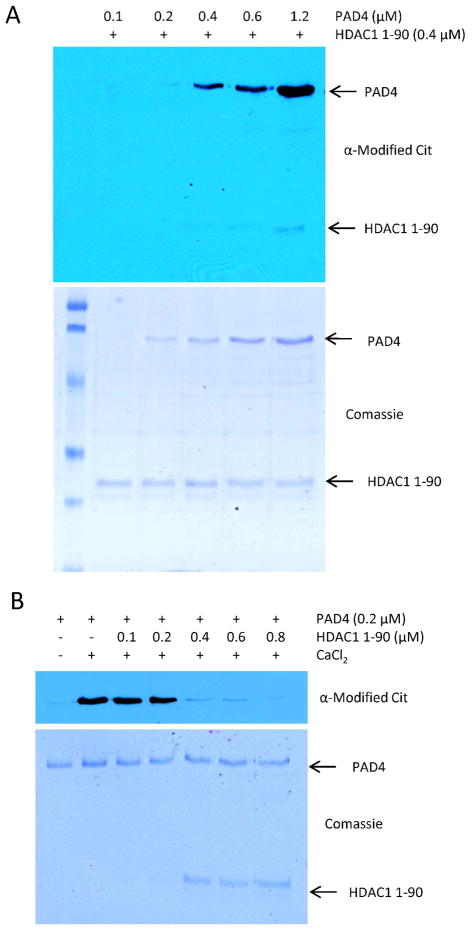
Figure 5.
(A) HDAC1 is a poor PAD4 substrate. HDAC1 (0.4 μM) was incubated with increasing amounts of PAD4 (0.1–1.2 μM) for 30 min at 37 °C. Modification of HDAC1 by PAD4 was monitored by western blot using an anti-modified citrulline antibody. (B) HDAC1 protects PAD4 from autodeimination. PAD4 was incubated for 60 min at 37 °C with increase amounts of HDAC1 1–90 (0.1–0.8 μM). Autodeimination of PAD4 was monitored by western blot using the anti-modified citrulline antibody.
Given the fact that the autodeimination of PAD4 decreases its ability to bind to a subset of the aforementioned proteins, we reasoned that one or more of these proteins could protect the enzyme from autodeimination. To test this hypothesis, we determined whether HDAC1-1-90 could inhibit PAD4 autodeimination. Note that we focused on this enzyme because it is a weak PAD4 substrate (see Figure 5A), whereas both H3 and PRMT1 (unpublished results) are PAD4 substrates. Thus, if autodeimination was prevented in the presence of HDAC1-1-90 it would most likely be due to the binding of PAD4 and not due to the addition of a competing substrate. For these studies, increasing amounts of HDAC1-1-90 were incubated in the presence of PAD4 and the autodeimination reaction was allowed to proceed for 1 h. The results of these studies indicated that HDAC1-1-90 does indeed prevent PAD4 autodeimination (Figure 5B). The fact that PAD4 autodeimination is almost completely blocked by the addition of a 2-fold molar excess of HDAC1-1-90, suggests that HDAC1-1-90 binds a PAD4 monomer as a dimer.
