Abstract
Glutamate-mediated neurotransmission through ligand-gated, ionotropic glutamate receptors is the main form of excitatory neurotransmission in the vertebrate central nervous system where it plays central roles in learning, memory and a variety of disorders. Acting as auxiliary subunits, transmembrane α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor (AMPAR) regulatory proteins (TARPs) are essential regulators for glutamate-mediated neurotransmission in the central nervous system. Here we report the first electron crystallographic reconstructions of full-length mouse stargazin (γ-2) at ~20Å resolution in a membrane bilayer environment. Formation of ordered arrays required anionic lipids and was modulated by cholesterol and monovalent cations. Projection structures revealed that the C-termini of stargazin monomers closely interacted with the bilayer surface in an extended conformation that placed the C-terminal PDZ-binding motif ~100Å away from the transmembrane domain and in close proximity to a membrane re-entrant region. The C-termini interaction with the bilayer was modulated by the ionic strength of the solution and overall protein secondary structure increased when membrane-bound. Our data suggest that stargazin interactions with and within the membrane play significant roles in TARP structure and directly visualize TARP functional mechanisms essential for AMPAR trafficking and clustering.
Keywords: Three dimensional electron microscopy; two-dimensional crystallization, electron crystallography; transmembrane AMPA receptor regulatory protein; stargazin
1. Introduction
TARPs are essential for AMPAR maturation, functional modulation and scaffolding at the postsynaptic density ([Nicoll et al., 2006], [Osten and Stern-Bach, 2006], [Ziff, 2007], [Sager et al., 2009a], [Coombs and Cull-Candy, 2009], and [Tomita, 2010]). Since the discovery of the prototypical TARP stargazin (Letts et al., 1998), six TARP isoforms have been identified biochemically, each with distinct expression patterns: the canonical type I TARPs γ-2 (stargazin), γ-3, γ-4 and γ-8 and type II TARPs γ-7 as well as γ-5 ([Tomita et al., 2003], [Fukaya et al., 2006], [Milstein et al., 2007], [Cho et al., 2007], [Kato et al., 2007], and [Kato et al., 2008]). Moreover, the recently identified cornichon family of proteins was also shown to associate with AMPARs further increasing the repertoire and structural complexity of AMPAR physiology ([Schwenk et al., 2009] and [Shi et al., 2010]).
At a molecular level the role of TARPs in AMPAR regulation has been established. Stargazin increases AMPAR affinity for glutamate and rate of opening (Tomita et al., 2005b). Moreover, TARPs regulate synaptic targeting of AMPARs by two separate mechanisms: (1) surface delivery of AMPARs requires interactions with the extracellular loop and part of stargazin's intracellular C-terminal domain (Tomita et al., 2005a), while (2) interactions of stargazin's PDZ-binding motif with membrane associated guanylate kinases (MAGUKs) like postsynaptic density protein-95 (PSD-95) are essential for targeting of AMPARs to the postsynaptic density ([Chen et al., 2000] and [Schnell et al., 2002]). Despite this tremendous progress, a lack of direct structural information limits mechanistic understanding of TARP function to a map of functionally important regions and an appreciation of TARP functional regulation through phosphorylation ([Choi et al., 2002], [Chetkovich et al., 2002], [Tomita et al., 2005a], [Tomita et al., 2005b], [Turetsky et al., 2005], and [Sager et al., 2009b]). In contrast to the recent advance in our understanding of AMPAR structure at atomic resolution (Sobolevsky et al., 2009), structural information about TARPs is limited to 35-40Å reconstructions of native AMPARs ([Nakagawa et al., 2005] and [Nakagawa et al. 2006]).
To address this scarcity of structural information, we sought to obtain a structure of stargazin, the prototypical type I TARP. Here, we present the results of an electron crystallographic analysis of negatively stained two-dimensional arrays of membrane embedded full-length stargazin. Stargazin is a 36kDa integral membrane protein consisting of an N-terminal region that contains four transmembrane domains with an extracellular loop between the first and second transmembrane domains (together 22kDa), and an intracellular C-terminal tail domain of ~14kDa. We show that stargazin's C-terminus extensively interacts with the lipid bilayer in the presence of anionic lipid head groups, which was consistent with our observation that reconstitution of stargazin into these membranes increased secondary structure. Moreover, the presence of cholesterol modulated packing interactions between the membrane-embedded domains of stargazin crystal subunits, which may have implications for the trafficking behavior of stargazin through the secretory pathway. Taken together, our data provide the first explicit visualization of a TARP and suggest that its interactions both with and within the bilayer likely play important functional roles in TARP assembly with AMPARs, AMPAR trafficking, and clustering at the postsynaptic membrane.
2. Materials and Methods
2.1 Stargazin expression construct and yeast transformation
A full-length mouse stargazin (UniProtKB/Swiss-Prot O88602; CCG2_MOUSE) construct was obtained from Susumu Tomita (Yale University) in a pGEMHE expression vector. A hexahistidine tag was inserted after the initiation methionine to achieve 6xHis-Stg as an open reading frame and this construct was subcloned into the pPIC3.5K vector for expression in the methylotrophic yeast Pichia pastoris. Host strain SMD1163 was transformed by electroporation and transformants were selected according to the manufacturers specifications first on His-deficient plates, followed by screening for G418 resistance (Multi-Copy Pichia Expression Kit; Invitrogen).
2.2 Purification of 6xHis-Stg
200mL cultures of P. pastoris expressing 6xHis-Stg were inoculated from overnight cultures and grown for three days at 30°C to an OD of 10 in buffered minimal glycerol (100 mM potassium phosphate, pH 6.0; 1.34% yeast nitrogen base with ammonium sulfate; 4x10-5% biotin; 1% glycerol). A 4L fermentor (BIOFLO 110 Fermentor/Bioreactor; New Brunswick Scientific) was then inoculated with 200mL of starter culture and grown for 60–70 hours with 4% glycerol as the sole carbon source at 30°C and 30% dissolved oxygen. 6xHis-Stg expression was induced by rapid influx of 30–40mL methanol concomitant with cessation of glycerol feed. Following rapid methanol influx, induction was maintained for six hours through feeding methanol at a rate of 5ml/h per liter of culture. The yeast cells were then harvested and lysed at 40,000 psi using one pass through a cell disrupter (Z Plus Series; Constant Cell Disruptions Systems, Daventry, U.K.) at -10°C. Cell debris was separated by a low-speed centrifuge spin (600xg, 10°C for 20 minutes in a Beckman JA-10 rotor) from cell membranes, which were obtained by ultracentrifugation at 40,000 rpm for 30 minutes at 10°C using a Beckman type Ti45 rotor. Membranes were homogenized in breaking buffer (10 mM MOPS, 50 mM NaCl, 10 mM MgCl2, 10 mM imidazole, 1 mM TCEP, pH 7.4) using a dounce glass homogenizer on ice. Protein concentration (Coomassie Stain Kit, Pierce) was adjusted to 1mg/ml and solubilization was carried out for 3 hours at 4°C in the presence of 3% Triton X-100. Unsolubilized material was pelleted by ultracentrifugation (40,000 rpm; 15 minutes; 10°C; Beckman type Ti45 rotor) and the supernatant was incubated overnight at 4°C in batch on Ni2+-NTA resin (GE Healthcare) for purification by metal affinity chromatography. The column was washed with twenty column-volumes of exchange buffer (breaking buffer plus 0.03% w/v dodecyl-β-D-maltopyranoside (Anatrace)) and 6xHis-Stg was eluted with four column-volumes exchange buffer containing 300mM imidazole. Remaining impurities were removed by passing the eluate over a 1mL cation-exchange chromatography column at 4°C (HiTrap™ SP HP; GE Healthcare) at a rate of 1 mL/min. To the flow through containing 6xHis-Stg, NaCl and glycerol were added to final concentrations of 500mM and 10%, respectively. 6xHis-Stg was then concentrated to ~10mg/mL by centrifugation (Amicon Ultra (Millipore); 30,000 MW cutoff, 4°C) and subjected to size exclusion chromatography (Superose 6 10/300 GL; Amersham Pharmacia Biosciences) at 10°C in a running/equilibration buffer containing 10mM MOPS, 300mM NaCl, 10mM MgCl2, 1mM TCEP, 0.02% w/v NaN3, 0.03% w/v dodecyl-β-D-maltopyranoside, pH 7.4. A gel filtration standard (Gel Filtration Standard; Bio-Rad) was subjected to size exclusion chromatography as a 6xHis-Stg molecular weight reference.
