Abstract
We have previously discovered the tubulin-binding anti-cancer properties of noscapine and its derivatives (noscapinoids). Here, we present three lines of evidence that noscapinoids bind at or near the well studied colchicine binding site of tubulin: 1) In silico molecular docking studies of Br-noscapine and noscapine yield highest docking score with the well characterised colchicine-binding site from the co-crystal structure; 2) the molecular mechanics-generalized Born/surface area (MM-GB/SA) scoring results ΔΔGbind-cald for both noscapine and Br-noscapine (3.915 and 3.025 kcal/mol) are in reasonably good agreement with our experimentally determined binding affinity (ΔΔGbind-expt of 3.570 and 2.988 kcal/mol, derived from Kd values); and 3) Br-noscapine competes with colchicine binding to tubulin. The simplest interpretation of these collective data is that Br-noscapine binds tubulin at a site overlapping with, or very close to colchicine-binding site of tubulin. Although we cannot rule out a formal possibility that Br-noscapine might bind to a site distinct and distant from the colchicine-binding site that might negatively influence the colchicine binding to tubulin.
Keywords: Noscapine, Br-noscapine, colchicine, binding affinity, docking, MM-GB/SA
1. Introduction
Chemotherapy remains the current modality of treatment for metastatic cancers. Microtubule (MT) dynamics has proven to be extremely useful target, in that along with DNA binding drugs MT-interacting drugs are now included in the first-line therapeutic modalities in many types of cancers. These include the haematological cancers, metastatic ovarian cancers, breast cancer, and lung cancer. The MT-targeting drugs currently approved fall in two classes: ones that depolymerise microtubules (such as vinca alkaloids), and the others that over polymerize MTs and bundle them (such as taxanes and epothilone) [1–4]. Because some human tissues also maintain frequent cell division, such as the erythropoetic bone marrow, the inner lining of the intestine, the hair-stem cells in hair follicles; these treatment strategies often lead to side effects including neurocytopenia, diarrhea, nausea, and hair loss [1]. Furthermore, even in post-mitotic neurons which require intact MT-organization for the essential axonal transport, current clinically infused intravenous MT-drugs such as vinca alkaloids and taxanes cause therapy limiting neurotoxicities. Hence, new types of MT-targeting drugs are needed that can avoid these side-effects of the existing MT-drugs.
In order to discover MT-interacting small orally available molecules that do not cause major reorganization of cellular microtubule arrays, we screened colchicine and podophyllotoxin like compounds that led to the discovery of noscapine [5]. Noscapine binds stoichiometrically to tubulin and alters tubulin conformation, suppresses the dynamics of microtubule assembly, blocks cell cycle progression at mitosis, and then causes apoptotic cell death in many cancer cell types [5, 6–9]. Furthermore, we have done extensive structure-activity relationship studies that led us to generate a battery of analogs among which Br-noscapine has shown potent activity against cancer cells without detectable toxicities to normal tissues [8]. Br-noscapine bound to tubulin with higher affinity (Kd = 54.9 ± 9.1 μM) than the lead compound noscapine (Kd = 144 ± 2.8 μM) [8]. Our extensive computational modelling efforts presented here elucidate the noscapinoid binding pocket of tubulin. The best way to understand the ligand binding site is to obtain a co-crystal structure. However, the low resolution co-crystal structures of colchicine, podophyllotoxin, taxane, and epothilone have been possible by the co-crystallization of these drugs with tubulin/stathmin-like-domain complexes based on low-energy electron crystallography studies [10–12]. Structural similarities between Br-noscapine, colchicine and podophyllotoxin allowed us to investigate if Br-noscapine competed with colchicine for tubulin binding. The molecular modelling calculations tally reasonably well with the experimental data.
2. Materials and methods
A. Experimental methods
2.1. Extraction and purification of tubulin
Microtubule proteins were isolated from goat brain in presence of 1M glutamate and 10% v/v DMSO by two cycles of polymerization and depolymerisation as described previously [14]. From the extracted microtubule proteins, tubulin was then purified using phosphocellulose chromatography [8,15]. Tubulin concentration was determined by Bradford method [16] using bovine serum albumin (BSA) as the standard. The purified tubulin was quickly frozen as drops in liquid nitrogen and stored at − 80 °C until further use.
2.2. Competition binding of Br-noscapine and colchicine to tubulin
Colchicine is nonfluorescent in aqueous medium, but fluoresces after binding to tubulin around 430 nm [17]. This has formed the basis for the reliable quantitative standard assay for colchicine/tubulin interaction [13], which is routinely used in determining the competitive interactions of other ligands at the colchicine-binding site of tubulin [18]. Here we used this assay to determine if Br-noscapine competitively inhibits colchicine-tubulin interaction. In brief, Br-noscapine (25, 50 and 100 μM) were incubated with 6 μM tubulin in PEM buffer consisted of 25 mM PIPES (pH 6.8), 1 mM EGTA and 3 mM MgSO4 for 20 min at 25 °C. Additionally, 100 μM noscapine was used in the similar experimental conditions to compare its effect with that of Brnoscapine. After 20 min of incubation 12 μM colchicine was added to the reaction mixture and incubated for an additional 45 min at 37 °C. Fluorescence intensity of tubulin-colchicine complex was then monitored in a JASCO FP-6500 spectrofluorometer (JASCO, Tokyo, Japan) by exciting the samples at 350 nm and emission was recorded in the range of 400–470 nm. A 0.3 cm path-length cuvette was used to minimize the inner filter effects caused by the absorbance of these agents at higher concentrations. Fluorescence intensities were corrected for inner filter effect using the formula Fcorrected = Eobserved × antilog [(Aexcitation + Aemission)/2]. Spectra of different concentrations of Br-noscapine with 6 μM tubulin (considered as blanks) were also taken. Fluorescence intensity spectra of tubulin-colchicine complex in the absence and presence of Br-noscapine was plotted after subtracting the corresponding blanks. Noscapine was purchased from Sigma-Aldrich. Br-noscapine was synthesized in our lab [8].
