Summary
Bactofilins are fibre-forming bacterial cytoskeletal proteins. Here, we report the structural and biochemical characterization of MXAN_7475 (BacM), one of the four bactofilins of Myxococcus xanthus. Absence of BacM leads to a characteristic ‘crooked’ cell morphology and an increased sensitivity to antibiotics targeting cell wall biosynthesis. The absence of the other three bactofilins MXAN_4637–4635 (BacN-P) has no obvious phenotype. In M. xanthus, BacM exists as a 150-amino-acid full-length version and as a version cleaved before Ser28. In the cell, native BacM forms 3 nm wide fibres, which assemble into bundles forming helix-like cytoplasmic cables throughout the cell, and in a subset of cells additionally a polarly arranged lateral rod-like structure. Isolated fibres consist almost completely of the N-terminally truncated version, suggesting that the proteolytic cleavage occurs before or during fibre formation. Fusion of BacM to mCherry perturbs BacM function and cellular fibre arrangement, resulting for example in the formation of one prominent polar corkscrew-like structure per cell. Immunofluorescence staining of BacM and MreB shows that their cellular distributions are not matching. Taken together, these data suggest that rod-shaped bacteria like M. xanthus use bactofilin fibres to achieve and maintain their characteristic cell morphology and cell wall stability.
Introduction
Cytoskeletal proteins were long thought to be exclusively, even defining, elements of eukaryotic cells (Löwe and Amos, 2009). However, in recent years, improved bioinformatics, structure determination and advanced visualization techniques (Morris and Jensen, 2008; Gitai, 2009) have helped to identify a growing number of proteins in bacteria that are either analogues of eukaryotic cytoskeletal proteins or novel bacteria-specific cytoskeletal elements (reviewed in Gitai, 2005; Shih and Rothfield, 2006; Thanbichler and Shapiro, 2008; Kühn et al., 2010). These bacterial cytoskeletal proteins play important roles in various cellular processes including morphogenesis, division, polarity determination, and DNA segregation (Michie and Löwe, 2006; Graumann, 2007; Graumann and Knust, 2009; Kühn et al., 2010). Moreover, like their eukaryotic counterparts, bacterial cytoskeletal proteins organize the cytoplasm and provide versatile scaffolds for the recruitment of other protein components (Löwe and Amos, 2009).
Probably the best-studied bacterial cytoskeletal protein is the highly dynamic tubulin homologue FtsZ (Bi and Lutkenhaus, 1991), which forms a ring-shaped complex of protofilament bundles (Z-ring) at the future division site (Margolin, 2005; Adams and Errington, 2009). The Z-ring is used to assemble the divisome, a multiprotein complex necessary for cell division (Goehring and Beckwith, 2005). Another highly conserved and widely distributed bacterial cytoskeletal protein is MreB with its Bacillus subtilis isoforms Mbl and MreBH (Jones et al., 2001; Carballido-López and Errington, 2003; Carballido-López et al., 2006). These actin homologues form helically arranged cables underneath the cell membranes of rod-shaped bacteria and are important for cell morphology. It has been suggested that MreB, together with integral membrane protein MreD and membrane-anchored periplasmic MreC, controls the topology of peptidoglycan synthesis by positioning the components of the bacterial morphogenetic complex (Kruse et al., 2005; White et al., 2010). In some bacteria, MreB and MreB-like proteins have also been implicated in other cellular processes such as cell polarity and DNA segregation (Carballido-López, 2006). While the bacterial homologues of tubulin (FtsZ) and actin (MreB and its isoforms) are relatively well understood, considerably less is known about bacterial intermediate filament-like proteins. An example of this class of cytoskeletal elements is crescentin, a protein that is essential for the crescent shape of Caulobacter crescentus (Ausmees et al., 2003; Charbon et al., 2009). In this bacterium, crescentin assembles into a unilateral ribbon that appears to mechanically limit cell wall extension, thereby leading to a bent and ultimately crescent-shaped morphology.
In 1999, a transposon-based genetic screen of Proteus mirabilis identified a protein that showed no similarity to any of the three types of cytoskeletal elements (Hay et al., 1999). Cells that lacked this protein were mildly bent, while cells producing a C-terminally truncated version displayed severely deformed morphologies characterized by irregularly curved cells with variable diameters. Consequently, its gene was named ccmA, for curved cell morphology. CcmA contains a highly conserved domain of unknown function DUF583 (Marchler-Bauer et al., 2009). A recent report defined DUF583-containing proteins as a novel class of filament-forming bacterial cytoskeletal elements characterized by spontaneous nucleotide-independent polymerization (Kühn et al., 2010). An analysis of sequenced bacterial genomes revealed that this class of proteins, termed bactofilins, is widespread among Gram-negative bacteria, and that many species including C. crescentus and M. xanthus possess more than one homologue. A detailed analysis of the two bactofilins of C. crescentus (Kühn et al., 2010), BacA and BacB, showed that they colocalize forming a cluster at the anterior base of the tubular stalk, a characteristic appendage of the cell body allowing C. crescentus to adhere to surfaces (Tsang et al., 2006). Lack of BacA and BacB results in shortened stalks, a phenotype that may be explained by the observation that both proteins interact with a specific peptidoglycan synthase (PbpC; CC3277) that appears to be involved in stalk morphogenesis. By contrast, in M. xanthus, fusion proteins of bactofilins with mCherry were found to form either slender filaments that are restricted to the medial parts of the cell (MXAN_4635–7-mCherry), or more irregular structures that stretch throughout the whole cell (mCherry-MXAN_7475). Furthermore, an mCherry fusion of the only bactofilin in Shewanella oneidensis (SO1662-mCherry) formed a fluorescent band at the cell division site, suggesting that bactofilins may possess species-specific cellular distributions. However, the localizations of the endogenous proteins were not investigated in M. xanthus or in S. oneidensis (Kühn et al., 2010).
None of the bactofilins studied appears to be essential, indicating that the cells either possess other proteins with redundant functions, or that these proteins are involved in non-essential processes, or both. In C. crescentus, other than a somewhat shortened stalk, no apparent phenotype has been reported for any of the bactofilin knockouts. The same study reported that M. xanthus cells tolerate the loss of all four bactofilins without any morphological consequences (Kühn et al., 2010). This observation is in sharp contrast to the loss of helical cell shape in Helicobacter pylori mutants lacking the organism’s only bactofilin, HPG27_1480 (Sycuro et al., 2010), and to the various deformations observed for P. mirabilis ccmA mutants. It has been suggested that two different versions of CcmA, which both accumulate during cell differentiation, are generated via alternate start site selection during transcription of the ccmA gene in P. mirabilis (Hay et al., 1999). The longer and the shorter version, respectively, behave like an integral and a peripheral membrane protein. Until now, no such alternative bactofilin versions have been reported for C. crescentus or M. xanthus, further highlighting the seeming paradox that this class of highly conserved proteins appears to be rather variant with respect to processing, cellular distribution and function.
Here, we report a detailed analysis of MXAN_7475 (BacM, for Bactofilin important for cell Morphology), one of four bactofilins of M. xanthus. The other three, MXAN_4637, MXAN_4636 and MXAN_4635, were termed BacN, BacO and BacP respectively. We find that BacM assembles into fibres in vivo and is required for wild-type cell morphology and wild-type tolerance against cell wall-targeting antibiotics but plays no obvious role in motility or cell division. BacN-P cannot complement a BacM deletion phenotype, suggesting that the various bactofilins in M. xanthus are functionally non-redundant. Like CcmA, BacM exists in two forms in the cell, a full-length and an N-terminally truncated form. Immunofluorescence microscopy shows that the assembled BacM fibres form helix-like cables throughout the cytoplasm and lateral polar rod-like structures in a subset of cells. The C-terminal attachment of mCherry to BacM drastically alters the cellular distribution and the morphology of the polymers. Furthermore, immunofluorescence microscopy experiments suggest that the distributions of BacM and MreB in M. xanthus are not matching.