Coimmunoprecipitation of PAD4 mutants with binding partners
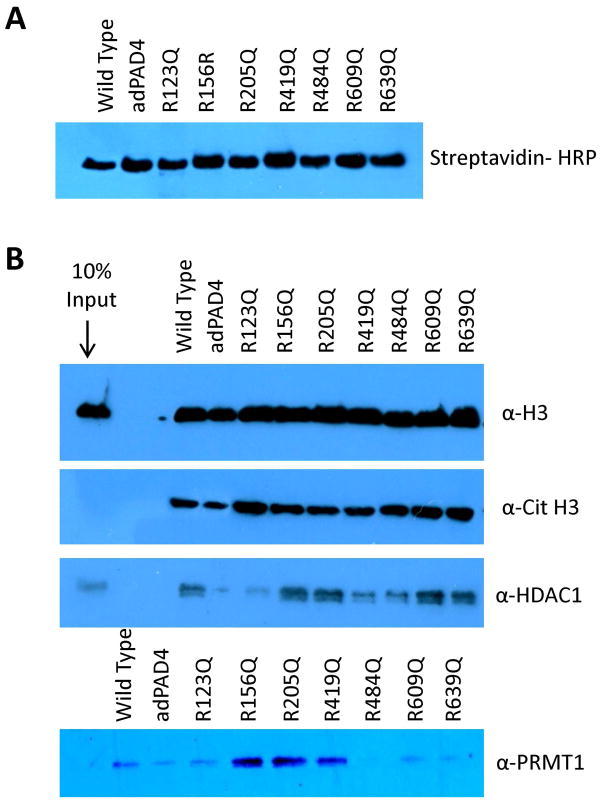
To identify the key residues responsible for mediating the interaction between PAD4 and Cit H3, HDAC1 and PRMT1, we investigated whether the glutamine substitution mutants could coimmunoprecipitate these proteins. Note that we focused on using the glutamine mutants, as opposed to the lysine mutants, because deimination reduced the binding of these proteins to PAD4 and this substitution mimics a constitutively deiminated arginine residue. Also note that H3 was used as a control. For these studies, the R123Q, R156Q, R205Q, R419Q, R484Q, R609Q, and R639Q substitution mutants were labeled with BFA (Figure 6A), incubated with MCF-7 WCE, and their ability to isolate the aforementioned proteins evaluated by western blot. As expected, the affinity for H3 is unchanged by the substitution mutants for no effect was seen with adPAD4. Similar results were obtained for Cit H3, suggesting that additive, or even synergistic, interactions from multiple autodeiminated arginine residues lowers the affinity of PAD4 for Cit H3 (Figure 6B). In contrast to these results, significant differences in the binding of these mutant enzymes to HDAC1 and PRMT1 were observed. For example, the affinity of HDAC1 for the R123Q, R419Q and R484Q mutants is decreased by 3.3-, 1.4- and 1.4 fold, respectively. For PRMT1, the effects are more dramatic, with the affinity of the R123Q, R484Q, R609Q, R639Q mutants for PRMT being reduced by 2-, 10-, 3.3-, and 5-fold, resepctively (Figure 6). These results indicate that these sites of deimination are likely critical for forming high affinity interactions between PAD4 and these proteins. With respect to HDAC1, these results are at least partially consistent with the results of Fuks and colleagues, who showed that HDAC1 interacts mainly with the C-terminus of PAD4 (residues 400–631), where R419 and R484 are located (33).
Figure 6.
Enrichment of PAD4 binding proteins using adPAD4 substitution mutants. (A) BFA labeled WT PAD4 and adPAD4 mutants were subjected to western blotting using streptavidin HRP to demonstrate equal protein loading. (B) adPAD4 substitution mutants pre-labeled with BFA were incubated with MCF-7 WCE overnight at 4 °C to enrich for PAD4 binding proteins, i.e. H3, Cit H3, HDAC1, and PRMT1. Enrichment of these proteins on streptavidin agarose was monitored by western blot analysis.
Characterization of PAD4 SNPs
Four exonic single nucleotide polymorphisms (SNPs) are present in the PAD4 gene and these SNPs have been shown to confer an increased risk of developing RA, at least in Asian populations (34, 35). Three of these SNPs results in amino acid substitutions (i.e., S55G, A82V, and A112G) and the fourth is silent. Note that all three residues are present in N-terminal subdomain 1 and because no electron density was observed for S55, its position in the crystal structure is estimated (Figure 2). Initially, we sought to examine the effects of these SNPs on protein stability and activity by generating PAD4 constructs containing the individual SNP site and a construct containing all three substitutions (Triple SNP). To examine the stability of these mutants, partial proteolysis experiments were performed. For these studies, the S55G, A82V, and A112G mutants, as well as the Triple SNP, were incubated with subtilisin over a range of time points. For the single mutants, incubation with subtilisin produced a pattern of degradation that was similar to the wild type protein. In contrast, the Triple SNP mutant appeared to be slightly less stable than wild-type, as it was more susceptible to proteolysis into smaller fragments (Figure 7A).