B. Molecular modeling calculations
2.3. Preparation of protein
We have used the co-crystal structure of colchicine-tubulin (PDB ID: 1SA0; 3.58 Å resolution) [10] for the preparation of colchicine binding site. We retained a complex of colchicine-tubulin complex consisting of only `A' and `B' chains. All water molecules were removed form the complex. Hydrogen atoms were added to the model using Maestro interface (version 8.5; Schrodinger LLC, New York) based on an explicit all atom model. Final preparation of tubulin-colchicine complex was done using the multi step Schrödinger's protein preparation tool (PPrep) followed by energy minimization using OPLS 2005 force field with Polack-Ribiere Conjugate Gradient (PRCG) algorithm [19]. The minimization was stopped either after 5,000 steps of minimizations or after the energy gradient converged below 0.001 kcal/mol.
2.4. Ligand preparation
Molecular structures of noscapine and Br-noscapine were built using the builder feature in Maestro (Schrödinger package) whereas molecular structure of colchicine was extracted from the PDB ID:1SA0. Each structure was assigned an appropriate bond order using ligprep (version 2.3, Schrodinger Inc.). Ligprep utility produces a number of structures from each input structure with various ionization states, tatutomers, stereochemistries, and ring conformations. The program automatically generated all possible stereoisomers (default value of 32 was used) for each ligand. Furthermore, a unique low-energy ring conformation for each stereoisomer with correct chirality was generated with the help of Ligprep. All structures were subsequently subjected to molecular mechanics energy minimization using Impact (version 5.6, Schrodinger Inc.) with default settings: maximum cycles 100, conjugate gradient minimizer, initial step size 0.05, maximum step size 1.0, gradient criteria 0.01. Partial atomic charges were assigned to the molecular structures using the 2005 implementation of the OPLS-AA force field. To ensure that the geometry of the structure is fairly reasonable, we performed complete geometric optimization of these structures using Jaguar (version 7.7; Schrödinger Inc.). We have used hybrid density functional theory with Becke's three-parameter exchange potential and the Lee-Yang-Parr correlation functional (B3LYP) [20,21] and 3-21G* basis set [22–24] for geometrical optimization. These geometrically optimized structures were used for Glide (grid-based ligand docking with energetics) docking.
2.5. Molecular docking of colchicine, noscapine and Br-noscapine to colchicine binding site
All docking calculations were performed using the “Extra Precision” (XP) mode of Glide docking [25,26] (version 4.5, Schrodinger Inc.) with the 2005 implementation of the OPLS-AA force field. The detailed algorithm of Glide docking has been described previously [25,26]. Briefly, Glide approximates a systematic search of positions, orientations, and conformations of the ligand in the receptor binding site using a series of hierarchical filters. The shape and properties of the receptor are represented on a grid by several different sets of fields that provide progressively more accurate scoring of the ligand pose. The binding site is defined in terms of two concentric cubes: the bounding box, which must contain the mass center of any acceptable ligand pose, and the enclosing box, which must contain all the atoms of a ligand pose for successful docking into the binding site. Glide also performed conformational searches for each input structure during docking process. A set of initial ligand conformations is generated through exhaustive search of the torsional minima and the conformers are clustered in a combinatorial fashion. Each cluster, characterized by a common conformation of the core and an exhaustive set of side-chain conformations, is docked as a single object in the first stage. The search begins with a rough positioning and scoring phase that significantly narrows the search space and reduces the number of poses to be further considered to a few hundred. These selected poses are energy minimized on precomputed OPLS-AA van der Waals and electrostatic grids for the receptor. In the final stage, the 5–10 lowest-energy poses obtained in this fashion are subjected to a Monte Carlo sampling in which nearby torsional minima are examined, and the orientation of peripheral groups of the ligand is refined. The minimized poses are then rescored using the GlideScore function.
In this work the bounding box of size 10Å × 10Å × 10Å was defined in tubulin and centered on the mass center of the crystallographic colchicine to confine the mass center of the docked ligand. The larger enclosing box with an edge length of 10 Å was also defined (which occupied all the atoms of the docked poses) in terms of the co-crystallized colchicine. The scale factor of 0.4 for van der Waals radii was applied to atoms of protein with absolute partial charges less than or equal to 0.25. Five thousand poses per ligand were generated during the initial phase of the docking calculation, out of which best 1000 poses per ligand were chosen for energy minimization. Energy minimization protocol included dielectric constant of 4.0 and 1000 steps of conjugate gradient minimizations. Upon completion of each docking calculation, 100 poses per ligand were generated and the best docked structure was chosen using a GlideScore (Gscore) function. GlideScore is a more sophisticated version of ChemScore [27] with force field–based components and additional terms accounting for solvation and repulsive interactions. The choice of the best pose is made using a model energy score (Emodel) that combines the energy grid score, Gscore, and the internal strain of the ligand. Glide docking is widely used by pharmaceutical industries and academic institutes to study drug-target interactions and to design new drug candidates with improved activities because of its better accuracy [28–31]. The resulted docked complexes of noscapine, Br-noscapine, and colchicine were then subjected to further refinement using quantum mechanical-polarized ligand docking protocol as explained below.
Furthermore, we have implemented the “blind docking” approach to calculate the Gscore of noscapinoids with other predicted binding sites of tubulin. Different binding sites of tubulin were predicted using SiteMap (version 2.4, Schrodinger Inc.) and the receptor-grid files were generated (van der Waals scaling 0.4, grid box size of 10Å × 10Å × 10Å each for the bounding and enclosing box) at the centroid of each predicted binding site. Both noscapine and Br-noscapine were then docked into each binding site using Glide XP methods mentioned above.
2.6. Quantum mechanical-polarized ligand docking (QPLD) protocol
QPLD protocol is an improved docking method that incorporates quantum mechanical and molecular mechanical (QM/MM) calculations [32]. Briefly, this protocol applies three basic steps: 1) Glide algorithm generates the best candidate poses for ligand docking; 2) replaces the partial charges on ligand-atoms with charges derived from QM calculations of ligand in the field of the receptor for each ligand-receptor complex (the QM charges are calculated from the electrostatic potential energy surface of the ligand that is generated from a single-point calculation using BLYP density function for the QM region); 3) Glide then re-docks each ligand with updated atomic charges and returns the most energetically favourable pose. The grid file generated from the previous Glide standard docking was used for QPLD refinement. The level of quantum-mechanical treatment was set as Fast mode.