Results
The bactofilin BacM of Myxococcus xanthus assembles into fibres in vivo and is essential for wild-type cell morphology
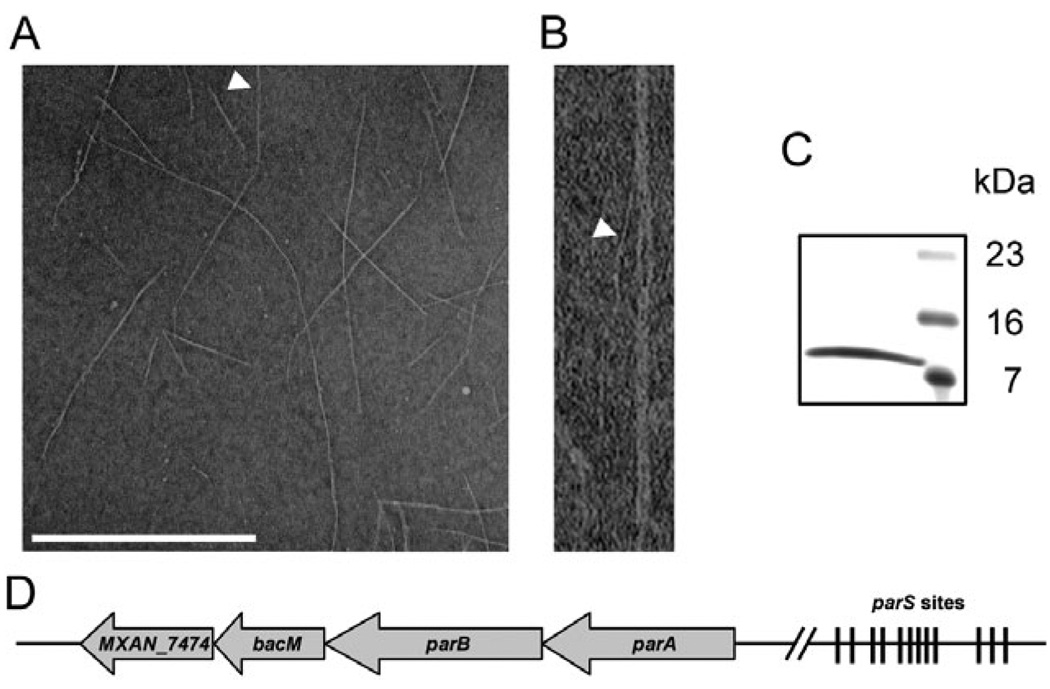
We initially discovered that BacM forms fibres while using electron microscopy to screen cellular lysates of M. xanthus for macromolecular structures. BacM fibre bundles ranged in length from 0.2 µm to more than 1.5 µm, and consisted of individual fibres of about 3 nm width (Fig. 1A and B). As these fibres were resistant to a number of treatments such as detergents, chaotropic agents and high salt, we were able to develop a purification strategy that resulted in preparations devoid of any cellular debris. Upon analysis by SDS-PAGE, these purified fibre preparations produced a single protein band at an apparent molecular weight (MW) of about 11 kDa (Fig. 1C). As the majority of M. xanthus proteins we had previously tried to sequence were N-terminally blocked (unpublished data) and the complete genome sequence was not available at the start of this investigation, we digested the protein band with trypsin and used HPLC-purified peptides for sequence analysis via Edman degradation. With this approach we identified the fibre protein as MXAN_7475 (Goldman et al., 2006), a hypothetical protein of 150 amino acids, which is mostly comprised of the highly conserved domain of unknown function DUF583 (Fig. S1; Marchler-Bauer et al., 2009). As MXAN_7475 has recently been identified as a member of a new conserved class of bacterial cytoskeletal proteins termed bactofilins (Kühn et al., 2010; Fig. S2), we named it BacM. The apparent MW of BacM obtained from the isolated fibres was substantially lower than its calculated mass of 16 kDa. We therefore determined the N-terminus of the purified protein via Edman degradation and identified the amino acid sequence SGEVHTLLGK, which corresponds to residues 28–37 of BacM. This indicated that the isolated fibres were composed of an N-terminally truncated version of BacM.
Fig. 1. Characterization of isolated BacM fibres and bacM-containing operon.
A. Electron micrograph of isolated purified BacM fibres. The arrowhead points to an individual fibre of about 3 nm width as it exits a fibre bundle. Bar represents 500 nm.
B. Magnified picture of the individual fibre marked in (A).
C. SDS-PAGE gel of these purified fibres shows a single band of about 11 kDa (right lane, MW marker).
D. Schematic of the operon containing bacM. The four open reading frames encode ATPase ParA (MXAN_7477), DNA-binding protein ParB (MXAN_7476), BacM (MXAN_7475) and putative lipoprotein MXAN_7474. Between 4.4 and 6.6 kilo base pairs (kbp) upstream of parA are twelve parS sites implicated in ParB binding. The regions comprising 0.5 kbp downstream and 8.7 kbp upstream of the operon do not encode known proteins. Note that the schematic is not drawn to scale.
The DNA sequence surrounding bacM contains three more open reading frames that are encoded on the same strand and flanked by longer regions of non-coding sequences (Fig. 1D). Using cDNA, we could link all four genes via PCR amplification, indicating that the genes MXAN_7477–7474 are transcribed as an operon (Fig. S3). Besides bacM, the operon contains genes encoding a putative lipoprotein of unknown function (MXAN_7474), and the highly conserved chromosomal partitioning proteins ParA (MXAN_7477) and ParB (MXAN_7476). In the region between 4463 and 6529 base pairs upstream of the parA start codon, we identified 12 sequence elements that showed the hallmarks of parS sites, short sequences to which ParB proteins bind during chromosomal segregation (reviewed in Thanbichler and Shapiro, 2008). Analysis of the region immediately upstream of the bacM start codon (GTG) revealed a sequence that qualifies as a ribosomal binding site (AGGAAGGAGTAGGACGTG). This region contains the Shine–Dalgarno consensus sequence AGGAGG three times with one nucleotide mismatch each (Shine and Dalgarno, 1975). This suggests robust binding of ribosomes to this region of the corresponding mRNA and efficient translation of the bacM transcript.
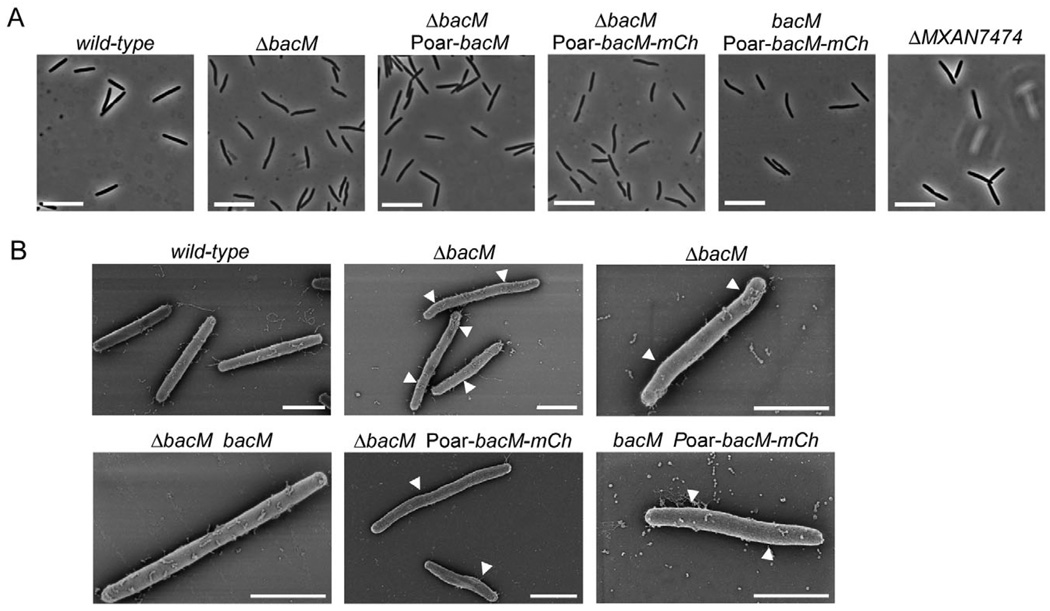
To explore the biological function of BacM, we generated strains that carry in-frame deletions or mCherry fusions of the corresponding gene. Contrary to an earlier report (Kühn et al., 2010), we found that the loss of BacM resulted in a characteristic crooked cell morphology. Initial observation of motile cells of bacM deletion strain EH301 on agar pads did not reveal this phenotype. However, when the cells were grown in liquid culture the morphology defect was easily detectable, independent of the growth phase (Fig. 2; Fig. S4A and B). Compared with the straight rod-shaped wild-type cells, the ΔbacM cells showed characteristic kinks and bends and a slight unevenness in cell width, suggesting that BacM is crucial for proper cell shape maintenance in M. xanthus. In order to confirm that the morphological deformations were indeed the result of the absence of BacM, we performed genetic complementation experiments. For this purpose bacM was reintroduced into the knockout strains either at its original chromosomal position, or under the control of the oar promoter (Martinez-Canamero et al., 1993) at the attB phage attachment site (Wu and Kaiser, 1995). In both cases, the complementation restored wild-type morphology confirming that the absence of BacM was indeed the cause of the deformation. It furthermore demonstrated that moderate overexpression of bacM from the oar promoter did not lead to a distorted cell morphology. Expression from this promoter produced an increase in cellular BacM to approximately twice the wild-type level, according to quantifications of immunoblot chemiluminescence like those described in Fig. S5. We then tested whether the absence of MXAN_7474, the putative lipoprotein encoded downstream of bacM, would produce a similar morphological defect. However, the deletion of MXAN_7474 had no detectable influence on cell shape, indicating that the distorted morphology was a direct consequence of the absence of BacM, rather than being caused by a polar effect on MXAN_7474. Subsequently, we generated strains expressing or moderately overexpressing a bacM– mCherry gene fusion in a ΔbacM background to study the effect of the fusion on cell morphology. The BacM– mCherry fusion protein was unable to rescue the morphological defect, indicating that it cannot replace the physiological function of the native BacM protein. Moreover, cells that contained both proteins simultaneously showed morphological defects similar to although slightly less pronounced than those observed with the ΔbacM strain, demonstrating that the mCherry fusion to the BacM C-terminus had a dominant negative effect on endogenous BacM. The morphologies of cells expressing bacM– mCherry from the endogenous bacM locus (Fig. S4C) were indistinguishable from those of cells that moderately overexpressed bacM–mCherry from the oar promoter (Poar; Fig. 2).