Figure 7.
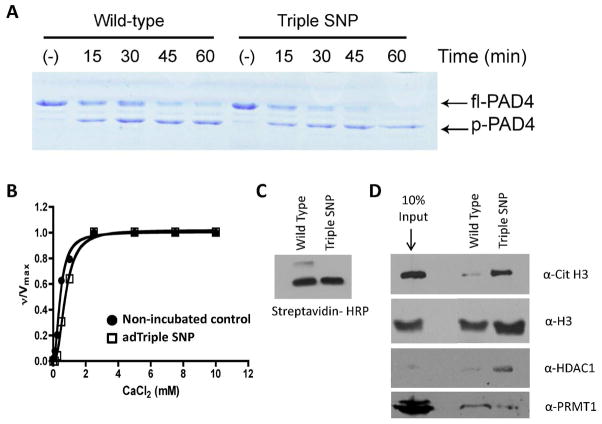
(A) Partial proteolysis of recombinant PAD4 and Triple SNP. Recombinant PAD4 and Triple SNP, 2 μg, were incubated with subtilisin (3.3 μg/ml) on ice over a range of time points (0–60 min). Samples were boiled in SDS loading buffer, ran on a 10% SDS PAGE gel, and stained with coomassie. (B) Calcium dependence of autodeiminated Triple SNP. Triple SNP (2 μM) was incubated in the presence of 10 mM calcium for 1 h (open squares) at 37 °C, followed by dialysis to remove calcium, then a determination of the calcium dependence in comparison to non-incubated positive control (closed circles) at 37 °C over a range of calcium concentrations (0–10 mM) using BAEE as the substrate. Graph shows a Hill plot of ν/Vmax versus calcium concentration. (C) Steptavidin-HRP western blot demonstrating equal loading and equal labeling of wild-type PAD4 and Triple SNP with BFA. (D) Triple SNP and wild-type PAD4 pre-labeled with BFA were incubated with MCF-7 WCE overnight at 4 °C to enrich for H3, Cit H3, HDAC1 and PRMT1. Enrichment of these proteins on streptavidin agarose was observed by western blot.
To investigate the effects of these SNPs on PAD4 activity, the steady-state kinetic parameters were determined for the S55G, A82V, and A112G single mutants, as well as the Triple SNP, using both BAEE and histone H4 as the substrates. The results of these studies indicated that mutation of these residues has only minor effects on the activity of PAD4; the kcat/Km values are either not affected (as in the case of the S55G mutant) or are decreased by only 1.7-2.5-fold and 2.3-3.2-fold for histone H4 and BAEE, respectively (Table 4). The effects of these mutations on the calcium dependence is similarly small (Table 5).
Table 4.
Steady-state kinetic parameters of PAD4 SNPs
| H4 | BAEE | |||||
|---|---|---|---|---|---|---|
| kcat (s−1) | Km (mM) | kcat/Km (s−1M−1) | kcat (s−1) | Km (mM) | kcat/Km (s−1M−1) | |
| wild-type | 2.1 ± 0.64 | 0.21 ± 0.10 | 10000 | 2.8 ± 0.58 | 0.58 ± 0.12 | 4800 |
| S55G | 1.1 ± 0.18 | 0.20 ± 0.05 | 5500 | 2.8 ± 0.21 | 0.44 ± 0.15 | 6400 |
| A82V | 1.4 ± 0.47 | 0.35 ± 0.16 | 4000 | 2.7 ± 0.17 | 1.3 ± 0.27 | 2100 |
| A112G | 1.0 ± 0.24 | 0.17 ± 0.07 | 5900 | 1.5 ± 0.12 | 0.80 ± 0.23 | 1900 |
| Triple SNP | 1.3 ± 0.54 | 0.24 ± 0.15 | 5400 | 2.9 ± 0.40 | 2.0 ± 0.74 | 1500 |
Kinetic parameters determined by incubating enzyme (0.2 μM) with substrates: H4 and BAEE over a range of concentrations at 37 °C for 6 min. Triple SNP contains all three mutations (S55G, A82V, and A112G).