2.3 2D crystallization of 6XHis-Stg
25μg of 6xHis-Stg, with concentrations ranging from 0.3–0.7 mg/mL (Coomassie Stain Kit, Pierce), was mixed with 10μg of lipid (1 mg/mL of 80% dimyristoyl phosphatidylcholine, 15% 1-stearoyl-2-oleoyl phosphatidylserine, 5% cholesterol (Avanti)) in 100μL gel filtration buffer for a final lipid-to-protein ratio of 0.4 (w/w) and incubated overnight at 26°C with mild shaking. Crystals were grown by transferring this mixture to dialysis cups (10,000 MWCO Slide-A-Lyzer Mini Dialysis Units; Pierce or in-house fabricated dialysis units) and dialyzing against 1L of crystal buffer (10mM MOPS, 50mM NaCl, 10mM MgCl2, 1mM TCEP, 0.02% w/v NaN3) at 26°C for 72 hours. Lattices formed under these conditions had mostly c222 symmetry. Crystals belonging to double-sided plane group p6 were obtained by omitting cholesterol from the lipid mix (80%:20% PC:PS).
2.4 Electron microscopy
Continuous carbon film was deposited on 400 mesh Cu/Rh grids (Electron Microscopy Sciences, Hatfield, PA). Crystals were applied after 30 seconds glow discharge at 30mA discharge current in a sputter coater (SCD005; BAL-TEC, Tucson, AZ) and preserved in 1% uranyl acetate. All microscopy was performed on a Tecnai 12 TEM with a LaB6 filament operating at an acceleration voltage of 120kV. Images were digitally collected on a 1K × 1K CCD (Gatan, Inc.) with a nominal magnification of 30,000 at defocus values of -5,000 to -10,000Å. Unstained specimens were similarly prepared on a continuous carbon support applied to C-FLAT holey carbon grids (2μm hole size 2μm hole spacing; Electron Microscopy Sciences) at 26°C and ~80% humidity, blotting time of 6 seconds, by rapid plunge into liquid ethane (Cryoplunge™3; Gatan, Inc.). Images were digitally collected on a 4Kx4K CCD (Gatan, Inc.) at liquid nitrogen temperature (-172°C) under low dose conditions (~10e-/Å2) on a Philips Tecnai F20 electron microscope.
2.5 Image processing
Crystal patches of approximately 0.1–0.25μm2 size were identified by calculating a forward Fourier transform using the program XIMDISP (Smith, 1999). The MRC suite of programs was used to correct for translational lattice disorder and the contrast transfer function following standard conventions (Crowther et al., 1996). The program ALLSPACE was used to test the image data for the presence of crystallographic symmetry, and data were merged after shifting the images to their appropriate crystallographic phase origins. In the case where data were compiled in plane group symmetry c222, measurements for which (h+k) = odd were deleted from the raw data files prior to calculations of phase origins and merging because these reflections are absent from the molecular transform. This data manipulation step was carried out to avoid the inclusion of a large number of very weak and noisy measurements in the calculations of origins and lattice lines (LATLINE) and seemed justified because ultimately, (h+k) = odd reflections would have to be removed if c222 symmetry was to be applied (Supplementary Figure S1 for global comparison of signal-to-noise ratios for (h+k) = even and (h+k) = odd). 3D volumes were calculated using the CCP4 suite of programs (CCP 4, 1994), and resolution estimates were obtained from the calculated pointspread function of the final data set (Unger, 2000). Phase statistics for the projection data were plotted using PLOTALL (Supplementary Figure S2). The volume data were rendered in 3D using Chimera for all viewing and figure preparation purposes (Huang et al., 1996).
2.6 Single Particle Analysis
25 nM of 6xHis-Stg were applied to glow-discharged holey carbon grids with a thin layer of carbon over the holes. After 1 minute, the sample was stained sequentially in 3 droplets of 2% uranyl acetate, excess stain was removed by gentle blotting, and allowed to air-dry. The specimen was observed as described in Section 2.4 using a nominal magnification of 30,000 (equal to pixel size of 3.8Å at specimen level). Images were recorded on a 4K × 4K CCD camera (Gatan, Inc.). The defocus range used to collect the image was -1.1 μm to -1.4 μm.
2.7 Circular dichroism analysis
Far-UV CD spectra of detergent purified 6xHis-Stg in 0.03% dodecyl-β-D-maltopyranoside and lipid-reconstituted, crystalline 6xHis-Stg were collected on an Aviv model 202 spectrometer. The 6xHis-Stg concentration used was 3.5μM. The spectra (195–260nm; stepsize 1nm, 3 second integration) were recorded at 25°C using a 0.1cm pathlength quartz cuvette. The average of three replicate scans was used for each trial. CD intensities were expressed in per residue molar absorbance units (M-1 cm-1). Overall secondary structure was determined using the CDPro suite of programs using the reference data set containing the most membrane proteins (SMP56; IBasis = 10) and based on agreement between the three analysis programs CONTIN, CDSSTR, and SELCON3 (Sreerama and Woody, 2004).
2.8 Chemical crosslinking of 6xHis-Stg with EGS
4μg monomeric 6xHis-Stg in 50μL buffer (10mM MOPS, 300mM NaCl, 10mM imidazole, 1mM TCEP, 0.2% decyl-β-D-maltopyranoside, pH 7.4) was crosslinked for up to several minutes on ice with 1mM ethylene glycolbis(succinimidylsuccinate) (Pierce) dissolved in DMSO. The reaction was quenched with 5μL of Tris buffer to give a final Tris concentration of 100mM. Samples were resolved by SDS-PAGE.
3. Results
3.1 Purification of a Stable Stargazin Oligomer
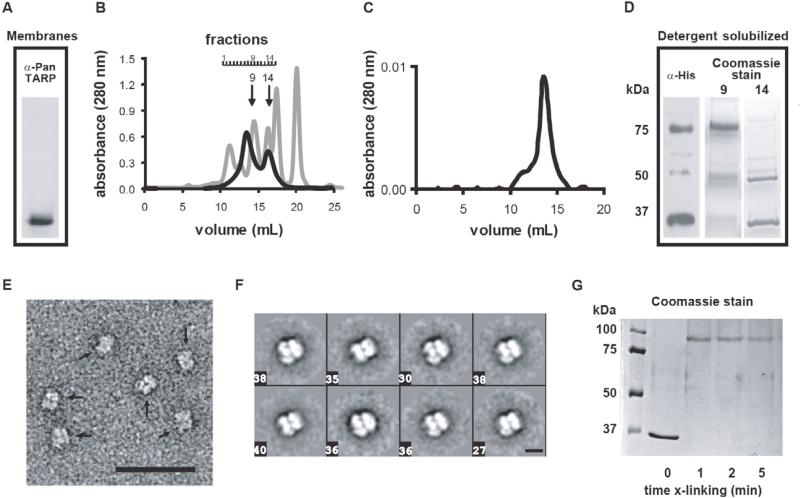
N-terminally histidine-tagged stargazin (6xHis-Stg) was heterologously expressed in the methylotrophic yeast P. pastoris. Prior to membrane solubilization for purification, host membrane proteins were separated by SDS-PAGE and recombinant 6xHis-Stg was detected by Western blot as a monomer (Fig. 1A). However, after solubilization of the host membranes with Triton X-100 detergent two distinct oligomeric species were observed during the final size exclusion step of the 6xHis-Stg purification (Fig. 1B). Based on the protein standard used for molecular weight estimation and accounting for the contribution of the detergent micelle (~70kDa), the apparent molecular weight of ~150kD for the larger species, which eluted at 13.5 ml, suggested that a significant fraction of stargazin was a dimer or, less likely, a tetramer in solution. The putative 6xHis-Stg dimer was stable when subjected to a second size exclusion column (Fig. 1C). Based on its Coomassie and Western blotting profiles (Fig. 1D), the putative dimer was distinctly different from the smaller species, which eluted at 16.2 ml (Fig. 1B). This latter fraction seemed to consist mostly of the 36kDa monomeric stargazin and a larger, ~50kDa species that was not recognized well by either the pan-TARP or hexahistidine primary antibodies.
Figure 1. 6xHis-Stg expression and purification.
A) Western blot of membranes of a recombinant strain of P. pastoris probed with an anti-pan-TARP antibody identified a single protein band at a molecular weight consistent with monomeric 6His-Stg (36kDa). B) Gel filtration profile of 6xHis-Stg (black) demonstrated the presence of two protein populations; a putative dimer (fraction B9) and a monomer (B14). A standard protein mixture (gray) was separated for 6xHis-Stg molecular weight reference: 11.0 – 12.5mL peaks; void volume and thyroglobulin (670kDa), 14.3mL; γ-globulin (158kDa), 16.1mL; ovalbumin (44kDa), 17.3mL; myoglobin (17kDa), 20mL; vitamin B12 (1.4kDa). C) Refiltration of the putative dimer revealed a stable oligomeric species (~75kDa). D) Western blot of pre-gel filtration 6xHis-Stg load, probed with an anti-histidine antibody, identified two major protein bands, presumably representing stargazin monomer and dimer. Coomassie stained gels (10μL load) of fractions B9 and B14 from the gel filtration showed that gel filtration separated dimeric stargazin from monomeric stargazin. A band migrating at ~50kDa was sometimes observed in fraction B14. E) High-magnification (30,000X) view of fraction B9 stargazin stained with 2% uranyl acetate (scale bar = 500 Å) revealed a homogeneous population of particles (marked by arrows). F) Representative two-dimensional class averages from specimens of 6xHis-Stg fraction B9 preserved and imaged as in E) revealed that most particles display a distinct conformation containing several punctate densities. Boxed numbers specify the number of particles that were averaged from the total data set (n = 2548) to obtain the corresponding class average (scale bar = 100 Å). G) Treatment of samples from fraction B14 with the cross-linker EGS quantitatively converted monomeric 6xHis-Stg to a molecular weight comparable to that observed with the putative dimer by SDS-PAGE (Coomassie gel shown).