Fig. 2. Cell morphology of bactofilin mutants. Relevant genotypes are indicated.
A. Phase-contrast light micrographs of cells from liquid CTT cultures in mid-logarithmic growth phase. Bars represent 10 µm.
B. Scanning electron micrographs of cells grown as in (A). White arrowheads mark kinks along the cell body. Bars represent 2 µm. The following strains were analysed: DK1622 (wild-type), EH301 (ΔbacM), EH344 (ΔbacM Poar-bacM), EH364 (ΔbacM Poar-bacM-mCh), EH362 (bacM Poar-bacM-mCh), EH332 (ΔMXAN_7474), EH309 (ΔbacM bacM).
BacM exists in the cell in a full-length and an N-terminally processed form
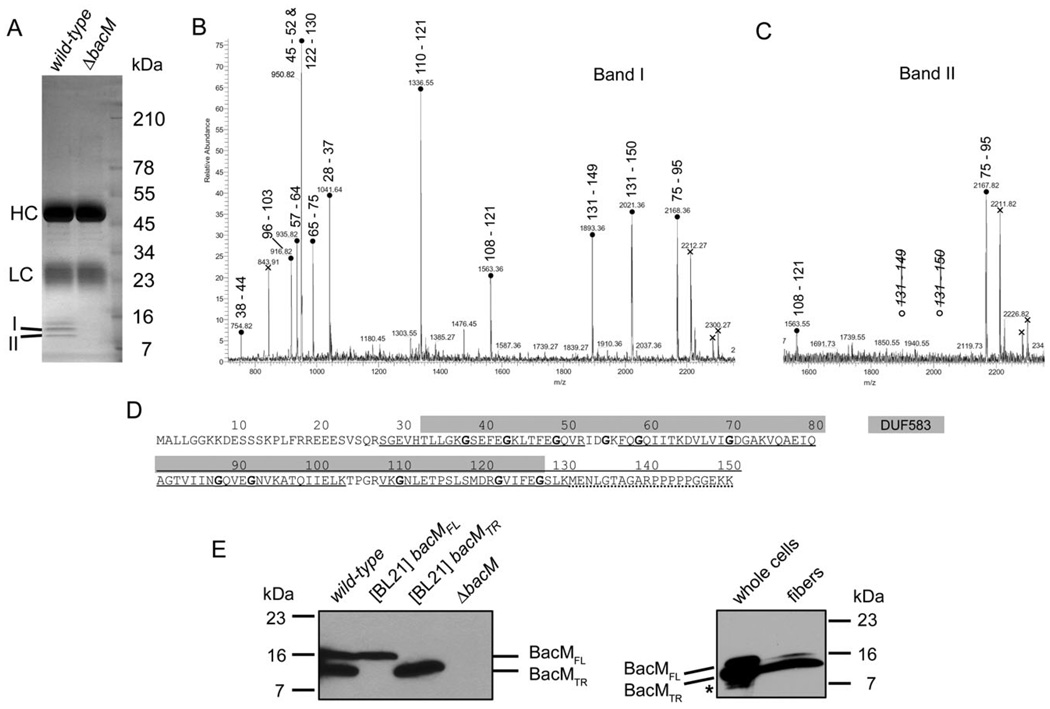
The finding that BacM in the isolated fibres was N-terminally truncated suggested that this protein, similar to its homologue CcmA of P. mirabilis (Hay et al., 1999), would exist in the cells in more than one form. To further investigate the processing of BacM, we generated polyclonal antibodies against recombinant BacM protein and subsequently purified them via incubation with fixed ΔbacM cells. Using these purified anti-BacM antibodies we performed immunoprecipitations with lysates of wild-type, and of ΔbacM cells as a control, to obtain purified cellular BacM for mass spectrometric analysis (Fig. 3A). After separation of the proteins on a 4–20% SDS-PAGE gel, three bands were present in the wild-type but not in the ΔbacM lane, and all of them reacted with anti-BacM antibody on an immunoblot (data not shown). Analysis of the excised bands via matrix-assisted laser desorption/ ionization tandem mass spectrometry (MALDI-MS/MS) provided peptide mass fingerprint (PMF) spectra of sufficient quality for a detailed peak analysis only for the two lower bands (I, 13 kDa and II, 11 kDa; Fig. 3B – D). The peak analysis revealed that the 13 kDa band contained an N-terminally truncated version of BacM still containing Ser28, whereas the 11 kDa band contained a BacM that was additionally truncated at its C-terminus. With Lys130 as the last identified residue and an estimated MW difference between the two BacM forms of approximately 2 kDa, the observed truncation has most likely occurred either immediately after Lys130 or very close to it. Interestingly, both truncation events left the conserved DUF583 domain intact (residues 33–127), and solely removed the two ends of the protein that are not part of the domain (residues 1–27 at the N-terminus and putatively 131–150 at the C-terminus). The finding that the first residue identified via MS in the amino acid sequences of the 13 and 11 kDa BacM forms was Ser28 was consistent with our previous identification of Ser28 as the N-terminal residue of isolated purified fibres.
Fig. 3. Processing of BacM.
A. SDS-PAGE gel of the protein A sepharose eluate of an immunoprecipitation with purified anti-BacM antibodies. Used lysates were from strains DK1622 (wild-type) and EH301 (ΔbacM). Band I (13 kDa) and band II (11 kDa) were subjected to MALDI-MS/MS analysis. HC and LC mark the locations of IgG heavy and light chains respectively.
B and C. PMF spectra of tryptic peptides from bands I and II, respectively, with residue numbers attributed to the peaks. Attribution of two different peptides of almost identical mass to the peak at 950.8 Da was confirmed by MS/MS analysis. Crosses indicate peaks originating from trypsin autolysis products. Note the absence of peaks for the C-terminal BacM peptide for band II.
D. BacM amino acid sequence with DUF583 domain (grey box) and sequence coverage of the PMF analysis indicated. Underlined peptides are present in both gel bands, dotted underlining marks presence only in band I. Glycine residues highlighted bold are well-conserved in species from a variety of different taxa (Figs. S1 and S2).
E. Anti-BacM immunoblots showing the different forms of BacM. Left: Cells of wild-type strain DK1622; E. coli BL21-derived strains expressing bacMFL (codon 1–151), or bacMTR (codon 28–151) respectively; and ΔbacM strain EH301. All cells were frozen in liquid nitrogen immediately after harvesting and subsequently dissolved by boiling in Laemmli buffer. BacMFL and BacMTR mark the positions of the full-length, and the truncated version of BacM respectively. Right: whole cellular protein from cells grown on large agar plates. The weak additional bands (asterisk) below the band of BacMTR probably result from proteolysis of BacM due to microscopically confirmed cell lysis on the agar plates prior to harvesting; BacM fibres isolated from these cells in the presence of protease inhibitors produce a strong 13 kDa and a weak 16 kDa band.
To investigate which BacM forms are present in M. xanthus wild-type cells in vivo, we generated anti-BacM immunoblots of total cellular protein. These blots showed only two bands at the apparent MWs of the full-length and the N-terminally truncated BacM, when the cells were harvested at mid-logarithmic growth phase (Fig. 3E) or from fresh agar plates (data not shown), and then immediately frozen in liquid nitrogen prior to boiling in Laemmli buffer. When analysing in the same manner total protein of Escherichia coli BL21 cells, which produced recombinant full-length BacM or a version lacking the first 27 amino acids, the observed apparent MWs were identical to those of the two BacM forms observed for M. xanthus. Furthermore, we found no processing of the full-length form in E. coli, suggesting that the protease performing the N-terminal truncation is M. xanthus-specific. In this context, it is important to note that E. coli does not encode a bactofilin homologue (Fig. S2). It therefore might be expected to lack a corresponding processing-specific protease. The absence of BacM forms smaller than 13 kDa in M. xanthus cells at mid-logarithmic growth phase suggested that the presence of smaller forms in other preparations was due to non-specific proteolysis after cell lysis. Such proteolysis indeed occurred in the absence of protease inhibitors, decreasing the amounts of the 13 and 16 kDa form and generating an additional 11 kDa band (Fig. S6).