Table 5.
Calcium dependence of PAD4 SNPs
| N | K0.5 (μM) | |
|---|---|---|
| wild-type | 1.5 ± 0.19 | 500 ± 91 |
| S55G | 2.3 ± 0.43 | 240 ± 90 |
| A82V | 3.2 ± 0.48 | 204 ± 66 |
| A112G | 1.8 ± 0.42 | 550 ± 180 |
| Triple SNP | 2.0 ± 0.56 | 180 ± 108 |
Values were obtained by incubating protein (0.2 μM) and BAEE (10 mM) over a range of calcium concentrations (0–10 mM) at 37 °C for 10 min. Triple SNP contains all three substitutions (S55G, A82V, and A112G).
Autodeimination of the Triple SNP mutant
It has previously been shown that autodeimination of the Triple SNP mutant and wild type PAD4 occurs with similar kinetics and that the relative activities of these two enzymes are similar (22, 36), however, detailed kinetic analyses have not been performed. Therefore, the steady state kinetic parameters were determined for the autodeiminated form of the Triple SNP mutant using BAEE and histones H3 and H4 as the substrates. The results of these experiments revealed that autodeimination of the Triple SNP mutant leads to only modest effects on the kinetic parameters (Table 6). The loss in activity compared to the non-incubated control is likely due to non-specific time dependent inactivation of the enzyme, as described above. The calcium dependence of the autodeiminated Triple SNP mutant was also examined and the results indicate that the K0.5 is increased by 2.9-fold (K0.5 = 520 ± 67 μM, n = 2.4 ± 0.2 ) (Figure 7B). In total, these data indicate that autodeimination does not have a significant effect on either the steady-state kinetic parameters or the calcium dependence of the Triple SNP mutant.
Table 6.
Steady-state kinetic parameters of autodeiminated Triple SNP
| kcat (s−1) | Km (mM) | kcat/Km (s−1M−1) | |
|---|---|---|---|
| H4 | |||
| Triple SNP control | 1.3 ± 0.54 | 0.24 ± 0.15 | 5400 |
| adTriple SNP | ND | ND | 4600 |
| conTriple SNP | ND | ND | 1700 |
|
| |||
| H3 | |||
| Triple SNP control | ND | ND | 3700 |
| adTriple SNP | ND | ND | 3000 |
| conTriple SNP | ND | ND | 2200 |
|
| |||
| BAEE | |||
| Triple SNP control | 2.9 ± 0.40 | 2.0 ± 0.74 | 1500 |
| adTriple SNP | 0.90 ± 0.03 | 0.89 ± 0.12 | 1000 |
| conTriple SNP | 0.74 ± 0.09 | 1.2 ± 0.48 | 620 |
Recombinant protein, 0.2 μM, incubated in the absence (non-AD) or presence (AD) of 10 mM calcium for 1 h at 37 °C, followed by assay. Kinetic parameters were determined by incubating enzyme with substrates: H4, H3, and BAEE over a range of concentrations at 37 °C for 6 min. Triple SNP control = non-incubated protein; ad = autodeiminated; con = non-autodeiminated; ND = not determined
Coimmunoprecipitation of the Triple SNP mutant and PAD4 binding proteins
To investigate whether the Triple SNP mutant alters interactions with known PAD4 binding proteins, coimmunoprecipitation experiments were performed. For these studies, the Triple SNP mutant and wild type PAD4 were labeled with BFA (Figure 7C), incubated with MCF-7 WCE, and their ability to isolate H3, Cit H3, HDAC1 and PRMT1 was evaluated by western blot. Interestingly, the Triple SNP had a higher affinity for H3, Cit H3, and HDAC1. In contrast, there was little to no change in affinity of the Triple SNP mutant for PRMT1 (Figure 7D). Although speculative, these results could partially explain some of the observed differences between the RA haplotype and wild type PAD4. For example, if the Triple SNP has a higher affinity to H3, it could potentially lead to higher rates of histone deimination in cellulo which would be expected to promote NET formation. Given that PAD4 and HDAC1 are known to work together to repress gene transcription(33), the higher affinity of the Triple SNP mutant for HDAC1 could also lead to the enhanced repression of genes required to resolve or constrain the inflammatory response.