When subjected to electron microscopy, negatively stained aliquots of the purified putative dimer protein revealed a virtually homogenous population of particles (Fig. 1E). This was consistent with the well-defined gel filtration peak profile observed and further confirmed by single particle analysis, which yieleded well-defined class-averages showing particles with dimensions of ~10×10nm (Fig. 1F; Supplementary Figure S3 for all class averages). Despite differences in how the two stargazin fractions behaved on SDS gels, quantitative amino acid analysis of the major bands confirmed the protein to be stargazin (not shown) and crosslinking of the 36kDa species converted it into a product that migrated by SDS-PAGE similarly to the putative dimer species (Fig. 1G). Attempts to determine whether glycosylation significantly contributed to the molecular heterogeneity observed by SDS-PAGE resulted in protein aggregation upon treatment with N-glycosidase F (not shown). The glycosylation state of the purified protein was not pursued further as previous studies have established that glycosylation is not required for stargazin function, specifically regulation of AMPAR gating or trafficking (Tomita et al., 2004). Despite the fact that 6xHis-Stg dimerization occurred during purification previous studies have reported stargazin self-association independent of heterologous protein expression ([Price et al., 2005] and [Bedoukian et al., 2006]).
3.2 Two-Dimensional Crystallization of Stargazin Requires Anionic Lipids
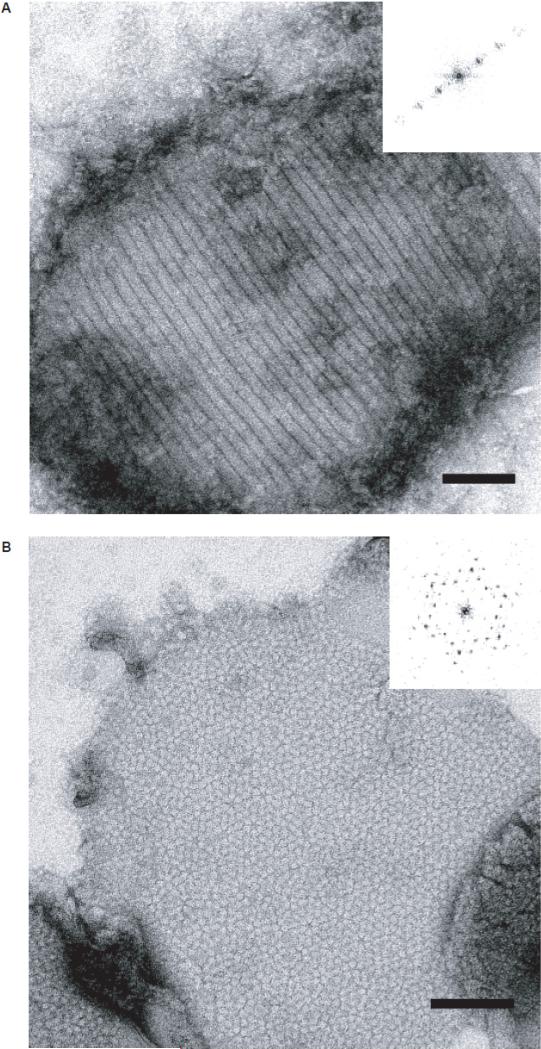
Initial crystallization experiments were carried out using dimyristoyl phosphatidylcholine (DMPC). These experiments resulted in proteoliposomes that displayed an anisotropic striated pattern (Fig. 2A). Experimentation with the lipid composition, specifically substitution of up to 15% w/w of DMPC with lipids that contained an anionic head group (e.g., dimyristoyl phosphatidylserine (DMPS)), resulted in formation of small, ~300×300 nm2 arrays in which the 75kDa stargazin fraction packed hexagonally with apparent six-fold symmetry (Fig. 2B). Similar crystals were obtained regardless of the type of anionic headgroup and for a variety of chain lengths of the acyl moiety (not shown): 1-palmitoyl-2-oleolyl phosphatidylcholine (POPS), 1-stearoyl-2-oleolyl phosphatidylserine (SOPS), dimyristoyl phosphatidic acid (DMPA), dimyristoyl phosphatidylglycerol (DMPG), and phosphatidylinositol 4,5-bisphosphate (PIP2). These results demonstrated that two-dimensional crystallization of stargazin solely depended on the presence but not the nature of anionic lipid head groups and suggested that some region of stargazin engaged the bilayer through nonspecific electrostatic interactions.
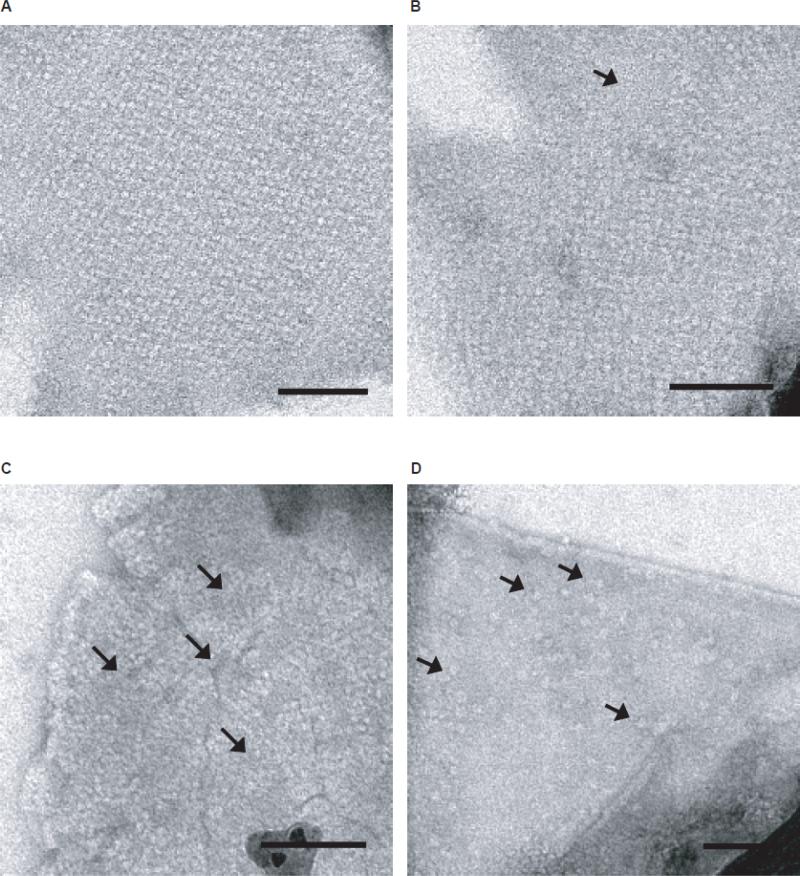
Figure 2. 6xHis-Stg two-dimensional crystallization requires anionic head group lipids.
High-magnification (30,000X) views of 6xHis-Stg dimer reconstituted in 100% DMPC (A) and 80% DMPC/20% DMPS, w/w (B). Both samples were stained with 1% uranyl acetate (scale bar = 1000 Å). Insets: calculated diffraction patterns supported that dimers formed a 1D-lattice in the presence of DMPC (A), while the presence of anionic lipids allowed the dimers to form a well-defined 2D-lattice with hexagonal symmetry (B).
3.3 Cholesterol and Monovalent Cations Modulate Stargazin Crystal Packing
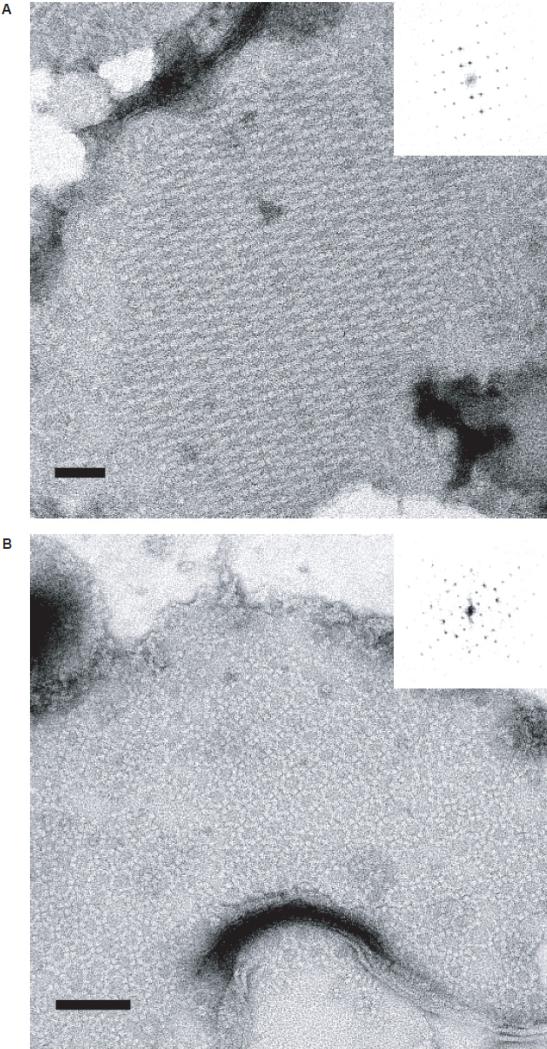
Introduction of 5% w/w of cholesterol to the PC/PS mixes that yielded hexagonal lattices caused dramatic changes in 6xHis-Stg packing, resulting in two different rectangular crystal forms (Fig. 3). Of these two, the first form contained interdigitated rows of stargazin dimers (Fig. 3A). This crystal form was both more abundant and more reproducible than a second, more complex crystal form (Fig. 3B), which was observed if potassium ions were present in the crystallization buffer (50mM). In some cases, the two crystal forms co-existed within the same proteoliposome, which precluded an unambiguous determination whether potassium ions were the sole reason for the formation of the more complex crystal packing.