The finding that our initial fibre purifications solely produced a BacM form, which started at Ser28 but had a MW of approximately 11 kDa (Fig. 1C), suggests that the C-terminus of BacM was cleaved off by proteases during the isolation process. Interestingly, this proteolysis did not interfere with the integrity of the BacM fibres. Subsequently, when the fibres were isolated in the presence of protease inhibitors PMSF and EDTA, immunoblots showed predominantly a 13 kDa form of BacM, consistent with a truncation before Ser28, in addition to only a small amount of the full-length 16 kDa form (Fig. 3E). A comparison of the band intensity ratios on these blots shows that the fraction of total BacM present as the 16 kDa form is smaller in the fibre preparation compared with whole cells, suggesting that a considerable amount of full-length BacM is present in the cells while not being part of the fibres. Quantifications on immunoblots showed that the overall levels of BacM and the relative amounts of the 16 and 13 kDa forms in wild-type cells remained constant throughout different growth phases. Approximately 15 ± 3% were present as the 16 kDa form and 85% ± 5% as the 13 kDa form, with a total number of approximately 12 000 molecules of BacM per average cell (Fig. S5).
BacM forms in vivo helix-like cytoplasmic cables, and rod-like structures which localize near the cell membrane
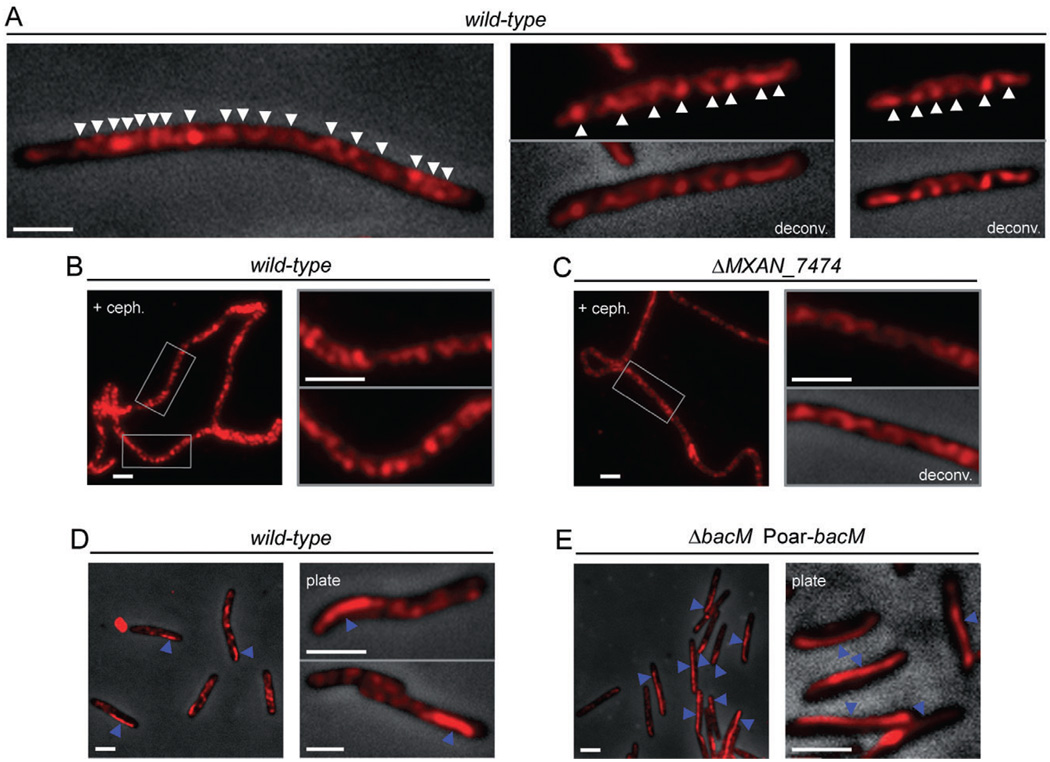
To investigate the cellular localization of BacM, we fixed CTT-grown cells and probed with affinity-purified anti-BacM antibody. Immunofluorescence microscopy revealed that under these conditions BacM formed helix-like cables running through the entire cell (Fig. 4A). These cables showed apparent periodicity in some regions of the cell, but were arranged less regularly in others. The distances between apparent turns of the cables usually varied between 0.5 and 1 µm. In order to test whether cell length had any influence on the formation and appearance of the BacM cables, cells were incubated with the cell division inhibitor cephalexin for 14 h and probed with antibody. As shown in Fig. 4B and C, the dramatic increase of cell length had no significant influence on the arrangement of the BacM cables.
Fig. 4.
Subcellular localization of BacM. Immunofluorescence micrographs of M. xanthus cells treated with anti-BacM antibody and Alexa594-coupled anti-rabbit IgG as secondary antibody. Shown are either only the fluorescent signals or their overlays onto phase-contrast pictures. Fluorescent signals were deconvoluted when indicated (deconv.). Cells were grown in CTT medium to mid-logarithmic growth phase, or, when indicated, collected from CTT agar 8 days after plating (plate).
A. Cells of wild-type strain DK1622. White arrowheads indicate apparent turns of intracellular helix-like cables of BacM (variable spacing, generally between 0.5 and 1.0 µm).
B. Elongated DK1622 cells after growth for 14 h with cephalexin are shown at lower magnification and boxed regions are magnified in the adjacent pictures.
C. Cells deleted in MXAN_7474 (strain EH332) were treated with cephalexin and displayed as described for wild-type.
D. In addition to the cables, approximately 25% of DK1622 cells contain a mostly lateral polar rod-like structure (blue arrowheads).
E. Cells (strain EH344) deleted in bacM and subsequently complemented with the bacM gene under control of the oar promoter (Poar) generally contain lateral rod-like structures that can extend through the whole cell (blue arrowheads). White bars represent 2 µm.
In addition to the cables, approximately 25% of the cells displayed a rod-like structure, which mostly localized laterally near a cell pole. In wild-type cells, these structures were generally between 1 and 3 mm long, thereby spanning one quarter or even half of the cell, while the cells still contained cables (Fig. 4D). When bacM was moderately overexpressed from the oar promoter (Martinez-Canamero et al., 1993) in a ΔbacM background, the BacM level was about twice that of wild-type, and BacM formed similar lateral rod-like structures that could span the entire length of the cell (Fig. 4E and Fig. S7).
Finally, we studied whether the putative lipoprotein MXAN_7474 had any influence on the cellular arrangement of the BacM cables or rods. For that purpose, we used the anti-BacM antibody to probe ΔMXAN_7474 cells, which had been exposed to cephalexin to increase their length for better visualization of the cables (Fig. 4C). Consistent with our findings that a deletion of MXAN_7474 did not influence the cell morphology, it also did not change the cellular arrangement of the BacM cables, and ΔMXAN_7474 cells showed rod structures indistinguishable from those of the wild-type (data not shown). Therefore, we have no indication that MXAN_7474 is necessary for the physiological function or cellular arrangement of BacM.
The fusion of mCherry to BacM severely disrupts the function and morphology of the fibre bundles
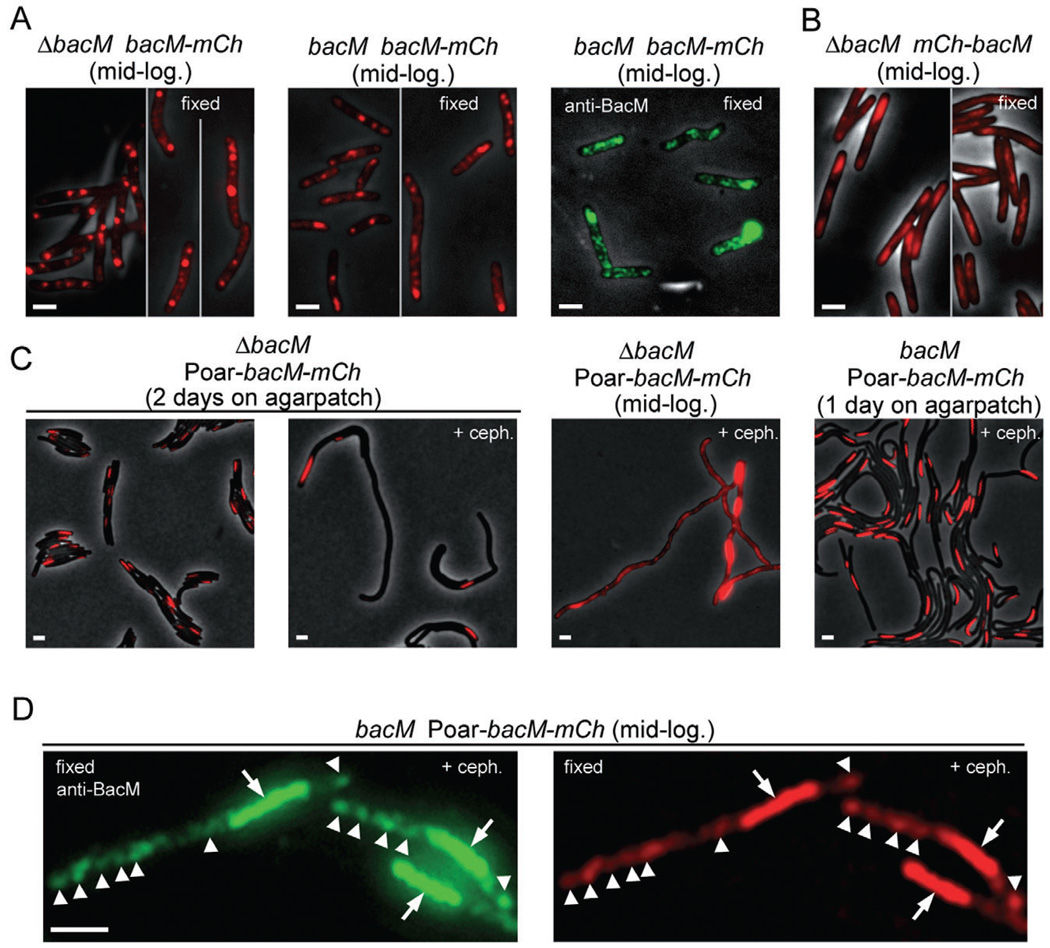
Fusions of target proteins to fluorescent proteins like GFP or mCherry have dramatically increased our understanding of protein function and cellular distribution in vivo (reviewed in VanEngelenburg and Palmer, 2008). After finding that BacM undergoes N-terminal processing, we generated various M. xanthus strains which produced BacM fused at its C-terminus to mCherry to study the cellular distribution of the fusion protein via fluorescence microscopy. First, we investigated cells expressing the bacM–mCherry gene fusion either as a replacement of the endogenous bacM gene or in addition to it. Instead of the expected cables and polar rods, several bright fluorescent clusters of various sizes were visible throughout the cytoplasm and some weak diffuse fluorescence existed between the clusters (Fig. 5A).