Conclusions
Enzymes in eukaryotic cells are regulated by a number of multilayered and interconnected mechanisms. From transcription of the new mRNA, to alternative RNA splicing, to translation, to further modification by select PTMs, these mechanisms work to continuously fine-tune a protein’s activity in a variety of cellular environments. Of the various regulatory mechanisms, PTMs serve a particular niche in being highly dynamic, largely reversible, and by modifying functionally important residues, can have profound effects on physiological processes. Given that the enzymes involved in regulating gene transcription possess multiple PTMs that regulate their activity, PAD4, which is a transcriptional corepressor, is likely to also possess PTMs that regulate its activity. As such, understanding the mechanisms that regulate PAD4 activity under physiological and pathological conditions is a high priority because the activity of this enzyme is aberrantly increased in several human diseases (e.g., RA, Cancer, and Colitis).
As we (this manuscript) and others have reported (22), PAD4 autodeiminates in vitro and in vivo. Given the potential role of this PTM in regulating PAD4 activity, we investigated the effect of autodeimination on the activity and calcium dependence of PAD4. In contrast, to a previous report (22), we found that autodeimination does not have pronounced effects on the activity of the enzyme. Note that this observation was independently verified by three different researchers. Although it is difficult to speculate on the specific reasons for these starkly different results, we did observe a non-specific time dependent partial loss in activity when PAD4 was incubated at 37 °C for 2 h. However, in our hands this effect is independent of autodeimination. It is also possible that the different results are due to differences in the methodology used to assess PAD4 activity. For example, we directly determined the steady state kinetic parameters of PAD4 and adPAD4 using a highly defined and well established enzyme assay system with highly purified substrates. In contrast, Andrade et. al used HL60 cell lysates as the source of substrate proteins, and quantified changes in activity by western blotting using an anti-modified citrulline antibody. Thus, it is possible that a protein, ion, or small molecule present in the HL60 cell extracts could promote the autodeimination of a different pattern of arginine residues (including for example R372) that favors enzyme inactivation.
To further confirm our findings, we identified six sites in PAD4 that were autodeiminated (i.e., R123, R156, R205, R419, R484, and R639), and used to site directed mutagenesis to establish the roles of these residues in controlling the activity and calcium dependence of PAD4. Consistent with our finding that autodeimination of PAD4 does not affect the activity or calcium dependence of the enzyme, we did not observe any pronounced effects on either parameter when these residues were mutated to either a glutamine or lysine, which respectively mimic constitutively deiminated and non-deiminated arginine residues. We also investigated whether the RA-associated PAD4 haplotype alters the activity of this isozyme. Consistent with recent reports (22, 36), only minor effects on the activity and calcium dependence of PAD4 were observed.
Given that autodeimination of PAD4 and the Triple SNP mutant do not strongly affect the activity and calcium dependence of PAD4, we hypothesized that these modifications might affect interactions between PAD4 and PAD4 binding proteins. Consistent with this possibility is our observation that the interaction between adPAD4 and PRMT1, citH3, and HDAC1 is significantly reduced. In contrast, the Triple SNP mutant shows enhanced binding to H3, Cit H3 and HDAC1. Although these interacting proteins do not significantly alter the activity or calcium dependence of PAD4 (not shown), it is likely that by modulating the ability of PAD4 to interact with these proteins, it is possible to alter downstream cell signaling pathways. For example, autodeimination of PAD4 may destabilize a corepressor complex consisting of PAD4 and HDAC1, thereby providing a mechanism to decrease the corepressor activity of this complex. With respect to the Triple SNP mutant, the enhanced binding affinity for HDAC1 could oppose the effects of autodeimination, thereby leading to the stabilization of this complex, which would be expected to enhance gene repression. As it pertains to RA, this effect may lead to the enhanced repression of genes required to resolve or constrain the inflammatory response. Future studies to define how autodeimination affects these interactions in vivo will undoubtedly increase our understanding of the role of PAD4 in both physiological and pathological processes.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grant GM079357 to PRT.