2.7. Post-scoring with MM-GB/SA
We have used noscapine, Br-noscapine and colchicine that have been pre-positioned with tubulin from Glide docking to study the association of these ligands with the receptor using the automated mechanism of multi-ligand biomolecular association with energetics (Embrace, Macromodel version 9.0; Schrödinger, LLC, New York). The complexes were energy minimized to perform a conformational search for the ligands in the unbound state with Embrace, specifying OPLS 2005 force field (by default), GB/SA continuum solvation treatment, and electrostatic treatment (dielectric constant of 1.0) [33–36]. Monte Carlo multiple minimum (MCMM) method (Macromodel version 9.8; Schrodinger, LLC, New York) was used for the conformational search of ligands. To ensure that the stochastic search was exhaustive and approached convergence, we employed the extended protocol for the torsion sampling and energy minimization to a low gradient norm. We retained all conformers within 5.0 kcal/mol from the lowest-energy conformer. A Boltzmann distribution was assumed to calculate the probabilities for each conformer (Pi) and the Boltzmann-averaged intra-molecular energy and solvation free energy in the unbound state were obtained for every compound. Conformational entropies (Sconf) were computed from the probabilities using equation 1, where kB is the Boltzmann constant.
| (1) |
To better account for the protein flexibility, the best pose for each molecule was energy-minimized in the bound state using conjugate gradient minimization scheme applying PRCG algorithm with a very tight convergence threshold. No constraints were applied to all receptor atoms from complete residues within 5 Å of the ligand during the energy minimization. A second shell of 3 Å around the first shell was defined, and constraints of 50 kcal/mol.Å2 were applied to the residues within it. Because the large portion of protein structure beyond 8 Å from the ligand does not significantly influence either the structure of the minimized ligand or its vicinity, the calculations were performed within the local 8 Å regions as previously reported [34–36]. The residues beyond the 8 Å boundary were restrained to lie close to the requested positions, but the moving atoms are allowed to interact with all other moving and fixed atoms in the usual way. Among the fixed atoms, however, only stretch interactions are considered. This method provides buffer zones between fully moving and ignored regions reducing significantly the computational time. When modelling large systems, such as the tubulin molecule, it is desirable to focus calculations on important regions and ignore any large portions of the system from which no significant influence is expected.
After accomplishment of energy minimization, the strain imposed by each ligand to the protein (Eptn) conformation was noted. We also calculated other energy parameters such as intramolecular (ΔEintra) and desolvation (ΔEsolv) penalty for the ligands upon binding, the protein-ligand intermolecular van der Waals (EVDW) and electrostatic (Eelect) interaction energies. The binding energy (ΔGbind) was calculated including the conformational entropy penalty as follows.
| (2) |
where, −TΔSconf is the ligand conformational entropy penalty, which was multiplied by temperature to convert into free energy. The relative binding energy (ΔΔGbind-cald) for noscapine and Br-noscapine was obtained by using colchicine as a reference by following relation.
| (3) |
The experimental free energy of binding for both noscapine and Br-noscapine were calculated from their respective dissociation constant (Kd) values using the relation:
| (4) |
where R is gaseous constant (0.001986 kcal/mol) and T is temperature (298 K).
The Kd values of 144 ± 2.8 μM for noscapine and 54 ± 9.1 for Br-noscapine, binding to tubulin were obtained from our earlier published work [8, 37] by measuring concentration dependent tubulin-binding curves derived from escalating concentrations of the compounds (until saturation) followed by Schatchard plots. This method has previously been used by many independent groups including Sherline et al, [38] to derive Kd for colchicine-tubulin interaction (0.35μM). The relative binding energy (ΔΔGbind-expt) of noscapine and Br-noscapine were then calculated from the experimental Kd values with reference to colchicine using the relation:
| (5) |
3. Results and discussion
3.1. Competitive interaction of Br-noscapine with colchicine binding
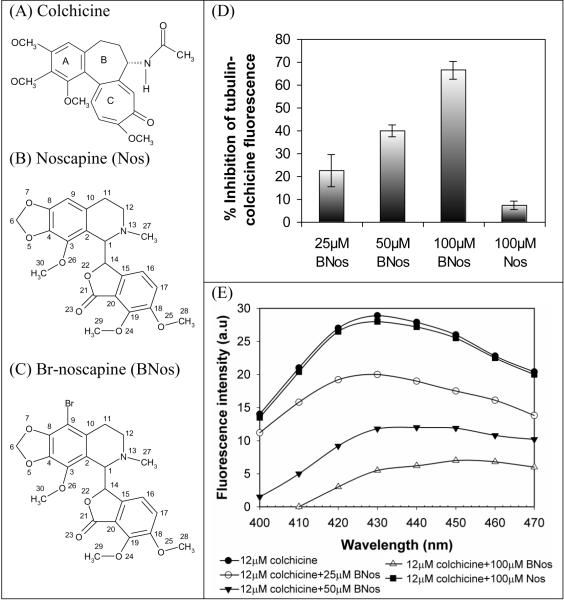
Although colchicine is nonfluorescent by itself, it shows clear novel fluorescent properties when bound to tubulin (excitation at 350 nm and emission at 430 nm) [13,39,40]. The actual binding reaction of colchicine is a slightly complex biphasic reaction. It first binds to tubulin in a quick diffusion limited step into an extremely low affinity site (phase I), and then it settles deep into a practically inexchangable site that is some times referred to as “irreversible” site (phase II). For this reason, the competition of structurally related ligands to colchicine-binding site are designed to inhibit the binding of colchicine onto tubulin pre-incubated with the test-ligands [18,39,41–43]. We initially performed such colchicine competition experiment with Br-noscapine at three different concentrations beginning at high concentration of 100 μM. Our previous efforts of noscapine at even up to 100 μM concentrations showed little (merely 5%) inhibition of colchicine binding. We observed that Br-noscapine at 100 μM concentration produced a robust competition of 66 ± 5% reduction with colchicine-binding. This is perhaps due to higher affinity of Br-noscapine to tubulin in comparison to noscapine [8]. Encouraged by this, we carried out competition binding of Br-noscapine with two additional lower concentrations: 25 μM that produced a modest inhibition of colchicine binding (23 ± 7%) and 50 μM that produced 40 ± 4% inhibition (Fig. 1d). We can not reliably measure less than 5% inhibition with any meaningful statistical significance thus the lower concentrations of noscapine were not further tested. The fluorescence emission of tubulin-colchicine complex is inhibited by Br-noscapine in a concentration dependent manner ranging from 25 μM to 100 μM (Fig. 1e). The Br-noscapine data indicate that the interaction of Br-noscapine with tubulin most likely inhibited binding of colchicine.