Fig. 5.
Fluorescence microscopy of M. xanthus cells which produce a protein fusion of BacM and mCherry. Relevant genotypes are listed. Red, fluorescence from mCherry; green, immunofluorescence after fixation and incubation with anti-BacM and secondary antibody. Visibility of black cells on grey background results from overlay of the phase-contrast image. Cells were visualized after harvesting from CTT cultures at mid-logarithmic growth phase (mid-log.), or directly on thin agar patches after growing there for 1–2 days. Growth was optionally in the presence of 100 µM cephalexin (+ceph.); cells were fixed when indicated.
A. Cells of strains EH358 and EH351, which express bacM–mCherry from the endogenous promoter in the absence or presence of native BacM respectively.
B. Cells of strain MT299, which express mCherry–BacM (Kühn et al., 2010). Regions in these cells that are devoid of mCherry signal colocalize with DNA stain, as shown in Fig. S8A.
C. Cells of strains EH364 and EH362, which express bacM–mCherry from the oar promoter (Poar) in the absence or presence of endogenous BacM respectively. Longer exposure of four cells at mid-logarithmic growth phase reveals one screw per cell plus additional unevenly distributed fluorescence throughout the cells.
D. Cells of strain EH362, harvested after cephalexin treatment, fixed, and incubated with anti-BacM and secondary antibody. Comparison of immuno- and mCherry-fluorescence shows colocalization of screws (arrows), and additionally of regions of weaker fluorescence throughout the cells (white arrowheads point to most intense signals in these regions for easier comparison of the images). Bars represent 2 µm.
Upon fixation of the cells, fluorescent cables became visible between the clusters, similar to the cables seen via immunofluorescence in wild-type cells. To confirm that the fluorescent clusters and cables contained the BacM portion in addition to the mCherry portion of the fusion protein, we performed immunofluorescence microscopy with these cells using anti-BacM as primary antibody. The experiments generally showed weakly fluorescing cable structures in addition to high-intensity clusters. This pattern was consistent with the epifluorescence results, indicating that the fusion protein formed cables as well as clusters. However, the pattern was very different from that observed for wild-type cells, in which such clusters were never observed but lateral rod-like structures were present in a fraction of the cells (Fig. 4D). The fusion of the two proteins therefore caused a dramatic change in the cellular distribution of BacM, consistent with the fusion protein’s inability to complement the morphology defect of ΔbacM cells, and with its dominant negative effect on endogenous BacM. The finding that the BacM–mCherry cables became easily visible only after fixation might be explained by motion as observed for the clusters in live cells under conditions of reduced photobleaching (Videos S1 and S2). Whether wild-type BacM cables display such motion within the cytoplasm is currently not known but an interesting possibility.
To compare the effects of C-terminal fusion of mCherry to BacM to those of N-terminal fusion, we obtained strain MT299, which had been generated in an earlier study to encode the N-terminal fusion protein mCherry–BacM in place of the native BacM protein (Kühn et al., 2010). When investigating this strain under the same conditions as our strains, we initially detected irregular cytoplasmic fluorescence consistent with the earlier study (Fig. 5B). We also found that the weak mCherry fluorescence present in many parts of the cells was absent from regions stained by the DNA stain DAPI (Fig. S8A), and that cable structures were not visible. After fixation to decrease the rate of photobleaching and to eliminate motion of cytoplasmic components, we recorded optical sections through these cells (Fig. S8B and Video S3). The observable positional shift of fluorescence signal when zooming through the cells was consistent with what would be expected for cables winding through cells. The structures detected after fixation resembled the helix-like BacM cables seen in wild-type cells by immunofluorescence. This was in line with the finding that MT299 cells had almost wild-type morphology and therefore experienced only a mild disruption of the physiological BacM function. However, immunoblot quantification demonstrated that the encoded mCherry–BacM fusion protein comprised less than 8% of the total BacM present in these cells, whereas the 16 and 13 kDa forms of BacM together comprised almost 80% (Fig. S5E). The facts that only a small fraction of BacM was present as the encoded fusion protein, and/or that in this strain mCherry was fused to the N-terminus of BacM, might explain why the cables still resembled those of wild-type cells, and why the cells did not become crooked contrary to cells producing the C-terminal fusion protein.
Since fusing mCherry to the C-terminus of BacM had a significant impact on its cellular distribution, we studied whether an increased expression level would additionally affect the localization of the fusion protein. We moderately overexpressed the bacM–mCherry fusion under the control of the oar promoter in a ΔbacM (strain EH364) and in a wild-type background (strain EH362), which produced an approximately twofold increase in the level of fusion protein compared with expression from the wild-type promoter. In both cases, in vivo epifluorescence showed that this moderate overexpression resulted in another distinct cellular distribution (Fig. 5C and D; Fig. S9). The fusion protein formed a tightly wound structure reminiscent of a corkscrew that was present at one cell pole and stretched through roughly a third to half of the cell (usually between 2 and 4 µm). In cells grown on agar plates for several days, screws could reach lengths greater than 9 µm.
Even in drastically elongated cells grown in the presence of cephalexin only a single screw was present per cell (Fig. 5C). While the screw localization was exclusively polar in the absence of cephalexin (N=340 cells), the elongated cells contained 70% polar and 30% non-polar screws (N=156 cells). The helical pitch of the highly regular right-handed screws (Fig. S10) was 570 ± 30 nm (N=60 helices). Upon extended exposure it became apparent that in addition to its presence in the screws, some of the fusion protein was unevenly distributed throughout the remainder of the cell. The screw positions in cells grown in the presence or absence of cephalexin were consistent with those of the rod-like structures in wild-type cells. Those structures were also usually polarly located but could frequently be detected in the cell centre after cephalexin treatment (Fig. S7A).
To compare the cellular distributions of the moderately overproduced BacM–mCherry fusion protein and native BacM, we investigated cells of strain EH362 with anti-BacM immunofluorescence microscopy (Fig. 5D and Fig. S7C). The comparison of epi- and immunofluorescence signals in individual cells showed that native BacM and the fusion protein had identical cellular localizations, suggesting that they copolymerized and/or that fibre bundles consisted of fibres formed by one or alternatively by the other. In EH362 cells, the fraction of total BacM present as the encoded fusion protein was 23% and therefore almost three times higher than in MT299 cells (Fig. S5). The 16 and 13 kDa forms together constituted only about 60%, as compared with 80% in MT299. An increasingly defective morphology with increasing proportions of fusion protein from strain MT299 to strain EH362 to strain EH364 suggests that an increase in the proportion of fusion protein progressively changes the fibre suprastructure and disrupts normal BacM function.
The cellular localization of the BacM cables does not match that of the MreB helix
The helix-like appearance of the BacM cables was somewhat reminiscent of the cell wall-associated helical cables reported for MreB in rod-shaped bacteria like E. coli and B. subtilis (Jones et al., 2001; Kruse et al., 2003; Graumann, 2007). To address if the patterns were related, we first analysed whether an antibody raised against B. subtilis MreB (Jones et al., 2001) would show sufficient cross-reaction with MreB (MXAN_6789) from M. xanthus. An immunoblot of SDS-PAGE-separated total cell lysate of DK1622 wild-type cells with this anti-MreB antibody produced a major band at an apparent MW of approximately 37 kDa, which was in good agreement with the calculated MW of 36.5 kDa for MreB from M. xanthus (Fig. 6A). The blot additionally produced bands at approximately 16 and 14 kDa. These bands were also present in a ΔbacM strain (data not shown), indicating that they did not result from a cross-reaction with the similarly sized BacM. The identities of the proteins causing the additional bands are not known at this time.