ABBREVIATIONS
- PAD
protein arginine deiminase
- Cit
citrulline
- RA
rheumatoid arthritis
- BAEE
benzoyl-L-arginine ethyl ester
- DTT
dithiothreitol
- TCEP
tris-2-carboxyethyl phosphine
- EDTA
ethylenediaminetetraacetic acid
- PRMT
protein arginine methyltransferase
- HDAC1
Histone Deacetylase 1
- Cit H3
citrullinated histone H3
- BFA
Biotin-conjugated F-amidine
- PTMs
post translational modifications
- SNP
single nucleotide polymorphism
Footnotes
Supporting Information Available
Supplementary Tables S1–S4 and Supplementary Figures S1–S3. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Jones JE, Causey CP, Knuckley B, Slack-Noyes JL, Thompson PR. Protein arginine deiminase 4 (PAD4): Current understanding and future therapeutic potential. Curr Opin Drug Discov Devel. 2009;12:616–627. [PMC free article] [PubMed] [Google Scholar]
- 2.Lee YH, Rho YH, Choi SJ, Ji JD, Song GG. PADI4 polymorphisms and rheumatoid arthritis susceptibility: a meta-analysis. Rheumatol Int. 2007;27:827–833. doi: 10.1007/s00296-007-0320-y. [DOI] [PubMed] [Google Scholar]
- 3.Gandjbakhch F, Fajardy I, Ferre B, Dubucquoi S, Flipo RM, Roger N, Solau-Gervais E. A Functional Haplotype of PADI4 Gene in Rheumatoid Arthritis: Positive Correlation in a French Population. J Rheumatol. 2009;36:881–886. doi: 10.3899/jrheum.080398. [DOI] [PubMed] [Google Scholar]
- 4.Luo Y, Arita K, Bhatia M, Knuckley B, Lee YH, Stallcup MR, Sato M, Thompson PR. Inhibitors and inactivators of protein arginine deiminase 4: functional and structural characterization. Biochemistry. 2006;45:11727–11736. doi: 10.1021/bi061180d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Willis V, Gizinski A, Knuckley B, Causey CP, Luo Y, Banda N, Thompson PR, Holers VM. Efficacy of Cl-amidine in the collagen induced model of rheumatoid arthritis. J Immunology. 2011 in press. [Google Scholar]
- 6.Li P, Yao H, Zhang Z, Li M, Luo Y, Thompson PR, Gilmour DS, Wang Y. Regulation of p53 target gene expression by peptidylarginine deiminase 4. Mol Cell Biol. 2008;28:4745–4758. doi: 10.1128/MCB.01747-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao H, Li P, Venters BJ, Zheng S, Thompson PR, Pugh BF, Wang Y. Histone Arg modifications and p53 regulate the expression of OKL38, a mediator of apoptosis. J Biol Chem. 2008;283:20060–20068. doi: 10.1074/jbc.M802940200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu GY, Liao YF, Chang WH, Liu CC, Hsieh MC, Hsu PC, Tsay GJ, Hung HC. Overexpression of peptidylarginine deiminase IV features in apoptosis of haematopoietic cells. Apoptosis. 2006;11:183–196. doi: 10.1007/s10495-006-3715-4. [DOI] [PubMed] [Google Scholar]
- 9.Slack JL, Causey CP, Thompson PR. Protein arginine deiminase 4: a target for an epigenetic cancer therapy. Cell Mol Life Sci. 2011;68:709–720. doi: 10.1007/s00018-010-0480-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neeli I, Khan SN, Radic M. Histone deimination as a response to inflammatory stimuli in neutrophils. J Immunol. 2008;180:1895–1902. doi: 10.4049/jimmunol.180.3.1895. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Li M, Stadler S, Correll S, Li P, Wang D, Hayama R, Leonelli L, Han H, Grigoryev SA, Allis CD, Coonrod SA. Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J Cell Biol. 2009;184:205–213. doi: 10.1083/jcb.200806072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li P, Li M, Lindberg MR, Kennett MJ, Xiong N, Wang Y. PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J Exp Med. 2010;207:1853–1862. doi: 10.1084/jem.20100239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marcos V, Zhou Z, Yildirim AO, Bohla A, Hector A, Vitkov L, Wiedenbauer EM, Krautgartner WD, Stoiber W, Belohradsky BH, Rieber N, Kormann M, Koller B, Roscher A, Roos D, Griese M, Eickelberg O, Doring G, Mall MA, Hartl D. CXCR2 mediates NADPH oxidase-independent neutrophil extracellular trap formation in cystic fibrosis airway inflammation. Nat Med. 2010;16:1018–1023. doi: 10.1038/nm.2209. [DOI] [PubMed] [Google Scholar]
- 14.Cuthbert GL, Daujat S, Snowden AW, Erdjument-Bromage H, Hagiwara T, Yamada M, Schneider R, Gregory PD, Tempst P, Bannister AJ, Kouzarides T. Histone deimination antagonizes arginine methylation. Cell. 2004;118:545–553. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y, Roeder RG, Clarke S, Stallcup MR, Allis CD, Coonrod SA. Human PAD4 Regulates Histone Arginine Methylation Levels via Demethylimination. Science. 2004;306:279–283. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- 16.Knuckley B, Jones JE, Bachovchin DA, Slack J, Causey CP, Brown SJ, Rosen H, Cravatt BF, Thompson PR. A fluopol-ABPP HTS assay to identify PAD inhibitors. Chem Commun (Camb) 2010;46:7175–7177. doi: 10.1039/c0cc02634d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knuckley B, Luo Y, Thompson PR. Profiling Protein Arginine Deiminase 4 (PAD4): a novel screen to identify PAD4 inhibitors. Bioorg Med Chem. 2008;16:739–745. doi: 10.1016/j.bmc.2007.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo Y, Knuckley B, Bhatia M, Thompson PR. Activity Based Protein Profiling Reagents for Protein Arginine Deiminase 4 (PAD4): Synthesis and in vitro Evaluation of a Fluorescently-labeled Probe. J Am Chem Soc. 2006;128:14468–14469. doi: 10.1021/ja0656907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo Y, Knuckley B, Lee YH, Stallcup MR, Thompson PR. A Fluoro-Acetamidine Based Inactivator of Protein Arginine Deiminase 4 (PAD4): Design, Synthesis, and in vitro and in vivo Evaluation. J Am Chem Soc. 2006;128:1092–1093. doi: 10.1021/ja0576233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slack JL, Causey CP, Luo Y, Thompson PR. The Development and Use of Clickable Activity Based Protein Profiling Agents for Protein Arginine Deiminase 4. ACS Chem Biol. 2011 doi: 10.1021/cb1003515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knuckley B, Causey CP, Jones JE, Bhatia M, Dreyton CJ, Osborne TC, Takahara H, Thompson PR. Substrate specificity and kinetic studies of PADs 1, 3, and 4 identify potent and selective inhibitors of protein arginine deiminase 3. Biochemistry. 2010;49:4852–4863. doi: 10.1021/bi100363t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andrade F, Darrah E, Gucek M, Cole RN, Rosen A, Zhu X. Autocitrullination of human peptidyl arginine deiminase type 4 regulates protein citrullination during cell activation. Arthritis Rheum. 2010;62:1630–1640. doi: 10.1002/art.27439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mechin MC, Coudane F, Adoue V, Arnaud J, Duplan H, Charveron M, Schmitt AM, Takahara H, Serre G, Simon M. Deimination is regulated at multiple levels including auto-deimination of peptidylarginine deiminases. Cell Mol Life Sci. 2010;67:1491–1503. doi: 10.1007/s00018-010-0262-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kearney PL, Bhatia M, Jones NG, Luo Y, Glascock MC, Catchings KL, Yamada M, Thompson PR. Kinetic characterization of protein arginine deiminase 4: a transcriptional corepressor implicated in the onset and progression of rheumatoid arthritis. Biochemistry. 2005;44:10570–10582. doi: 10.1021/bi050292m. [DOI] [PubMed] [Google Scholar]