Figure 3. Cholesterol and potassium ions affect crystallographic packing of 6xHis-Stg dimers.
High-magnification (30,000X) views of 6xHis-Stg two-dimensional crystals formed upon reconstitution into 80% DMPC/15% DMPS/5% cholesterol (w/w/w) in the absence (A) and presence (B) of potassium ions (50mM KCl) in the dialysis buffer. Both samples were stained with 1% uranyl acetate. Forward Fourier transforms (insets) demonstrate distinct 6xHis-Stg packing arrangements. Of the two forms, the simpler of the two packing arrangements (A) was more common and more reproducible than the more complex form (B). Insets show the calculated diffraction patterns of the crystals. In some cases, both crystal forms were observed to exist within the same proteoliposome.
3.4 A Direct Visualization of Stargazin's Intracellular C-terminal Domain Interaction with a Membrane Bilayer
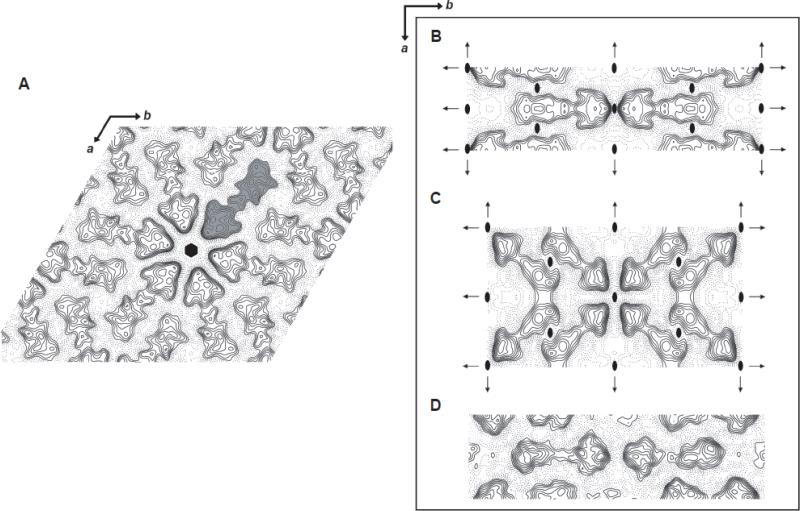
To obtain a quantitative description of membrane-embedded stargazin, we collected images of negatively stained crystals of all three crystal forms and calculated their projection structures at ~15-20Å resolution (Fig. 4; refer to Table 1 for image statistics and Supplementary Figure S2 for analysis of the projection structure phase statistics). Phase relationships in the projection data supported plane group symmetry p6 and c222 for the hexagonal and rectangular forms, respectively (Figs. 4A – 4C). To differentiate between crystal forms and reflect the crystallographic symmetry applied for structure determination, we termed the hexagonally-packed 6xHis-Stg crystals the “p6 form” (Fig. 2B). The lattices that formed upon addition of cholesterol is referred to as the “simpler c222 form” (Fig. 3A), while the third crystal arrangement observed in the presence of potassium is referred to as the “complex c222 form” (Fig. 3B). The presence of c222 symmetry implied that 6xHis-Stg formed antiparallel dimers with one subunit “up” relative to its partner's “down” orientation within the membrane. We confirmed this finding through independent 3D reconstructions for the p6 form and by reconstructing the simpler of the c222 crystal forms without imposition of the full symmetry (not shown). This particular dimer arrangement is not likely to exist in vivo, because it places the PDZ-binding motif present in the stargazin C-terminal domain on both the extracellular and cytoplasmic sides of the plasma membrane. Nevertheless, previous electron crystallographic studies have reported informative structures in the form of antiparallel dimers (Tate et al., 2001), and despite their intramembrane arrangement, the 6xHis-Stg crystal subunits revealed novel insights into TARP structural organization in a membrane environment.
Figure 4. Projection density maps of 6xHis-Stg 2D crystals.
A-C) Projection structures were generated from negatively stained 6xHis-Stg 2D crystals. A) A projection structure at 20Å resolution of a hexagonal crystal form unit cell from nine merged images with p6 symmetry applied. One 6xHis-Stg crystallographic dimer is colored gray and the six-fold symmetry center is shown (black hexagon). B) A projection structure at 15Å resolution of a “simple” rectangular crystal form unit cell from nine merged images with c222 symmetry applied. Two-fold axes perpendicular to the membrane plane (ovals) and two-fold axes in the plane of the membrane (arrows) are shown relative to the unit cell. C) A projection structure at 20Å resolution of a “complex” rectangular crystal form unit cell from two merged images with c222 symmetry applied. Symmetry elements are shown as in (B). D) A 15Å projection calculated from images of unstained, frozen hydrated crystals of the simpler of the two rectangular crystal forms. No symmetry (p1) was applied in this case to provide a completely unbiased view of the molecular arrangement. Common to all crystal forms, the crystallographic dimers are represented by a well-defined, triangular “head” domain of approximately 5625Å2 and a lower-density “tail” extending some 100Å across the bilayer. 6xHis-Stg C-terminal domains interact in an antiparallel fashion between crystallographic dimers as do the protein subunits per crystallographic dimer. The main differences between crystal forms were in the interactions of the head regions between dimers.
Table 1.
Image statistics (projection structure)
| Plane group symmetry | p6 | c222 (simple) | c222 (complex) | p1 |
| No. of images | 9 | 9 | 2 | 5 |
| Unit cell parameters | ||||
| a, Å | 335 | 156 | 264 | 148 |
| b, Å | 335 | 552 | 477 | 519 |
| γ, ° | 120 | 90 | 90 | 90 |
| Effective resolution cutoff, Å | 20 | 15 | 20 | 15 |
| Overall phase residuala | 34.7° | 26.4° | 26.5° | 34.8° |
| B factor | -1000 | -1000 | -1000 | 0 |
Averaged phase residual deviation between observed and theoretical 0°/180° where 45° is random for p6 and c222 and 90° is random for p1.
In projection, 6xHis-Stg displayed a “jellyfish”-like shape consisting of a ~5625Å2 triangular region that is connected to a less dense extended region of approximately 100Å in length (Fig. 4). Based on the stargazin sequence, these projections could only be explained if the triangular region was assigned to the transmembrane domains of the stargazin dimer, while the extended “tail” region corresponded to the C-terminal domains. The observation of a crystallographically ordered, extended conformation of the C-termini implied a direct interaction of the C-termini with the bilayer surface. This idea was supported by two lines of evidence: first, the “triangular footprint” of the membrane embedded region was observed in many of the single particle class averages of the purified dimer (Fig. 1F), whose putative tail regions were likely to be disordered and/or mobile in solution. Second, reconstitution into neutral lipids resulted in anisotropic packing consistent with a loose and disordered association of the membrane domains, but did not show defined densities that corresponded to the tail region (Fig. 2A). Interestingly, the interaction of the C-terminal domain with the bilayer was conserved between the different crystal forms, while the interactions between the membrane embedded regions differed, depending on whether cholesterol and potassium were present (Figs. 4B and 4C) or absent (Fig. 4A).
To assure that the interaction of the C-terminal domains was not a consequence of negative staining, we collected images of unstained, fully-hydrated crystals of the simpler c222 crystal form. Cryopreservation of the sample proved to be challenging because crystals did not adhere well to the carbon films under these conditions, and the high transition temperature of the bulk lipid DMPC required samples to be frozen from room temperature, which despite many efforts resulted in uneven ice across the grids. Because of these reasons, we were only able to collect a limited number of images of the unstained crystals (Table 1, “p1” column). Data merging without assuming any symmetry yielded a projection that looked virtually identical to the projections that were obtained from negatively stained crystals (Fig. 4D). From this result, we concluded that an analysis in negative stain was relevant, and that the putative interaction of the stargazin C-terminal domain with the bilayer was a genuine feature of our stargazin structure.