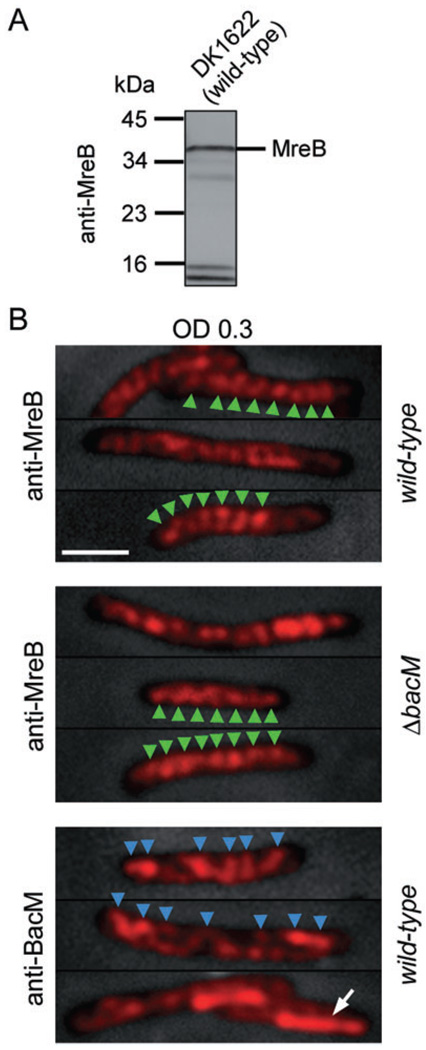
Fig. 6. MreB and BacM have different cellular localization patterns.
A. Immunoblot of DK1622 total proteins with anti-MreB antibody. A major band is present at approximately 36.5 kDa, the MW of M. xanthus MreB.
B. Anti-MreB fluorescence micrographs of wild-type (DK1622) and ΔbacM (EH301) cells from CTT cultures of OD 0.30. Bars represent 2 mm. After fixation, the cells were treated with either anti-MreB or anti-BacM primary and fluorophore-coupled secondary antibodies. Fluorescence signals are shown on top of phase-contrast pictures of the cells. In a selection of cells, green arrowheads mark the regular banding pattern present after staining with anti-MreB. When staining wild-type cells from the same batch with anti-BacM antibody, the observed pattern was less regular (blue arrowheads), and in about 25% of the cells, a lateral rod-like region near one of the cell poles (white arrow) showed stronger fluorescence compared with the rest of the cell.
As the immunoblot result was consistent with the recognition of M. xanthus MreB by the anti-MreB antibody, we then tested whether this antibody could be used to localize MreB in M. xanthus cells in immunofluorescence experiments to allow for a comparison of the cellular localizations of BacM and MreB (Fig. 6B). Wild-type cells were harvested at mid-logarithmic growth phase, fixed, and probed with anti-MreB or anti-BacM antibodies and with secondary antibody. The anti-MreB antibody generally produced a very regular ‘striated’ staining pattern consistent with a cell-spanning MreB helix with an apparent pitch of 520 ± 60 nm (N=20 cells) when assuming a single helix. This value is slightly higher but still in good agreement with the recently published value of 470 ± 100 nm for the apparent helical pitch of M. xanthus MreB (Mauriello et al., 2010). However, contrary to that study, we did not observe MreB-enriched polar clusters. A possible explanation for this apparent disparity in polar staining might be the use of different antibodies and/or different strains in the two studies. The generally highly regular anti-MreB staining pattern was similar in wild-type and ΔbacM cells. However, it was clearly different from the pattern obtained with anti-BacM antibody, in which the spacing between strong BacM signals was very variable ranging from approximately 0.5–1 µm. Furthermore, with anti-MreB antibody we did not observe polar rod-like structures like those seen with anti-BacM. These findings show that in M. xanthus cells the distribution of BacM does not match that of MreB, and that the MreB distributions in the presence or absence of BacM are indistinguishable.
Deletion of BacM has no detectable influence on chromosomal segregation and normal cell growth but lowers the tolerance to cell wall-targeting antibiotics
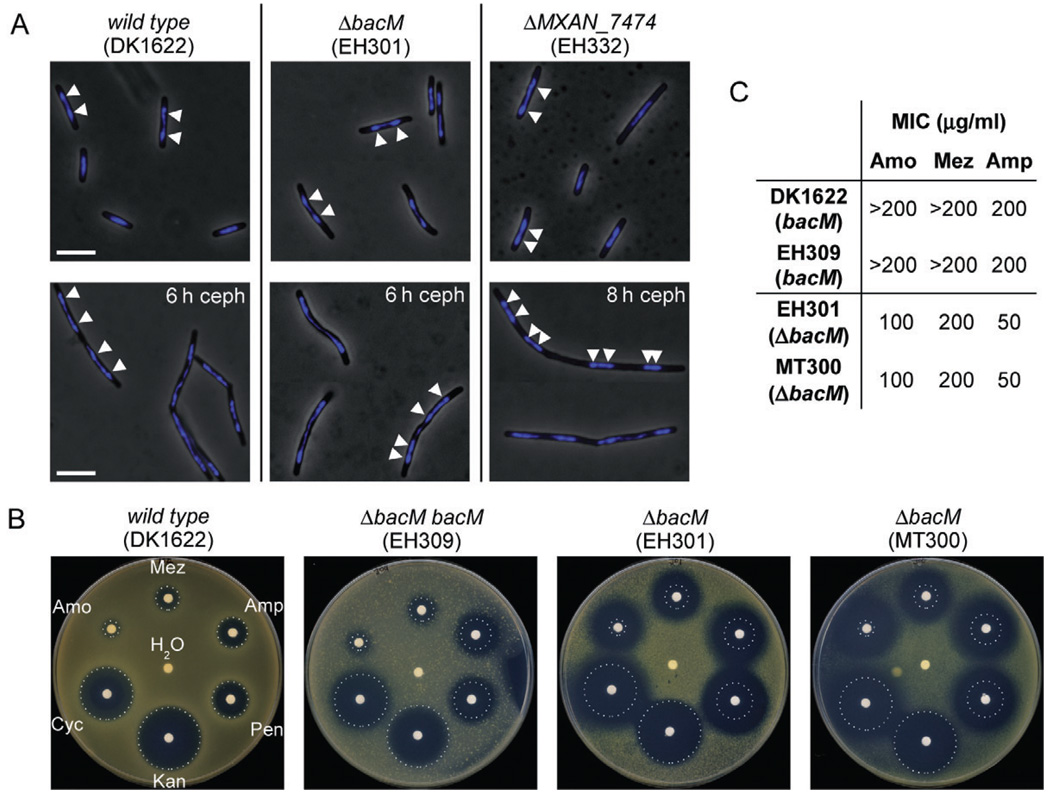
In many bacteria, the ATPase ParA and the parS binding protein ParB have been implicated in chromosomal segregation (Leonard et al., 2005). After finding that bacM and MXAN_7474 are cotranscribed with parA and parB, the question arose whether their deletions would have an effect on chromosomal segregation or cell growth. To investigate the localization of their chromosomal DNA, cells of different strains were grown to mid-logarithmic phase and their DNA was stained with DAPI. Fluorescence microscopy did not reveal differences in the localization of nucleoids when comparing wild-type cells to cells deleted in bacM or MXAN_7474 (Fig. 7A). Also when growing these cells in the presence of the cell division inhibitor cephalexin, the chromosomal distributions were indistinguishable between wild-type and deletion strains.
Fig. 7. Chromosome segregation and sensitivity to antibiotics targeting cell wall biosynthesis.
A. Overlays of phase-contrast and DAPI fluorescence micrographs of M. xanthus cells in mid-logarithmic growth phase, or after 6–8 h of growth to OD 0.6–0.8 in medium containing 100 µM cephalexin (ceph). Relevant genotypes are indicated. White bars represent 5 µm. White arrowheads on a selection of cells mark the regions of highest DAPI signal intensity, indicating the centres of the nucleoids.
B. Paper disc diffusion assay comparing antibiotic sensitivities of strains containing or lacking BacM. Antibiotic-containing paper discs were placed on top of CTT agar plates containing cells of the indicated strains. Plates were scanned after 100 h of incubation at 32°C. Sizes of growth inhibition zones of wild-type strain DK1622 are marked by white dotted circles. The discs contained cell wall antibiotics mezlocillin (Mez, 250 µg), ampicillin (Amp, 250 µg), penicillin G (Pen, 100 µg), d-cycloserine (Cyc, 600 µg) and amoxicillin (Amo, 250 mg). Control discs contained water and kanamycin (Kan, 50 µg).