Figure 1.
Inhibition of tubulin-colchicine fluorescence by Br-noscapine and comparision with noscapine. A, B, and C show the molecular structures of colchicine, noscapine (Nos) and Br-noscapine (BNos), respectively. The fluorescence emission of tubulin-colchicine complex is inhibited by Br-noscapine in a concentration dependent manner ranging from 25 μM to 100 μM (D). However, noscapine inhibited tubulin-colchicine fluorescence modestly even at concentration as high as 100 uM (>10%, D). E, the percentage inhibition of fluorescence from colchicine-tubulin complex (it can be viewed as the inhibition of colchicine binding to tubulin).
3.2. Computational determination of noscapinoid binding pocket of tubulin
Early reports have revealed that noscapinoids binds to tubulin and alters microtubule dynamics both in vitro and in vivo [3–9, 37]. However, where do noscapinoids bind specifically to tubulin is still not known. Therefore we have used an approach of “blind docking” to determine the probable site of interactions of noscapinoids with tubulin which could better correlate with the experimental binding affinity. In this approach we have used the different predicted binding sites (using SiteMap, Schrödinger Inc.) of tubulin for molecular docking and evaluation of binding sites. Eventually it turned out to be the colchicine binding site of tubulin that gave better docking score for noscapinoids (Table 1) – indicating a binding site for noscapinoids overlapping with colchicine binding site [18] or a site very close to it. However, there are minor differences in the molecular interactions of noscapinoids that perhaps account for their net affect on the microtubule assembly reaction that is different than that of colchicine.
Table 1.
Docking results (Glide XP) of noscapine and Br-noscapine with respect to different binding sites predicted by SiteMap (Schrödinger Inc.). Site 10 (correspond to colchicine binding site) is having better Glide score for both noscapine and Br-noscapine.
| Site ID | Site score | Volume | Glide XP score (kcal/mol) | |
|---|---|---|---|---|
| Noscapine | Br-noscapine | |||
| 1 | 1.022 | 1483.8 | −5.170 | −5.466 |
| 2 | 1.106 | 220.9 | −4.084 | −4.349 |
| 3 | 0.910 | 236.6 | −4.794 | −4.871 |
| 4 | 0.916 | 127.6 | −0.751 | −1.869 |
| 5 | 0.849 | 138.6 | −2.113 | −2.342 |
| 6 | 0.793 | 170.5 | −2.150 | −2.385 |
| 7 | 0.746 | 67.2 | −1.957 | −3.263 |
| 8 | 0.671 | 84.7 | −2.296 | −3.336 |
| 9 | 0.886 | 85.7 | −1.042 | −1.302 |
| 10 | 0.632 | 80.6 | −5.521 | −6.261 |
3.3. Validation of the docking method by reproducing the crystallized colchicin/tubulin structure
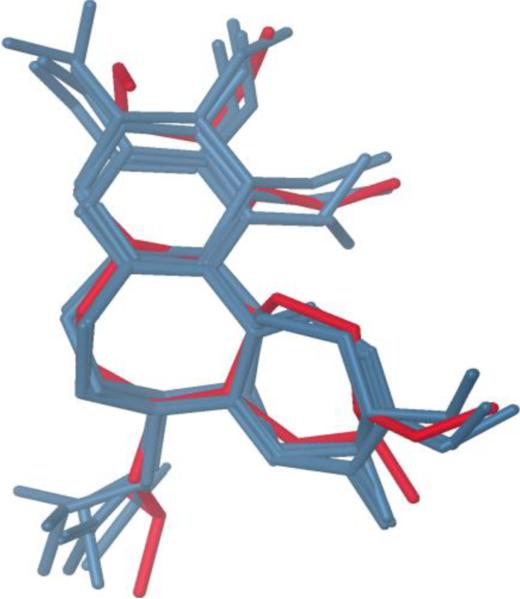
The original crystal structure of tubuline-colchicine complex (PDB ID: 1SA0) was used to validate the Glide-XP docking protocol. This was done by moving the crystallized colchicine ligand outside of active site and then docking it back into the active site. The top 6 configurations after docking were taken into consideration to validate the result (Table 2). The root mean square deviation (RMSD) for each configuration in comparison to the co-crystal colchicine was 0.19–1.06 Å. Whereas the RMSD value calculated from the accepted poses for each configuration was 0.35–0.82 Å. This revealed that the docked configurations have similar binding positions and orientations within the binding site and are similar to the crystal structure. The best docked structures, which are the configurations with the lowest Gscore, were compared with the crystal structure as shown in Fig. 2. These docking results illustrate that the best-docked colchicine complex agrees well with its crystal structure, and Glide (XP)-docking protocol successfully reproduces the crystal structure of colchicine. After this validation, the docking protocol was extended to both noscapine and Br-noscapine that docked reasonably well at the colchicine binding site (Fig. 3a,b).
Table 2.
The RMSD and docking score from the docking simulation of 6 lowest configurations of co-crystal colchicine with tubulin (1SA0).
| Configuration | Glide score | Δ Gscorea | RMSDb(Å) | RMSDc(Å) |
|---|---|---|---|---|
| 1 | −9.88 | 0 | 0.35 | 0.19 |
| 2 | −8.94 | 0.94 | 0.45 | 0.24 |
| 3 | −8.46 | 1.42 | 0.68 | 0.59 |
| 4 | −8.23 | 1.65 | 0.33 | 0.76 |
| 5 | −8.05 | 1.83 | 0.14 | 0.92 |
| 6 | −7.64 | 2.24 | 0.82 | 1.06 |
ΔGscore = Ei − Elowest;
RMSD, RMSD between docked poses corresponding to each configuration;
RMSD, RMSD between docked and co-crystal colchicine structure (only heavy atoms from core rings are considered).