3.5 Electrostatic Interactions Drive the Stargazin C-termini-Bilayer Interaction
From the projection structures, the 6xHis-Stg tail densities appeared to be the critical crystal packing contacts because this interaction looked the same in all three crystal form projections (Fig. 4A – 4C). To test our hypothesis that the extended densities in the projection structures were 6xHis-Stg C-terminal domains that interacted with the membrane through electrostatic interactions, we attempted to disrupt this interaction by increasing the concentration of sodium chloride in the crystallization buffer during two-dimensional crystallization (Figure 5). This experiment was feasible because we could routinely generate samples where ≥ 95% of the 6xHis-Stg observed in proteoliposomes formed the simpler c222 crystal form (Fig. 5A). Doubling the sodium chloride concentration of the crystallization buffer from 50mM to 100mM resulted in local gaps in 6xHis-Stg lattices; however the vast majority of the sample still formed robustly-packed lattices (Fig. 5B). At a concentration of 250mM sodium chloride the majority of proteoliposomes appeared mostly disordered and very few well-diffracting crystals were observed (Fig. 5C). At 500mM sodium chloride, bilayers with protein densities similar in appearance to the detergent-solubilized particles were visible within many proteoliposomes (Fig. 5D). Two-dimensional crystals were not observed and notably the tail densities were not visible in these negatively stained images. The latter observation ruled out that any part of the extended region of density accounted for a bona-fide part of the transmembrane domain of stargazin because the transmembrane helices would be too hydrophobic to be released from the bilayer through the addition of salt. Overall, our observations were consistent with the current model for stargazin C-terminal domain-lipid bilayer binding (Sumioka et al., 2010) and supported our hypothesis that the extended electron densities in the projection structures were 6xHis-Stg C-terminal domains.
Figure 5. Increasing NaCl concentration disrupts 6xHis-Stg crystal packing by disrupting C-terminal domain-bilayer interactions.
A) A concentration of 50mM NaCl in the dialysis buffer resulted in uniform, rectangular crystal packing on ≥ 95% of proteoliposomes. B) Doubling the NaCl concentration to 100mM slightly compromised 6xHis-Stg crystal packing as evidenced by small (~50×50nm) gaps in most lattices. C) At a concentration of 250mM NaCl, 6xHis-Stg subunits were difficult to resolve as individual “jellyfish” and lattice formation was severely compromised. More lattice gaps were also apparent (arrows). D) At 500mM NaCl 6xHis-Stg transmembrane domains are visible but lacking tail domains (arrows); no lattices were observed at this NaCl concentration. Samples preserved in 1% uranyl acetate negative stain. Magnification = 27,000X; scale bar = 100nm.
3.6 Stargazin Overall Secondary Structure Increases Concomitant with the Interaction Between its Intracellular C-terminal Domain and the Lipid Bilayer
Based on a secondary structure prediction using Jpred 3, the stargazin C-terminal domain peptide sequence is thought to be largely disordered (not shown; Cole et al., 2008). To reconcile this disparity with the observation that some portion of stargazin's C-terminal domain is ordered in our structure, we performed circular dichroism (CD) on 6xHis-Stg (Fig. 6A). CD measurements performed on 6xHis-Stg crystals and detergent-solubilized 6xHis-Stg indicated that reconstitution of stargazin into the lipid mix was accompanied by an overall increase in alpha-helical secondary structure and a concomitant decrease in disordered polypeptide (Figs. 6B and 6C). In contrast, no significant change in either 6xHis-Stg beta-sheet or turn secondary structure was observed (Figs. 6D and 6E). Importantly, the α-helical content (~35% total secondary structure) for stargazin in solution, determined from CD spectra analysis, agreed well with the percentage of amino acid residues predicted to reside in transmembrane helices (~28% by TMHMM algorithm; Krogh et al., 2001). From these data we concluded that the increase in secondary structure observed in stargazin crystals most likely reflected some component of the C-terminal domain interacting with the lipid bilayer, as demonstrated in our structure. Moreover, the CD data also suggested that binding of the C-terminal domain to the bilayer is accompanied by structuring of part of the polypeptide, which could not have been deduced from the recently reported biochemical data that identified anionic head group lipids and stargazin phosphorylation as regulators of synaptic AMPAR activity (Sumioka et al., 2010).
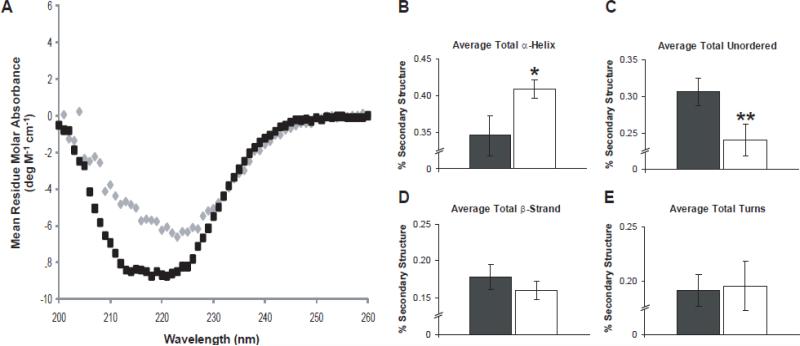
Figure 6. Interactions with anionic head group lipids induce structuring of 6xHis-Stg's C-terminal domain.
A) A secondary structure comparison of detergent-solubilized 6xHis-Stg dimer (black) and crystalline 6xHis-Stg (gray) by CD. B) – E) Bar graphs show the deconvolution of the CD spectra of detergent-solubilized 6xHis-Stg (black) and after reconstitution into the lipid mix that was used for crystallization (white) ± STD. Results are plotted as a fraction of the total where 1 equals 100%. Detergent-solubilized 6xHis-Stg: α-helix, 0.346 ± 0.027; unordered, 0.306 ± 0.019; β-strand, 0.178 ± 0.017; turns, 0.191 ± 0.015; n = 4. Crystalline 6xHis-Stg: α-helix, 0.409 ± 0.013; unordered, 0.240 ± 0.022; β-strand, 0.160 ± 0.012; turns, 0.195 ± 0.023; n = 3. Given P values are relative to detergent-solubilized 6xHis-Stg: * = p < 0.05, ** = p < 0.01 by Student's t test.
3.7 A Three-Dimensional Reconstruction of Stargazin Reveals a Membrane Anchor Close to the C-terminal PDZ-binding Motif
We next obtained three-dimensional reconstructions of membrane embedded stargazin. For this purpose, we focused on the simpler c222 crystal form because it formed the largest arrays and therefore gave the most reliable data. To compile the dataset, we collected and combined data from single-axis tilt series of ten crystals, which sampled different orientations of the lattices with respect to the tilt axis. We recorded images of crystals tilted up to 55° in five degree increments. The final reconstruction was calculated to a resolution of ≈22Å in the membrane plane and ≈31Å resolution perpendicular to the membrane plane. Effective resolution cutoffs were calculated from the point spread function of the fitted data set by comparison to an ideal data set with an in plane resolution cutoff of 16Å (Unger, 2000; refer to Table 2 for 3D reconstruction statistics, Supplementary Figure S4 for select lattice lines, and Supplementary Figure S5 for point spread function plots).
Table 2.
Summary of c222 (simple) 3D reconstruction statistics
| No. of images | 74 |
| Range of crystal tilts | 0° to 55° |
| Total no. of measurements | 21,986 |
| Total no. of fitted measurements | 13,026 |
| Effective resolution cutoff, Åa | |
| In-plane | x = 22.5 |
| y = 20.2 | |
| Vertical | 31.1 |
| Overall weighted phase residual | 24.1° |
Effective resolution cutoffs were determined from the point-spread functions of the data sets
The final reconstruction is shown in Figure 7. Viewed perpendicular to the membrane, the reconstruction showed all the features that were expected based on the projection maps (Fig. 7A). When viewed parallel to the membrane, the roughly triangular region that was observed in projection corresponded to a volume that spanned the entire bilayer and appeared to extend past the bilayer on either side of the membrane (Fig. 7B). Bracketing the bilayer on either side, part of the polypeptide extended along the bilayer surface, which explained the tail region of the projection and reinforced the interpretation that this part of the structure was formed by the 6xHis-Stg C-terminal domain. The arc-shaped C-terminal extension was most easily appreciated in contour plots of cross-sections taken at +/-24Å relative to the middle of the membrane (Fig. 7C).
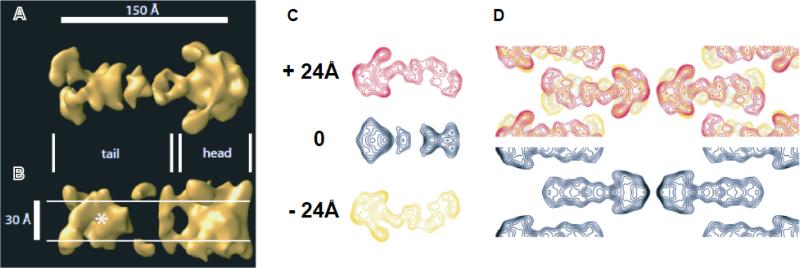
Figure 7. Three-dimensional TARP reconstruction at 20Å resolution.