C. Minimal inhibitory concentrations (MIC) of three of the above antibiotics for strains containing or lacking BacM, as determined in 96-well microtitre plate assays (Fig. S11).
To verify that the absence of these proteins had also no effect on the kinetics of chromosome segregation, we compared the growth rates of bacM deletion strains EH301 and EH302, complementation strains EH309 and EH310, and of the MXAN_7474 deletion strain EH322 to two different strains expressing endogenous bacM, wild-type strain DK1622 and ΔpilQ strain DK8615 (data not shown). No difference in growth rates was detected for these strains. The observed doubling time of 4.6 ± 0.2 h at 32°C for mid-logarithmic growth of all tested strains was in good agreement with previously published values (Janssen et al., 1977). We conclude that neither BacM nor MXAN_7474 is necessary for chromosome segregation or optimal cell growth under standard conditions.
The crooked cell morphology caused by the absence of BacM suggested that an underlying alteration in the cell wall structure might render the cells more sensitive to antibiotics targeting cell wall biosynthesis. In a paper disc diffusion assay we compared the growth inhibition zones around paper discs containing defined amounts of individual antibiotics. The discs were placed on agar plates containing cells either producing or lacking BacM (Fig. 7B). While the extent of growth inhibition was very similar for the wild-type and a bacM complementation strain, the bacM deletion strains EH301 and MT300 (Kühn et al., 2010) both showed significantly larger inhibition zones for cell wall-targeting antibiotics but not for the protein synthesis-targeting antibiotic kanamycin. To confirm these results in an independent assay, we determined the minimal inhibitory concentrations (MICs) of amoxicillin, mezlocillin and ampicillin for these strains in a 96-well plate format (Fig. 7C and Fig. S11). For both strains lacking BacM the MICs for individual antibiotics were identical and significantly lower than for the strains containing BacM. These results confirmed that in the absence of BacM the tolerance of M. xanthus cells to cell wall-targeting antibiotics is significantly lowered.
The four homologous bactofilins of M. xanthus are functionally non-redundant
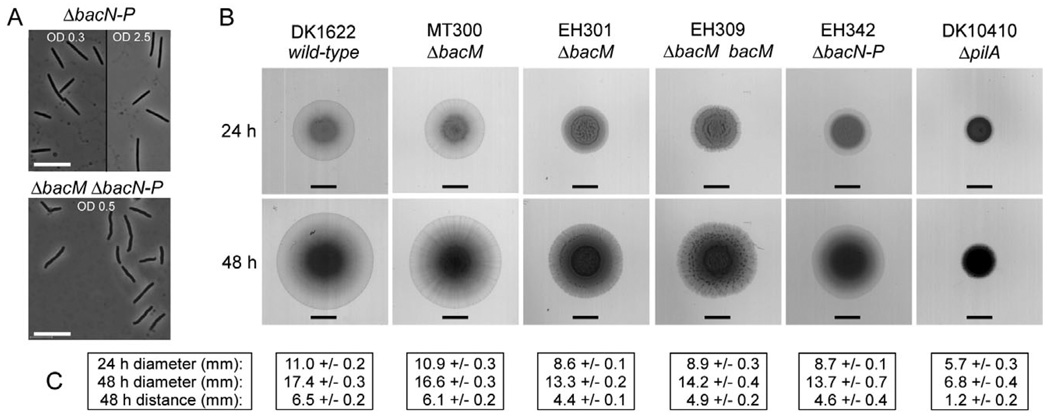
As outlined earlier, M. xanthus encodes a total of four bactofilins: BacM (MXAN_7475), and three homologues (BacN-P; MXAN_4637–4635), which are encoded by adjacent genes in a different operon. The crooked cell morphology phenotype that was always observed in the absence of BacM was neither compensated for by the presence of BacN-P nor exacerbated or caused by their absence. This suggested that the cellular function of BacM is distinct from those of BacN-P (Fig. 8A and Fig. S4).
Fig. 8. Morphology of cells lacking multiple bactofilins, and S-motility assay.
A. Morphology of cells lacking BacN-P (strain EH342) or all four bactofilins (strain EH341) after growth in liquid CTT medium to the indicated ODs. Bars represent 10 µm.
B. Swarming of M. xanthus strains on 0.4% agar containing 50% CTT medium. All strains (genotypes indicated) were grown to an OD of 1.0 in CTT medium, pelleted and resuspended in CTT medium prior to spotting of 4 µl cell suspension containing 2 × 107 cells. Plates were scanned after 24 and 48 h. Bars represent 4.5 mm, the initial diameter of the spots. The ΔpilA strain exemplifies a lack of S-motility.
C. Diameters of swarm areas after 24 and 48 h, and distances covered from the initial spot borders to the spot borders at 48 h. Listed are averages from three experiments and standard deviations.
While it had been suggested earlier that in C. crescentus the bactofilins BacA and BacB form mixed polymers (Kühn et al., 2010), in our preparation of purified M. xanthus fibres only BacM was identified and generated the only visible band on a gel (Fig. 1). This suggests that BacN-P do not copolymerize with BacM under standard growth conditions. Additional support that the similarly sized BacN and BacO proteins are absent in BacM fibres was provided by the use of highly sensitive mass spectrometry. With this method we identified only BacM peptides in gel bands present at the approximate MW of BacN and BacO after immunoprecipitation of cellular BacM with anti-BacM antibodies (Fig. 3A – C).
After discovering the previously undetected ‘crooked’ phenotype of ΔbacM cells, we decided to reinvestigate the role of all four bactofilins in S-motility (Fig. 8B). We could confirm earlier reports that the loss of BacM had no influence on the cells’ ability to swarm (Kühn et al., 2010), which is consistent with the protein’s established role in cell shape maintenance. However, in contrast to the published results, our strain deleted in bacN-P demonstrated wild-type S-motility. To resolve this discrepancy we obtained the strains used in the earlier study, and reinvestigated ΔbacN-P strains MT295 and JK328 (Kühn et al., 2010). Consistent with the results obtained with our strain, we found wild-type S-motility for these strains as well (Fig. S12). Consequently, we conclude that BacM is involved in cell shape maintenance but seems to have no function in motility, whereas for BacN-P no function has been established so far.
Like M. xanthus, but unlike C. crescentus, Stigmatella aurantiaca and P. mirabilis contain bactofilins which exist as a full-length version and additionally in a shortened form
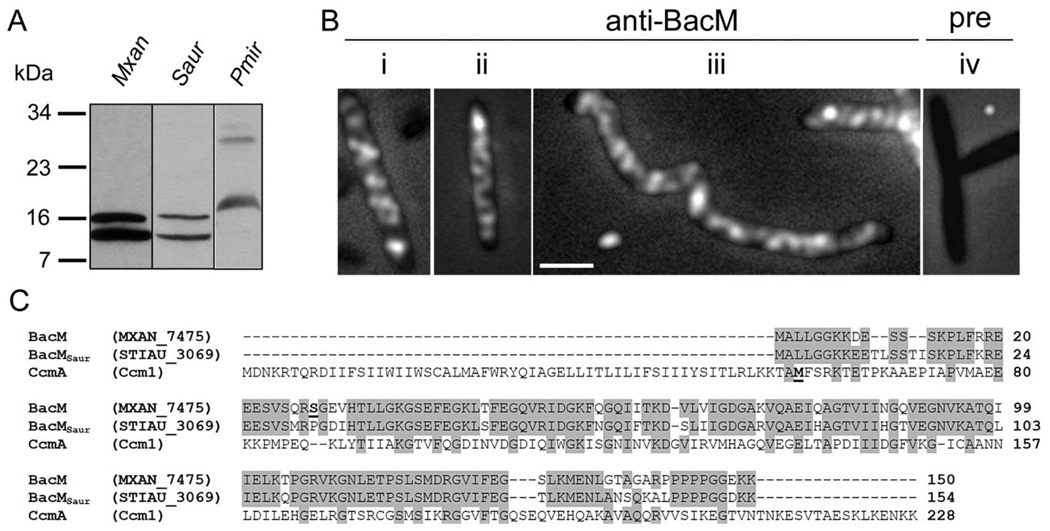
To extend our investigations of the cellular localization and processing of BacM to orthologues in other organisms, we chose to study S. aurantiaca (Fig. 9A – C). This species contains among its four encoded bactofilins a BacM orthologue with sufficient similarity to expect a cross-reaction with our anti-BacM antibody. At the same time it is genetically and physiologically sufficiently different from M. xanthus to extend our knowledge about bactofilins. An immunoblot with total cellular protein of S. aurantiaca, and of M. xanthus for comparison, showed that BacMSaur (‘Stiau_3069’, length-corrected from 125 to 154 amino acids based on homology to BacM) existed in cells as the full-length version with a calculated MW of 16.6 kDa, and additionally as a shorter version of approximately 13 kDa. The MW of the shorter version would be consistent with cleavage of BacMSaur at or near the site that corresponds to the cleavage site in BacM. After establishing that the anti-BacM antibody specifically recognized BacMSaur, we performed immunofluorescence microscopy using S. aurantiaca cells grown with or without cephalexin. Like BacM, BacMSaur formed cables throughout the entire cell which were indistinguishable from those observed in M. xanthus (Fig. 4).