Figure 2.
Superimposition of docked configurations onto the crystal structure of colchicine (red stick). The RMSD (heavy atoms from core rings) = 0.19 –1.06 Å.
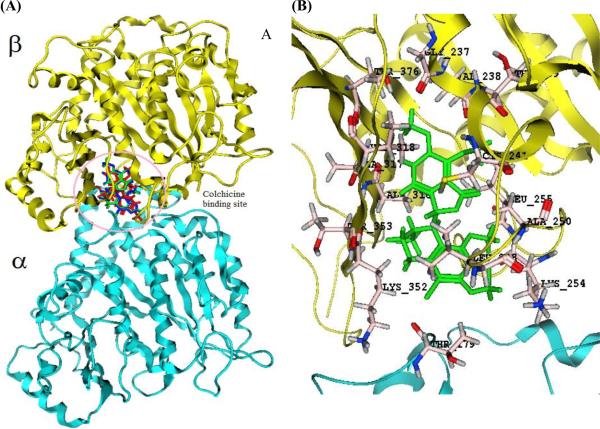
Figure 3. Comparison of colchicine, noscapine, and Br-noscapine with the colchicine binding site of tubulin.
(A) Showing the localization of docked configurations of these ligands primarily in β-tubulin near the α- tubulin/β-tubulin interface. The amino acid residues involved in the interaction of Br-noscapine at the binding site is represented in (B).
3.4. Architecture of the noscapinoid binding site
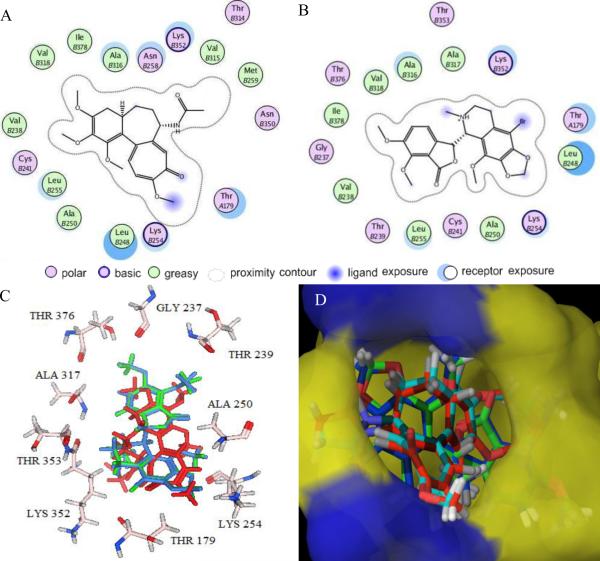
The computationally determined noscapinoid binding pocket of tubulin is considerably hydrophobic. The ligand plot generated from the tubulin-colchicine and tubulin-Br-noscapine docked complexes revealed identical sets of amino acids, interacting with both ligands (Fig. 4a & b). The binding modes and key protein-ligand interactions are shown in Fig. 4c. The molecular superposition of bound conformation of colchicine, noscapine, and Br-noscapine indicates that these compounds have more or less identical binding mode with tubulin. Macromodel surface representation of binding site according to residue charge (electropositive charge) is shown in Fig. 4d.
Figure 4. Results of Glide XP docking of colchicine, noscapine, and Br-noscapine into the colchicine binding site of tubulin.
2D representations of the binding site with amino acid residues involved in the interaction of (A) colchicine and (B) Br-noscapine reveal identical sets of amino acids. (C) 3D representations of the binding site amino acids within a 4Å distances from superimposed conformations of docked noscapine (blue) and Br-noscapine (green). The co-crystal structure of colchicine is represented in red color. Some of the amino acids (Leu248, Cys241, Val318, Ala316 and Val238) are not shown for better clarity. (D) Overlapped docking poses of colchicine, noscapine, and Br-noscapine obtained form Glide docking onto co-crystal of colchicine within the colchicine binding site of tubulin. Tubulin is represented as Macromodel surface according to residue charge (electropositive charge, blue; neutral, yellow) as implemented in Maestro.
3.5. QPLD refinement of the docking poses
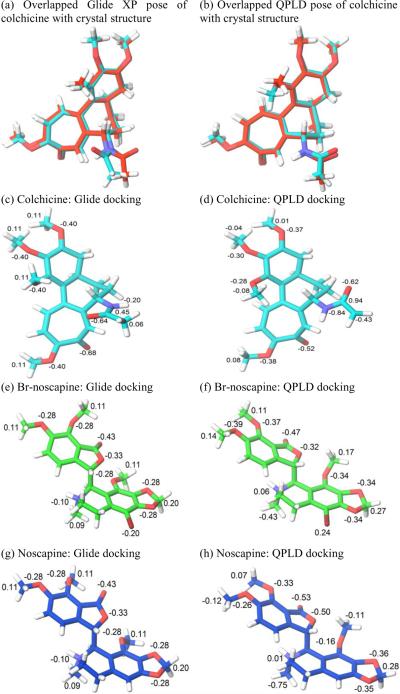
Obtaining accurate structural information on the binding pose of a ligand at a binding site is essential to the design of optimized lead compounds in computer-aided drug discovery. An accurate calculation of atomic partial charges of a ligand in the receptor field would result in improved docking results. We tested whether charges obtained from the QM/MM calculation for ligand/tubulin structure would provide a more precise binding pose compared to the standard docking method that relies on the default force-field charges. The results of QPLD docking runs for colchicine, noscapine, and Br-noscapine are shown in Fig 5. The root mean square deviation (RMSD) value between crystal and docking poses of colchicine was 0.760 Å from the standard docking run (Fig. 5a), while the QPLD method returned a significantly improved RMSD value of 0.595 Å (Fig. 5b). Atomic charge values in parts of the ligand structure were also represented in the figures (Fig. 5c–h). These results indicate that polarization effects induced by the field of the receptor have improved the docking predictions.
Figure 5.