Views of the 3D reconstruction perpendicular (A) and parallel (B) to the membrane revealed two prominent structural features that likely represented the bona fide transmembrane domain of stargazin dimers (the triangular “head”) and the extended C-termini (“tail”). The approximate boundaries of the hydrophobic core of the bilayer are indicated by white lines in the view parallel to the membrane. In this view, a second membrane-embedded region was apparent (marked with an asterisk) that was located at a distance of ~80Å from the transmembrane domains and likely represented a reentrant membrane anchor close to the C-terminal PDZ-binding motif. C) From top to bottom: contoured cross-sections taken at +24, 0, and -24Å with respect to the middle of the bilayer revealed more clearly than the rendered volume how the C-terminus extended from the transmembrane domain in an extended, arc-shaped manner. D) 6xHis-Stg structural features are explained by superposition of the three cross-sections (top) when compared to the observed projection structure of the simpler c222 projection structure (bottom).
In addition to the extended region of the C-terminus, the view parallel to the membrane revealed a second “membrane-embedded” region that was centered at a distance of ~80Å from the triangular body of the structure. This feature was very significant and easily recognizable in both surface rendered volumes (Fig. 7B, asterisk) and contour plots taken at the center of the bilayer (Fig. 7C, middle section). A superposition of the three representative cross-sections (Fig. 7D, top) illustrated how the different structural elements contributed to the observed projection structure (Fig. 7D, bottom). Notably, the extensions on both surfaces as well as the second re-entrant membrane region were also observed when the data set was reconstructed in the lower symmetry plane group p2 as well as in the p6 crystal form, neither of which imposes symmetry about the center of the bilayer (not shown). This validated the conclusion that 6xHis-Stg formed antiparallel dimers and justified application of the full c222 symmetry. More importantly, however, the conservation of the C-terminal domain extension between different crystal forms (one p6, two c222) suggested that the C-terminus adopted a well-defined structure in the presence of anionic lipid head group moieties.
4. Discussion
4.1 Stargazin Forms Dimers Advantageous for Structural Studies
Stargazin was purified as both a monomer and a putative dimer. The 6xHis-Stg fraction used for crystallization was purified as a homogenous and stable species based on gel filtration and two-dimensional class averages of the protein particles. Overall, the proportions of the class averages were consistent with the projection structures. However, the dimensions of the molecules in the class averages were smaller than the dimensions observed in the projection structures; specifically, the length (~100Å) of the particles, was shorter than the length (~150Å) of the antiparallel stargazin dimers observed in projection. We attributed this difference to flexibility and disorder of the C-terminal tail domain in solution. Based on previous structural studies of integral membrane proteins antiparallel dimerization is neither a unique phenomenon nor does it preclude study of a protein's structure and function ([Palczewski et al., 2000] and [Tate et al., 2001]). Notably, the antiparallel nature of the 6xHis-Stg dimer may have facilitated the formation of flat two-dimensional arrays because the presence of a re-entrant C-terminal anchor on only one side of the bilayer would induce membrane curvature in a way similar to what is observed in membrane tubulation by N-BAR domains (Frost et al., 2009).
4.2 Anionic Head Group Lipids Facilitate Two-Dimensional Crystallization via the Stargazin C-terminal Domain-Bilayer Electrostatic Interaction
Two-dimensional crystallization of stargazin required the presence of anionic head group lipids. Interestingly, previous studies have demonstrated that mammalian brain membranes contain 16% phosphatidylserine based on the total amount of phospholipid phosphorus present in these lipids (Dawson, 1960). This amount of PS is well within the range typically present in eukaryotic cellular membranes, and is closely aligned with our observation that 5–15% (w/w) of anionic head group lipid was necessary for two-dimensional crystallization of 6xHis-Stg. Taken together these findings suggest that the lipid composition we used mimicked the native bilayer composition encountered by stargazin in vivo. Moreover, scrutiny of the C-terminal domain sequence showed several positively charged regions which were likely to be responsible for the association of this cytoplasmic tail with the surface of the bilayer. This idea is supported by recent findings of Tomita's group who reported that the C-terminal domain can engage negatively charged bilayers even in the absence of full-length stargazin's transmembrane domains (Sumioka et al., 2010). Besides visualizing these interactions, our work extends the findings of Sumioka et al. by the discovery of a re-entrant membrane anchor that is proximal to the C-terminus and close to the PDZ-binding motif. Recent studies on the affects of PDZ-binding motif deletion on stargazin mobility at postsynaptic sites support our anchor finding (Opazo et al., 2010), but further studies will be necessary to determine if this is a bona fide feature of type I TARP tails. The low vertical resolution in our reconstructions precluded a determination of how deeply the putative membrane anchor inserted into the bilayer.
4.3 A Possible Role for the Cellular Cholesterol Gradient in Modulating In Vivo TARP Structure
Lastly, interactions between the transmembrane regions of stargazin dimers were modulated by the presence of cholesterol (Figs. 4B and 4C). Interestingly, use of 5% cholesterol (w/w) in our crystallization trials closely resembled the 6-8% of cholesterol reported to be globally present in the gray matter of human brains (O'Brien and Sampson, 1964). Moreover, the fact that the cholesterol content of bilayers in a eukaryotic cell typically increases from the endoplasmic reticulum (~10 mol% total phospholipid), where stargazin first interacts with AMPAR (Vandenberghe et al., 2005), to the plasma membrane (~45 mol% total phospholipid; van Meer, 1989), raises the intriguing possibility that cholesterol functions as a chemical chaperone in stargazin trafficking or as a modulator of TARP-AMPAR association. Though further studies will be required to determine the detailed role of lipids in aiding stargazin function, the data presented here provide novel insights into the complex nature of stargazin's interaction with and within the membrane bilayer.
5. Conclusion
TARPs are the primary integral membrane protein regulators of neuronal AMPAR trafficking and function ([Nicoll et al., 2006], [Payne, 2008], [Sager et al., 2009a], [Coombs and Cull-Candy, 2009], and [Tomita, 2010]). Here, we present the first direct visualization of the prototypical TARP stargazin. Extending conclusions drawn from recent biochemical studies, our reconstruction revealed that the C-terminal domain of stargazin adopts an extended, structured organization in the presence of anionic lipid head groups and that the far end of the C-terminus, close to the PDZ-binding motif, formed an additional membrane “anchor.” Our data were also in agreement with recent findings that immobilization of the stargazin C-terminus on the bilayer surface reduces accessibility of stargazin's PDZ-binding motif to MAGUK proteins (Sumioka et al., 2010). Moreover, the cholesterol content of the bilayer affected the interactions between the transmembrane regions of 6xHis-Stg subunits, which may be functionally relevant for stargazin assembly with AMPAR and trafficking of the complex to the cell surface.
Supplementary Material
Appendix A: Supplementary data
Supplementary Figure S1. Global Comparison of (h+k)=even versus (h+k)=odd
An analysis of phase relationships in the projection data of the rectangular crystal forms suggested the presence of c222 symmetry. In this two-sided plane group, reflections for which (h+k)=odd are absent everywhere in the molecular transform. However, due to noise and local breaks of symmetry, these absences are not always strictly observed in the images, which would preclude enforcement of the symmetry and adversely affect calculations of phase origins as well as lattice lines. To independently confirm that the symmetry was present in the 3D data, we analyzed the signal-to-noise ratio distribution in the two populations of reflections. Specifically, (h+k)=even (white bars) and (h+k)=odd (dark bars) were written out into separate data files. The files were then merged using the phase origins that had been calculated for the (h+k)=even set, and the quality of the signal-to-noise ratio (IQ) was tabulated for each of the two data sets by simply counting the IQ1-8 reflections in each data set. The average IQ was 5.7 for (h+k)=even and 7.2 (h+k)=odd. The histograms further illustrated that the vast majority of measurements with high to moderate signal-to-noise ratios were contained in the (h+k)=even half of the data. This was consistent with the presence of c222 symmetry. Moreover, the histogram distribution also justified the removal of the (h+k)=odd reflections, which ultimately have to be eliminated from the data at any rate if c222 symmetry were to be enforced.
Supplementary Figure S2. Phase errors for projection data
The program PLOTALL was used to plot the individual phase error for each structure factor after averaging of data from the various images. Phase errors are encoded as follows: 1, <8°; 2, <14°; 3, <20°; 4, <30°; 5, <40°; 6, <50°; 7, <80°; 8,<90°, where 90° is random; errors larger than 30° are indicated by decreasing box sizes. A) Phase error plot for data of unstained stargazin crystals belonging to the less complex of the two c222 forms. Data were averaged in p1, which is the reason why the full half of the transform is shown. While averages were significant to better than 10Å, the projection was only calculated to 15Å because completeness of the data beyond that resolution was low. Phase error plots for the simple (B) and complex (C) c222 forms. D) Phase error plot for the p6 crystal form. B)-D) all plots represent the combined phase error obtained after averaging and rounding to 0/180°.