- 25.Obianyo O, Osborne TC, Thompson PR. Kinetic mechanism of protein arginine methyltransferase 1. Biochemistry. 2008;47:10420–10427. doi: 10.1021/bi800904m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson PR, Kurooka H, Nakatani Y, Cole PA. Transcriptional coactivator protein p300. Kinetic characterization of its histone acetyltransferase activity. J Biol Chem. 2001;276:33721–33729. doi: 10.1074/jbc.M104736200. [DOI] [PubMed] [Google Scholar]
- 27.Knuckley B, Bhatia M, Thompson PR. Protein arginine deiminase 4: evidence for a reverse protonation mechanism. Biochemistry. 2007;46:6578–6587. doi: 10.1021/bi700095s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knipp M, Vasak M. A colorimetric 96-well microtiter plate assay for the determination of enzymatically formed citrulline. Anal Biochem. 2000;286:257–264. doi: 10.1006/abio.2000.4805. [DOI] [PubMed] [Google Scholar]
- 29.Leatherbarrow RJ. Grafit Ver 5.0. Erathicus Software; Staines, UK: 2004. [Google Scholar]
- 30.Weerapana E, Speers AE, Cravatt BF. Tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP)--a general method for mapping sites of probe modification in proteomes. Nat Protoc. 2007;2:1414–1425. doi: 10.1038/nprot.2007.194. [DOI] [PubMed] [Google Scholar]
- 31.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 32.Arita K, Shimizu T, Hashimoto H, Hidaka Y, Yamada M, Sato M. Structural basis for histone N-terminal recognition by human peptidylarginine deiminase 4. Proc Natl Acad Sci U S A. 2006;103:5291–5296. doi: 10.1073/pnas.0509639103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Denis H, Deplus R, Putmans P, Yamada M, Metivier R, Fuks F. Functional connection between deimination and deacetylation of histones. Mol Cell Biol. 2009;29:4982–4993. doi: 10.1128/MCB.00285-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki A, Yamada R, Chang X, Tokuhiro S, Sawada T, Suzuki M, Nagasaki M, Nakayama-Hamada M, Kawaida R, Ono M, Ohtsuki M, Furukawa H, Yoshino S, Yukioka M, Tohma S, Matsubara T, Wakitani S, Teshima R, Nishioka Y, Sekine A, Iida A, Takahashi A, Tsunoda T, Nakamura Y, Yamamoto K. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet. 2003;34:395–402. doi: 10.1038/ng1206. [DOI] [PubMed] [Google Scholar]
- 35.Kang CP, Lee HS, Ju H, Cho H, Kang C, Bae SC. A functional haplotype of the PADI4 gene associated with increased rheumatoid arthritis susceptibility in Koreans. Arthritis Rheum. 2006;54:90–96. doi: 10.1002/art.21536. [DOI] [PubMed] [Google Scholar]
- 36.Horikoshi N, Tachiwana H, Saito K, Osakabe A, Sato M, Yamada M, Akashi S, Nishimura Y, Kagawa W, Kurumizaka H. Structural and biochemical analyses of the human PAD4 variant encoded by a functional haplotype gene. Acta Crystallogr D Biol Crystallogr. 2011;67:112–118. doi: 10.1107/S0907444910051711. [DOI] [PubMed] [Google Scholar]
- 37.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.