Overlapped docking poses of colchicine obtained from (a) Glide docking (RMSD = 0.760 Å) and (b) QPLD (RMSD = 0.595 Å). In this figure the conformation of colchicine in the co-crystal structure is represent in red color. Comparison between force field charges and quantum mechanical charges for colchicine, Br-noscapine and noscapine from standard Glide docking and QPLD docking.
3.6. Glide docking and rescoring using MM-GB/SA of ligands
Molecular docking methods are widely used by academic institutes and pharmaceutical industries to study drug-target interactions in order to understand the basic electronic/steric features required for therapeutic action and to design new drug candidates with improved activities. These docking calculations provide insight into interactions of ligands with amino acids in the binding pocket of a target and to predict the corresponding binding affinities of ligands [44]. Table 3 presents Glide XP docking results of noscapine, Br-noscapine, and colchicine. Their Glide score values ranges from − 6.047 to −9.884 kcal/mol. The relative docking score (ΔGscore) of noscapine and Br-noscapine with reference to colchicine is 3.837 and 3.623 kcal/mol respectively (Table 3). These results demonstrate strong binding of Br-noscapine to tubulin in comparison to noscapine.
Table 3.
Docking results (Glide XP) for colchicine, noscapine and Br-noscapine.
| Compound | Glide score (Gscore) | Glide energy | Glide Emodel | Δ Gscore | Δ Δ Gbind-Expt |
|---|---|---|---|---|---|
| Colchicine | −9.884 | −56.09 | −88.34 | 0.0 | 0.0 |
| Br-noscapine | −6.261 | −25.32 | −25.56 | 3.623 | 2.988 |
| Noscapine | −5.521 | −17.84 | −18.99 | 4.363 | 3.570 |
All the energy parameters are expressed in kcal/mol. ΔGscore = Gscore-ligand – Gscore-colchicine and ΔΔGbind-Expt was obtained by calculating relative binding energies using colchicine, the molecule used as reference in competition binding assay. ΔΔGbind-Expt values were calculated from the experimental Kd values (noscapine = 144μM; Br-noscapine = 54 μM and colchicine = 0.35μM) using ΔΔGbind-Expt = − RT In (colchicineKd/ligandKd).
Although more computationally demanding, the MM-GB/SA scoring generally yields far superior correlations with experimentally determined activity than standard docking scoring functions [45–48]. The docked complexes were rescored with MM-GB/SA and the relative binding energy (ΔΔGbind-cald) of noscapine and Br-noscapine was calculated using colchicine as reference. Table 4 shows that the MM-GB/SA scoring yields a better correlation with the experimental relative binding energy (ΔΔGbind-exp) for both noscapine and Br-noscapine. This is indeed due to the flexibility of these molecules. The desolvation and intramolecular strain penalties show greater variation and the deficiencies of the docking functions are amplified. In comparison to colchicine, Br-noscapine yielded poorer desolvation and intramolecular penalty and hence strong interactions with tubulin; while noscapine displayed weaker interactions with tubulin (Table 4). The drop in calculated relative binding energy (ΔΔGbind-cald) for Br-noscapine revealed strong binding (3.025 kcal/mol) to tubulin in comparison to noscapine (3.915 kcal/mol). The calculated value of ΔΔGbind-cald for both noscapine and Br-noscapine (3.915 and 3.025 kcal/mol) was in reasonable agreement with the experimentally determined value (ΔΔGbind-expt is 3.570 and 2.988 kcal/mol).
Table 4.
MM-GB/SA results for colchicine, noscapine and Br-noscapine.
| Compound | Δ Δ Gbind-cald | Δ Δ Gbind-Expt | T Δ Sconf | Δ Esolv | Δ Eintra | Δ EVDW | Eelect | EPTN |
|---|---|---|---|---|---|---|---|---|
| Colchicine | 0.0 | 0.0 | 1.347 | 24.63 | 3.144 | −28.39 | 14.16 | −1298.8 |
| Br-noscapine | 3.025 | 2.988 | 1.435 | −52.04 | 4.841 | −17.05 | 83.30 | −1301.4 |
| Noscapine | 3.915 | 3.570 | 1.428 | −26.01 | 6.551 | −23.33 | 64.57 | −1303.3 |
All the energy parameters are expressed in kcal/mol. ΔΔGbind-cald and ΔΔGbind-Expt were obtained by calculating relative binding energies using colchicine, the molecule used as reference in competition binding assay. ΔΔGbind-expt values were calculated from the experimental Kd values (noscapine = 144 μM; Br-noscapine = 54 μM and colchicine = 0.35 μM) using ΔΔGbind-Expt = − RT In (colchicineKd/ligandKd). EPTN is the protein energy, EVDW and Eelect are the van der Waals and electrostatic interaction energies, and ΔSconf, ΔEintra, and ΔEsolv are the conformational entropy, intramolecular, and desolvation penalties for each ligand upon binding (see eq. 2).
4. Conclusion
In this manuscript we have presented three lines of evidence that noscapinoids bind at or near the colchicine binding site of tubulin: 1) Br-noscapine and noscapine yield highest docking score with the well characterised colchicine-binding site; 2) MM-GB/SA scoring results ΔΔGbindcald for both noscapine and Br-noscapine are in good agreement with experimentally determined binding affinity; 3) Br-noscapine competes with colchicine binding to tubulin. The most likely explanation of our collective data indicate to a binding site of noscapinoids very close to or overlapping with the colchicine binding site of tubulin.
Supplementary Material
Acknowledgement
We are thankful to Jaypee University of Information Technology, India for providing sabbatical leave to Pradeep K. Naik and the members of the Joshi laboratory for critically reading the manuscript. Grant support: NIH grants CA-095317-01A2 (H.C. Joshi) and BOYSCAST fellowship (SR/BY/L-37/09; Department of Science and Technology, Govt. of India) to Pradeep K. Naik.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary information: Consists of the coordinates of the refined poses (both protein and ligands) using QPLD.