Supplementary Figure S3. 6xHis-Stg 2D class averages
Particles were boxed out from seventeen raw micrographs using semi-automated particle picking in EMAN into boxes of 100×100 square pixels (Ludtke et al., 1999). The raw particles were normalized, high- and low-pass filtered, and centered. After discarding bad particles, 2548 particles were subjected to several iterations of reference-free 2D alignment and classification using multivariate statistical analysis and multi-reference alignment in IMAGIC to a final 100 classes (van Heel et al., 1996). The scale bar (lower right-hand corner) equals 100 Å.
Supplementary Figure S4. Select lattice lines at a range of resolutions
Only the phase part of the fit is shown in each case. Lines are plotted at 1/Å along the x-axis. Resolutions are for the respective (h,k,0) projection term. Lattice line (0,16) is shown to demonstrate that c222 symmetry constraints were obeyed in the 3D-transform.
Supplementary Figure S5. Point spread function plots for determining effective resolution cutoffs
Using a point spread function obtained for a simulated data set with an in-plane resolution cutoff of 16Å as a reference (A), effective resolution cutoffs were determined by comparison as in-plane cutoffs of x = 22.5Å, y = 20.2Å (B) and a vertical cutoff of 31.1Å (C) from the point spread function of the experimental data. Grid spacing corresponds to 10Å. Peaks were contoured at half height (solid line) in line with the Raleigh criterion for resolution.
References
S.J. Ludtke, P.R. Baldwin, and W. Chiu, EMAN: semiautomated software for high-resolution single-particle reconstructions, J. Struct. Biol. 128 (1999), pp. 82-97.
M. van Heel, G. Harauz, E.V. Orlova, R. Schmidt, and M. Schatz, A new generation of the IMAGIC image processing system, J. Struct. Biol. 116 (1996), pp. 17-24.
Acknowledgements
We thank Susumu Tomita for the gift of the full-length mouse stargazin pGEMHE construct and much constructive criticism. His evaluation of this work was essential for its completion and we are extremely grateful for his help. We thank Chen Xu of Brandeis University for use of cryo-EM facility equipment and his help in generating vitrified specimens. We also acknowledge the generous support of the Yale School of Medicine towards support of the local cryo-EM facility and especially thank Hongwei Wang for his help with single particle analysis. This work was supported by National Institutes of Health grants T32 GM007223 (Predoctoral Program in Cellular and Molecular Biology; to M.F.R.), F31 NS064748 (to M.F.R.), and DA024101 (to V.M.U.); D.W.T. is a National Science Foundation Graduate Research Fellow.
Abbreviations
- AMPAR
α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor
- TARP
transmembrane AMPAR regulatory protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bedoukian MA, Weeks AM, Partin KM. Different domains of the AMPA receptor direct stargazin-mediated trafficking and stargazin-mediated modulation of kinetics. J. Biol. Chem. 2006;281:23908–23921. doi: 10.1074/jbc.M600679200. [DOI] [PubMed] [Google Scholar]
- Chen L, Chetkovich DM, Petralia RS, Sweeney NT, Kawasaki Y, Wenthold RJ, Bredt DS, Nicoll RA. Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature. 2000;408:936–943. doi: 10.1038/35050030. [DOI] [PubMed] [Google Scholar]
- Chetkovich DM, Chen L, Stocker TJ, Nicoll RA, Bredt DS. Phosphorylation of the postsynaptic density-95 (PSD-95)/Discs Large/Zona Occludens-1 binding site of stargazin regulates binding to PSD-95 and synaptic targeting of AMPA receptors. J. Neurosci. 2002;22:5791–5796. doi: 10.1523/JNEUROSCI.22-14-05791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho C-H, St-Gelais F, Zhang W, Tomita S, Howe JR. Two families of TARP isoforms that have distinct effects on the kinetics properties of AMPA receptors and synaptic currents. Neuron. 2007;55:890–904. doi: 10.1016/j.neuron.2007.08.024. [DOI] [PubMed] [Google Scholar]
- Choi J, Ko J, Park E, Lee J-R, Yoon J, Lim S, Kim E. Phosphorylation of stargazin by protein kinase A regulates its interaction with PSD-95. J. Biol. Chem. 2002;227:12359–12363. doi: 10.1074/jbc.M200528200. [DOI] [PubMed] [Google Scholar]
- Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;38:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 The CCP4 suite: programs for protein crystallography. Acta. Crystallogr. D. Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Coombs ID, Cull-Candy SG. Transmembrane AMPA receptor regulatory proteins and AMPA receptor function in the cerebellum. Neuroscience. 2009;162:656–665. doi: 10.1016/j.neuroscience.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther RA, Henderson R, Smith JM. The MRC image processing programs. J. Struct. Biol. 1996;116:9–16. doi: 10.1006/jsbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- Dawson RMC. A hydrolytic procedure for the identification and estimation of individual phospholipids in biological samples. Biochem. J. 1960;75:45–53. doi: 10.1042/bj0750045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost A, Unger VM, De Camilli P. The BAR Domain Superfamily: Membrane-Molding Macromolecules. Cell. 2009;137:191–196. doi: 10.1016/j.cell.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaya M, Tsujita M, Yamazaki M, Kushiya E, Abe M, Akashi K, Natsume R, Kano M, Kamiya H, Watanabe M, Sakimura K. Abundant distribution of TARP gamma-8 in synaptic and extrasynaptic surface of hippocampal neurons and its major role in AMPA receptor expression on spines and dendrites. Eur. J. Neurosci. 2006;24:2177–2190. doi: 10.1111/j.1460-9568.2006.05081.x. [DOI] [PubMed] [Google Scholar]
- Huang CC, Couch GS, Pettersen EF, Ferrin TE. Chimera: an extensible molecular modeling application constructed using standard components. Pacific Symposium on Biocomputing. 1996;1:724. [Google Scholar]
- Kato AS, Zhou W, Milstein AD, Knierman MD, Siuda ER, Dotzlaf JE, Yu H, Hale JE, Nisenbaum ES, Nicoll RA, Bredt DS. New transmembrane AMPA receptor regulatory protein isoform, γ-7, differentially regulates AMPA receptors. J. Neurosci. 2007;27:4969–4977. doi: 10.1523/JNEUROSCI.5561-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato AS, Siuda ER, Nisenbaum ES, Bredt DS. AMPA receptor subunit-specific regulation by a distinct family of type II TARPs. Neuron. 2008;59:986–996. doi: 10.1016/j.neuron.2008.07.034. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Letts VA, Felix R, Biddlecome GH, Arikkath J, Mahaffey CL, Valenzuela A, Bartlett II FS, Mori Y, Campbell KP, Frankel WN. The mouse stargazer gene encodes a neuronal Ca2+-channel γ subunit. Nat. Genet. 1998;19:340–347. doi: 10.1038/1228. [DOI] [PubMed] [Google Scholar]
- Milstein AD, Zhou W, Karimzadegan S, Bredt DS, Nicoll RA. TARP subtypes differentially and dose-dependently control synaptic AMPA receptor gating. Neuron. 2007;55:905–918. doi: 10.1016/j.neuron.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Cheng Y, Ramm E, Sheng M, Walz T. Structure and different conformational states of native AMPA receptor complexes. Nature. 2005;433:545–549. doi: 10.1038/nature03328. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Cheng Y, Sheng M, Walz T. Three-dimensional structure of an AMPA receptor without associated stargazin/TARP proteins. Biol. Chem. 2006;387:179–187. doi: 10.1515/BC.2006.024. [DOI] [PubMed] [Google Scholar]
- Nicoll RA, Tomita S, Bredt DS. Auxiliary subunits assist AMPA-type glutamate receptors. Science. 2006;311:1253–1256. doi: 10.1126/science.1123339. [DOI] [PubMed] [Google Scholar]
- O'Brien JS, Sampson EL. Lipid composition of the normal human brain: gray matter, white matter, and myelin. J. Lipid Res. 1964;6:537–544. [PubMed] [Google Scholar]
- Opazo P, Labrecque S, Tigaret CM, Frouin A, Wiseman PW, De Koninck P, Choquet D. CaMKII triggers the diffusional trapping of surface AMPARs through phosphorylation of stargazin. Neuron. 2010;67:239–252. doi: 10.1016/j.neuron.2010.06.007. [DOI] [PubMed] [Google Scholar]
- Osten P, Stern-Bach Y. Learning from stargazin: the mouse, the phenotype and the unexpected. Curr. Opin. Neurobiol. 2006;16:275–280. doi: 10.1016/j.conb.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Payne HL. The role of transmembrane AMPA receptor regulatory proteins (TARPs) in neurotransmission and receptor trafficking. Mol. Membr. Biol. 2008;25:353–362. doi: 10.1080/09687680801986480. [DOI] [PubMed] [Google Scholar]
- Price MG, Davis CF, Deng F, Burgess DL. The alpha-amino-3-hydroxyl-5-methyl-4-isoxazolepropionate receptor trafficking regulator “stargazin” is related to the claudin family of proteins by Its ability to mediate cell-cell adhesion. J. Biol. Chem. 2005;280:19711–19720. doi: 10.1074/jbc.M500623200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager C, Tapken D, Kott S, Hollman M. Functional modulation of AMPA receptors by transmembrane AMPA receptor proteins. Neuroscience. 2009;158:45–54. doi: 10.1016/j.neuroscience.2007.12.046. [DOI] [PubMed] [Google Scholar]
- Sager C, Terhag J, Kott S, Hollmann M. C-terminal domains of transmembrane alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate (AMPA) receptor regulatory proteins not only facilitate trafficking but are major modulators of AMPA receptor function. J. Biol. Chem. 2009;284:32413–32424. doi: 10.1074/jbc.M109.039891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell E, Sizemore M, Karimzadegan S, Chen L, Bredt DS, Nicoll RA. Direct interactions between PSD-95 and stargazin control synaptic AMPA receptor number. Proc. Natl. Acad. Sci. USA. 2002;99:13902–13907. doi: 10.1073/pnas.172511199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk J, Harmel N, Zolles G, Bildl W, Kulik A, Heimrich B, Chisaka O, Jonas P, Schulte U, Fakler B, Klöcker N. Functional proteomics identify cornichon proteins as auxiliary subunits of AMPA receptors. Science. 2009;323:1313–1319. doi: 10.1126/science.1167852. [DOI] [PubMed] [Google Scholar]
- Shi Y, Suh YH, Milstein AD, Isozaki K, Schmid SM, Roche KW, Nicoll RA. Functional comparison of the effects of TARPs and cornichons on AMPA receptor trafficking and gating. Proc. Natl. Acad. Sci. USA. 2010;107:16315–16319. doi: 10.1073/pnas.1011706107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM. XIMDISP – a visualization tool to aid structure determination from electron microscope images. J. Struct. Biol. 1999;125:223–228. doi: 10.1006/jsbi.1998.4073. [DOI] [PubMed] [Google Scholar]
- Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreerama N, Woody RW. On the analysis of membrane protein circular dichroism spectra. Protein Sci. 2004;13:100–112. doi: 10.1110/ps.03258404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumioka A, Yan D, Tomita S. TARP phosphorylation regulates synaptic AMPA receptors through lipid bilayers. Neuron. 2010;66:755–767. doi: 10.1016/j.neuron.2010.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate CG, Kunji ERS, Lebendiker M, Schuldiner S. The projection structure of EmrE, a proton-linked multidrug transporter from Escherichia coli, at 7 Å resolution. EMBO J. 2001;20:77–81. doi: 10.1093/emboj/20.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita S, Chen L, Kawasaki Y, Petralia RS, Wenthold RJ, Nicoll RA, Bredt DS. Functional studies and distribution define a family of transmembrane AMPA receptor regulatory proteins. J. Cell Biol. 2003;161:805–816. doi: 10.1083/jcb.200212116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita S, Fukata M, Nicoll RA, Bredt DS. Dynamic interaction of stargazin-like TARPs with cycling AMPA receptors at synapses. Science. 2004;303:1508–1511. doi: 10.1126/science.1090262. [DOI] [PubMed] [Google Scholar]
- Tomita S, Adesnik H, Sekiguchi M, Zhang W, Wada K, Howe JR, Nicoll RA, Bredt DS. Stargazin modulates AMPA receptor gating and trafficking by distinct domains. Nature. 2005;435:1052–1058. doi: 10.1038/nature03624. [DOI] [PubMed] [Google Scholar]
- Tomita S, Stein V, Stocker TJ, Nicoll RA, Bredt DS. Bidirectional synaptic plasticity regulated by phosphorylation of stargazin-like TARPs. Neuron. 2005;45:269–277. doi: 10.1016/j.neuron.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Tomita S. Regulation of ionotropic glutamate receptors by their auxiliary subunits. Physiol. 2010;25:41–49. doi: 10.1152/physiol.00033.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turetsky D, Garringer E, Patneau DK. Stargazin modulates native AMPA receptor functional properties by two distinct mechanisms. J. Neurosci. 2005;25:7438–7448. doi: 10.1523/JNEUROSCI.1108-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger VM. Assessment of electron crystallographic data obtained from two-dimensional crystals of biological specimens. Acta. Crystallogr. D. Biol. Crystallogr. 2000;56:1259–1269. doi: 10.1107/s0907444900011252. [DOI] [PubMed] [Google Scholar]
- van Meer G. Lipid traffic in animal cells. Annu. Rev. Cell Biol. 1989;5:247–275. doi: 10.1146/annurev.cb.05.110189.001335. [DOI] [PubMed] [Google Scholar]
- Vandenberghe W, Nicoll RA, Bredt DS. Interaction with the unfolded protein response reveals a role for stargazin in biosynthetic AMPA receptor transport. J. Neurosci. 2005;25:1095–1102. doi: 10.1523/JNEUROSCI.3568-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziff EB. TARPs and the AMPA receptor trafficking paradox. Neuron. 2007;53:627–633. doi: 10.1016/j.neuron.2007.02.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix A: Supplementary data
Supplementary Figure S1. Global Comparison of (h+k)=even versus (h+k)=odd
An analysis of phase relationships in the projection data of the rectangular crystal forms suggested the presence of c222 symmetry. In this two-sided plane group, reflections for which (h+k)=odd are absent everywhere in the molecular transform. However, due to noise and local breaks of symmetry, these absences are not always strictly observed in the images, which would preclude enforcement of the symmetry and adversely affect calculations of phase origins as well as lattice lines. To independently confirm that the symmetry was present in the 3D data, we analyzed the signal-to-noise ratio distribution in the two populations of reflections. Specifically, (h+k)=even (white bars) and (h+k)=odd (dark bars) were written out into separate data files. The files were then merged using the phase origins that had been calculated for the (h+k)=even set, and the quality of the signal-to-noise ratio (IQ) was tabulated for each of the two data sets by simply counting the IQ1-8 reflections in each data set. The average IQ was 5.7 for (h+k)=even and 7.2 (h+k)=odd. The histograms further illustrated that the vast majority of measurements with high to moderate signal-to-noise ratios were contained in the (h+k)=even half of the data. This was consistent with the presence of c222 symmetry. Moreover, the histogram distribution also justified the removal of the (h+k)=odd reflections, which ultimately have to be eliminated from the data at any rate if c222 symmetry were to be enforced.
Supplementary Figure S2. Phase errors for projection data
The program PLOTALL was used to plot the individual phase error for each structure factor after averaging of data from the various images. Phase errors are encoded as follows: 1, <8°; 2, <14°; 3, <20°; 4, <30°; 5, <40°; 6, <50°; 7, <80°; 8,<90°, where 90° is random; errors larger than 30° are indicated by decreasing box sizes. A) Phase error plot for data of unstained stargazin crystals belonging to the less complex of the two c222 forms. Data were averaged in p1, which is the reason why the full half of the transform is shown. While averages were significant to better than 10Å, the projection was only calculated to 15Å because completeness of the data beyond that resolution was low. Phase error plots for the simple (B) and complex (C) c222 forms. D) Phase error plot for the p6 crystal form. B)-D) all plots represent the combined phase error obtained after averaging and rounding to 0/180°.
Supplementary Figure S3. 6xHis-Stg 2D class averages
Particles were boxed out from seventeen raw micrographs using semi-automated particle picking in EMAN into boxes of 100×100 square pixels (Ludtke et al., 1999). The raw particles were normalized, high- and low-pass filtered, and centered. After discarding bad particles, 2548 particles were subjected to several iterations of reference-free 2D alignment and classification using multivariate statistical analysis and multi-reference alignment in IMAGIC to a final 100 classes (van Heel et al., 1996). The scale bar (lower right-hand corner) equals 100 Å.
Supplementary Figure S4. Select lattice lines at a range of resolutions
Only the phase part of the fit is shown in each case. Lines are plotted at 1/Å along the x-axis. Resolutions are for the respective (h,k,0) projection term. Lattice line (0,16) is shown to demonstrate that c222 symmetry constraints were obeyed in the 3D-transform.
Supplementary Figure S5. Point spread function plots for determining effective resolution cutoffs
Using a point spread function obtained for a simulated data set with an in-plane resolution cutoff of 16Å as a reference (A), effective resolution cutoffs were determined by comparison as in-plane cutoffs of x = 22.5Å, y = 20.2Å (B) and a vertical cutoff of 31.1Å (C) from the point spread function of the experimental data. Grid spacing corresponds to 10Å. Peaks were contoured at half height (solid line) in line with the Raleigh criterion for resolution.
References
S.J. Ludtke, P.R. Baldwin, and W. Chiu, EMAN: semiautomated software for high-resolution single-particle reconstructions, J. Struct. Biol. 128 (1999), pp. 82-97.
M. van Heel, G. Harauz, E.V. Orlova, R. Schmidt, and M. Schatz, A new generation of the IMAGIC image processing system, J. Struct. Biol. 116 (1996), pp. 17-24.