References
- [1].Rowinski EK. The development and clinical utility of the taxane class of animicrotubule chemotherapy agents. Annu. Rev. Med. 1997;48:353–374. doi: 10.1146/annurev.med.48.1.353. [DOI] [PubMed] [Google Scholar]
- [2].Jordan MA, Wilson L. The use and action of drugs in analyzing mitosis. Methods Cell Biol. 1999;61:267–295. doi: 10.1016/s0091-679x(08)61986-x. [DOI] [PubMed] [Google Scholar]
- [3].Checchi PM, Nettles JH, Zhou J, Snyder JP, Joshi HC. Microtubule-interacting drugs for cancer treatment. Trends in Pharmacol. Sci. 2003;24:361–365. doi: 10.1016/S0165-6147(03)00161-5. [DOI] [PubMed] [Google Scholar]
- [4].Joshi HC, Salil A, Bughani U, Naik PK. Medicinal Plant Biotechnology. CABI South Asia Edition; UK: 2010. Noscapinoids: a new class of anticancer drugs demand biotechnological intervention; pp. 303–320. [Google Scholar]
- [5].Ye K, Ke Y, Keshava N, Shanks J, Kapp IA, Tekmal RR, Petros J, Joshi HC. Opium alkaloid noscapine is an antitumor agent that arrests metaphase and induces apoptosis in dividing cells. Proc. Natl. Acad. Sci. USA. 1998;95:2280–2286. doi: 10.1073/pnas.95.4.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhou J, Panda D, Landen JW, Wilson L, Joshi HC. Minor alteration of microtubule dynamics causes loss of tension across kinetochore pairs and activates the spindle checkpoint. J. Biol. Chem. 2002a;277:17200–17208. doi: 10.1074/jbc.M110369200. [DOI] [PubMed] [Google Scholar]
- [7].Zhou J, Gupta K, Yao J, Ye K, Panda D, Giannakakou P, Joshi HC. Paclitaxel-resistant human ovarian cancer cells undergo c-Jun NH2-terminal kinase-mediated apoptosis in response to noscapine. J. Biol. Chem. 2002b;277:39777–39785. doi: 10.1074/jbc.M203927200. [DOI] [PubMed] [Google Scholar]
- [8].Zhou J, Gupta K, Aggarwal S, Aneja R, Chandra R, Panda D, Joshi HC. Brominated derivatives of noscapine are potent microtubule-interfering agents that perturb mitosis and inhibit cell proliferation. Mol. Pharmacol. 2003;63:799–807. doi: 10.1124/mol.63.4.799. [DOI] [PubMed] [Google Scholar]
- [9].Landen JW, Lang R, McMahon SJ, Rusan NM, Yvon AM, Adams AW, Sorcinelli MD, Campbell R, Bonaccorsi P, Ansel JC, Archer DR, Wadsworth P, Armstrong CA, Joshi HC. Noscapine alters microtubule dynamics in living cells and inhibits the progression of melanoma. Cancer Res. 2002;62:4109–4114. [PubMed] [Google Scholar]
- [10].Ravelli RB, Gigant B, Curmi PA, Jourdain I, Lachkar S, Sobel A, Knossow M. Insight into tubulin regulation from a complex with colchicines and a stathmin-like domain. Nature. 2004;428:198–202. doi: 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
- [11].Nogales E, Wolf SG, Downing KH. Structure of the alpha beta-tubulin dimmer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- [12].Nettles JH, Li H, Cornett B, Krahn JM, Snyderr JP, Downing KH. The binding mode of epothilone A on alpha, beta-tubulin by electron crystallography. Science. 2004;305:866–869. doi: 10.1126/science.1099190. [DOI] [PubMed] [Google Scholar]
- [13].Bhattacharyya B, Wolff Promotion of fluorescence upon binding of colchicine to tubulin. J. Proc. Nat. Acad. Sci. USA. 1974;71:2627–2631. doi: 10.1073/pnas.71.7.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hamel E. Antimitotic natural products and their interactions with tubulin. Med. Res. Rev. 1996;16:207–231. doi: 10.1002/(SICI)1098-1128(199603)16:2<207::AID-MED4>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- [15].Panda D, Chakrabarti G, Hudson J, Pigg K, Miller HP, Wilson L, Himes RH. Suppression of microtubule dynamic instability and treadmilling by deuterium oxide. Biochemistry. 2000;39:5075–5081. doi: 10.1021/bi992217f. [DOI] [PubMed] [Google Scholar]
- [16].Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- [17].Arai T, Okuyama T. Purification and properties of colchicine-binding protein from the bovine brain. Seikagaku. 1973;45:19–29. [PubMed] [Google Scholar]
- [18].Peyrot V, Leynadier D, Sarrazin M, Briand C, Menendez M, Laynez J, Andreu JM. Mechanism of binding of the new antimitotic drug MDL 27048 to the colchicines site of tubulin: equilibrium studies. Biochem. 1992;31:1112–11132. doi: 10.1021/bi00160a024. [DOI] [PubMed] [Google Scholar]
- [19].Polak E, Ribiere G. Revue Francaise Inf Rech Oper. Serie Rouge. 1969;16:35–43. [Google Scholar]
- [20].Lee C, Yang W, Parr RG. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- [21].Becke AD. A new mixing of Hartree-Fock and local density-functional theories. J. Chem. Phys. 1993;98:1372–1377. [Google Scholar]
- [22].Binkley JS, Pople JA, Hehre WJ. Self-consistent molecular orbital methods. 21. Small split-valence basis sets for first-row elements. J. Am. Chem. Soc. 1980;102:939–947. [Google Scholar]
- [23].Gordon MS, Binkley JS, Pople JA, Pietro WJ, Hehre WJ. Self-consistent molecular-orbital methods. 22. Small split-valence basis sets for second-row elements. J. Am. Chem. Soc. 1982;104:2797–2803. [Google Scholar]
- [24].Pietro WJ, Francl MM, Hehre WJ, Defrees DJ, Pople JA, Binkley JS. Self-consistent molecular orbital methods. 24. Supplemented small split-valence basis sets for second-row elements. J. Am. Chem. Soc. 1982;104:5039–5048. [Google Scholar]
- [25].Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. Glide: a new approach for rapid, accurate docking and scoring. 1. method and assessment of docking accuracy. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- [26].Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- [27].Eldridge MD, Murray CW, Auton TR, Paolini GV, Mee RP. Empirical scoring functions: I. The development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes. J. Comput. Aided Mol. Des. 1997;11:425–445. doi: 10.1023/a:1007996124545. [DOI] [PubMed] [Google Scholar]
- [28].Li X, Li Y, Cheng T, Liu Z, Wang R. Evaluation of the performance of four molecular docking programs on a diverse set of protein-ligand complexes. J. Comput. Chem. 2010;31:2109–2125. doi: 10.1002/jcc.21498. [DOI] [PubMed] [Google Scholar]
- [29].Cross JB, Thompson DC, Rai BK, Baber JC, Fan KY, Hu Y, Humblet C. Comparison of several molecular docking programs: pose prediction and virtual screening accuracy. J. Chem. Inf. Model. 2009;49:1455–1474. doi: 10.1021/ci900056c. [DOI] [PubMed] [Google Scholar]
- [30].Zhou Z, Felts AK, Friesner RA, Levy RM. Comparative performance of several flexible docking programs and scoring functions: enrichment studies for a diverse set of pharmaceutically relevant tergets. J. Chem. Inf. Model. 2007;47:1599–1608. doi: 10.1021/ci7000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Perola E, Walters WP, Charifson PS. A detailed comparison of current docking and scoring methods on systems of pharmaceutical relevance. Proteins. 2004;56:235–249. doi: 10.1002/prot.20088. [DOI] [PubMed] [Google Scholar]
- [32].Cho AE, Guallar V, Berne BJ, Friesner R. Importance of accurate charges in molecular docking: Quantum mechanical/molecular mechanical (QM/MM) approach. J. Comput. Chem. 2005;26:915–931. doi: 10.1002/jcc.20222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Still WC, Tempczyk A, Hawley RC, Hendrickson T. Semianalytical treatment of solvation for molecular mechanics and dynamics. J. Am. Chem. Soc. 1990;112:6127–6129. [Google Scholar]
- [34].Guimaraes CRW, Cardozo M. MM-GB/SA rescoring of docking poses in structure-based lead optimization. J. Chem. Inf. Model. 2008;48:958–970. doi: 10.1021/ci800004w. [DOI] [PubMed] [Google Scholar]
- [35].Gabriela B, Guimaraes CRW, Tubert-Brohman I, Lyons TM, Tirado-Rives J, Jorgensen WL. Search for non-nucleoside inhibitors of HIV-1 reverse transcriptase using chemical similarity, molecular docking, and MM-GB/SA scoring. J. Chem. Inf. Model. 2007;47:2416–2428. doi: 10.1021/ci700271z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Guimaraes CRW, Mathiowetz AM. Addressing limitations with the MM-GB/SA scoring procedure using the water map method and free energy perturbation calculations. J. Chem. Inf. Model. 2010;50:547–559. doi: 10.1021/ci900497d. [DOI] [PubMed] [Google Scholar]
- [37].Aneja R, Vangapandu SN, Lopus M, Viswesarappa VG, Dhiman N, Verma A, Chandra R, Panda D, Joshi HC. Synthesis of microtubule-interfering halogenated noscapine analogs that perturb mitosis in cancer cells followed by cell death. Biochem. Pharmacol. 2006;72:415–426. doi: 10.1016/j.bcp.2006.05.004. [DOI] [PubMed] [Google Scholar]
- [38].Sherline PJ, Leung T, Kipnis DM. Binding of colchicine to purified microtubule protein. J. Biol. Chem. 1975;250:5481–5486. [PubMed] [Google Scholar]
- [39].Hastie SB, Puett D, Macdonald TL, Williams RC., Jr. Binding of tubulin of the colchicine analog 2-methoxy-5-(2′,3′,4′-trimethoxyphenyl)tropone: Thermodynamic and kineticaspect. J. Biol. Chem. 1984;259:7391–7398. [PubMed] [Google Scholar]
- [40].Bhattacharyya A, Bhattacharyya B, Roy SA. Study of colchicine tubulin complex by donor quenching of fluorescence energy transfer. Eur. J. Biochem. 1993;299:757–761. doi: 10.1111/j.1432-1033.1993.tb18195.x. [DOI] [PubMed] [Google Scholar]
- [41].Ray K, Bhattacharyya B, Biswas BB. Role of B-ring of colchicine in its binding to tubulin. J. Biol. Chem. 1981;256:6241–6244. [PubMed] [Google Scholar]
- [42].Chakrabarti G, Sengupta S, Bhattacharyya B. Thermodynamics of colchicinoid-tubulin interactions. Role of B-ring and C-7 substituent. J. Biol. Chem. 1996;271:2897–2901. doi: 10.1074/jbc.271.6.2897. [DOI] [PubMed] [Google Scholar]
- [43].Pyles EA, Rava RP, Hastie SB. Effect of B-ring substituents on absorption and circular dichroic spectra of colchicine analogues. Biochem. 1992;31:2034–2039. doi: 10.1021/bi00122a020. [DOI] [PubMed] [Google Scholar]
- [44].Krovat EM, Steindl T, Langer T. Recent advances in docking and scoring. Curr. Comput. Aided Drug. Des. 2005;1:93–102. [Google Scholar]
- [45].Bernacki K, Kalyanaraman C, Jacobson MP. Virtual ligand screening against Escherichia coli dihydrofolate reductase: Improving docking enrichment physics-based methods. J. Biomol. Screening. 2005;10:675–681. doi: 10.1177/1087057105281220. [DOI] [PubMed] [Google Scholar]
- [46].Huang N, Kalyanaraman C, Irwin JJ, Jacobson MP. Physics-based scoring of protein-ligand complexes: Enrichment of known inhibitors in large-scale virtual screening. J. Chem. Inf. Model. 2006;46:243–253. doi: 10.1021/ci0502855. [DOI] [PubMed] [Google Scholar]
- [47].Huang N, Kalyanaraman C, Bernacki K, Jacobson MP. Molecular mechanics methods for predicting protein-ligand binding. Phys. Chem. Chem. Phys. 2006;8:5166–5177. doi: 10.1039/b608269f. [DOI] [PubMed] [Google Scholar]
- [48].Lyne PD, Lamb ML, Saeh JC. Accurate prediction of the relative potencies of members of a series of kinase inhibitors using molecular docking and MM-GBSA scoring. J. Med. Chem. 2006;49:4805–4808. doi: 10.1021/jm060522a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.