Fig. 9. BacM orthologues in other species.
A. Anti-BacM immunoblot of whole cell protein from M. xanthus, S. aurantiaca and P. mirabilis obtained by immediate freezing of cells followed by boiling in Laemmli buffer.
B. Anti-BacM immunofluorescence micrographs of S. aurantiaca cells from cultures with ODs of 0.10 (i), or 0.85 (ii) or after 7 h 100 µM cephalexin (iii). Pre-immune serum control of OD 0.85 cells (iv). Bar represents 2 µm.
C. Alignment of BacM with its putative orthologues from S. aurantiaca, BacMSaur, and from P. mirabilis, CcmA. Alternative names are given in parentheses. The N-terminal residues of the shorter forms of BacM (Ser27) and of CcmA (M59, according to Hay et al., 1999) are highlighted bold and underlined. Residues are highlighted grey if identical in at least two of the sequences.
We furthermore chose P. mirabilis, to test whether our antibody would recognize a bactofilin that had only a low level of sequence identity with BacM (Fig. 9A and C). Surprisingly, anti-BacM generated immunoblot bands of the calculated MWs for the long and the short version of CcmA (25 kDa and 18 kDa) from P. mirabilis, despite the low primary sequence similarity. This might argue for some remaining tertiary structure in bactofilins despite boiling in Laemmli buffer prior to SDS-PAGE. The existence of BacM, BacMSaur and CcmA as full-length and processed versions is in contrast to BacA and BacB, which are present in C. crescentus cells only as unprocessed versions (Kühn et al., 2010).
Discussion
Bactofilins have been suggested to function in bacterial morphogenesis, cell division and motility (Kühn et al., 2010). Of these three functions, the morphogenetic one is well supported. Bactofilin loss reduces the length of stalks in C. crescentus (Kühn et al., 2010), while H. pylori (Sycuro et al., 2010) and P. mirabilis (Hay et al., 1999) cells become misshapen. Interestingly, the latter cells become even more deformed when producing a C-terminally truncated version of their bactofilin CcmA, indicating that modification of a bactofilin can be more detrimental to cells than its absence. Contrary to an earlier report (Kühn et al., 2010), the present study demonstrates that BacM clearly affects the cell morphology of M. xanthus. The deformations that result from the loss of BacM are similar to the ones reported for P. mirabilis cells lacking CcmA (Hay et al., 1999), suggesting that these bactofilins have similar morphogenetic functions.
In most bacteria, proper cell shape maintenance is achieved through the interplay of a shape-defining peptidoglycan (PG) sacculus and the activity of morphogenetic proteins. These proteins include PG synthesis and modification enzymes like the penicillin-binding proteins (Sauvage et al., 2008), as well as cytoskeletal proteins like MreB, FtsZ and crescentin. Cell morphology is generally disrupted once any of these proteins are depleted or removed. As none of the cytoskeletal proteins is directly involved in PG synthesis they must exert their morphogenetic effect indirectly either through interaction with the PG synthesis enzymes (Kawai et al., 2009; Takacs et al., 2010) or by altering the mechanical forces that guide cell wall formation (Gayda et al., 1992; Cabeen et al., 2009).
Several lines of evidence suggest that bactofilins exert their morphogenetic functions through the interaction with PG synthesis/modification enzymes. In C. crescentus, BacA and BacB interact with the penicillin-binding protein PbpC, an enzyme involved in polar morphogenesis (Kühn et al., 2010). In H. pylori, the bactofilin gene ccmA is part of an operon encoding LytM peptidoglycan endopeptidase homologues, termed Csd proteins. The deletion of either ccmA or any of these csd genes produces very similar changes in muropeptide cross-linking, in all cases resulting in the loss of helical cell shape (Sycuro et al., 2010). As CcmA has no recognizable peptidase motif, the similarity of the knockout phenotypes points to an interaction of CcmA with these PG modification enzymes. So far, no such interacting enzymes have been identified for the bactofilins of P. mirabilis and M. xanthus. Nevertheless, it is an attractive hypothesis that the morphogenetic effect of bactofilins is generally achieved through the interaction with proteins involved in PG synthesis and/or modification, similar to other cytoskeletal proteins, such as MreB.
A role for bactofilins in cell division has been suggested only for the bacterium Shewanella oneidensis, in which a bactofilin–mCherry fusion forms a fluorescent band at the septum of dividing cells (Kühn et al., 2010). However, this result must be interpreted with caution, because bactofilin fusion proteins, as shown in this study (Fig. 5), can have drastically altered cellular localizations and suprastructures compared with native BacM. Moreover, although bacM is part of an operon encoding putative chromosomal partitioning proteins, its deletion had no effect on growth or chromosomal segregation. Similarly, H. pylori cells lacking their only bactofilin CcmA show wild-type growth and septation (Sycuro et al., 2010). Consequently, there is no unambiguous evidence supporting a role of bactofilins in chromosomal segregation or cell division.
Bactofilins have also been implicated in motility. In M. xanthus, deletion of bacN-P has been linked to an S-motility defect (Kühn et al., 2010). However, our own ΔbacN-P strain (Fig. 8) and the carefully re-examined strains of Kühn et al. all demonstrated wild-type level S-motility (Fig. S12). Furthermore, the reported motility defects of P. mirabilis ccmA mutants may rather be the consequence of the morphogenetic function of this bactofilin (Hay et al., 1999). P. mirabilis cells must align into multicellular rafts in order to swarm. Morphologically deformed bactofilin mutants do not properly align and the severity of deformation correlates with the severity of the swarming defect. In M. xanthus ΔbacM strains, the cellular morphology defects are distinct when cells are grown in liquid culture. However, when these cells glide on agar, their deformations are less obvious and their motility is unaffected. Whether morphology changes are also responsible for the motility defects of B. subtilis yhbE and yhbF mutants is unclear, as these defects were detected in genetic studies that did not investigate cell morphology (Rajagopala et al., 2007). Consequently, only a function of bactofilins in cell morphogenesis is well supported; a finding that may indicate that these conserved proteins may have, after all, a limited spectrum of functions.
The analysis of the BacM localization in M. xanthus cells showed an arrangement into helix-like cables, and lateral polar rod-like structures present in a subset of cells. The helix-like arrangement raised the question whether BacM would colocalize with the cell membrane-associated helix formed by MreB. However, while immunofluorescence analysis of the MreB localization produced a regular striated pattern consistent with findings for MreB (Mauriello et al., 2010), the observed arrangement of BacM is distinctly less regular. Furthermore, contrary to BacM, MreB does not form lateral polar rod-like structures. These results indicate that the cellular localizations of BacM and MreB do not match. BacM therefore does not seem to be a component of the MreB-associated morphogenetic protein complex.
At present, it is not clear which role the different BacM suprastructures play with respect to the morphogenetic function of BacM. The cables and/or rod-like structures might be required for proper localization of accessory PG modification enzymes along the cell wall. Such a BacM-mediated localization might be needed to establish uniform cell wall synthesis, while not being necessary for cell growth in general. Consequently, an improper localization of such enzymes in the absence of BacM might lead to an uneven synthesis and/or modification of PG resulting in a perturbed cell wall structure. This would explain the observed kinks and bends along the cell body and the decreased tolerance of these cells against cell wall-targeting antibiotics. Given the conservation of the DUF583 domain and its wide distribution among species, bactofilins might be suitable secondary targets for antibiotic combination therapy to increase the efficiency of existing cell wall-targeting antibiotics.
The only bactofilins for which the intracellular localization of the endogenous protein had previously been shown are BacA and BacB from C. crescentus, which form a cluster of approximately 200 BacA and 20 BacB molecules at the stalked cell pole (Kühn et al., 2010). This localization is drastically different from that of BacM, which forms fibre bundles that are present throughout the length of the cell. Consequently, quantification of immunoblot signals results in the significantly higher number of approximately 12 000 BacM molecules per average M. xanthus cell (Fig. S5). This number is within the range of copy numbers previously reported for other cytoskeletal proteins, for example 15 000 copies of FtsZ in a log phase E. coli cell (Lu et al., 1998), or 8000 MreB and 12 000 to 14 000 Mbl molecules in a B. subtilis cell (Jones et al., 2001).
Bactofilins are tripartite proteins that contain as central part the DUF583 domain flanked by highly variable N- and C-terminal tails (Fig. S1 and Table S1). The DUF583 domain acts as a polymerization module in the presence and in the absence of these tails. Overproduced denatured full-length BacM forms fibres upon renaturation in vitro (Kühn et al., 2010), while native BacM fibres in M. xanthus cells consist predominantly of an N-terminally truncated 13 kDa form. Even the additional proteolytic removal of the C-terminal tail, which leaves almost exclusively the DUF583 domain, does not cause fibre disassembly (Fig. 1). In fact, BacM polymerizes even when fused at its N- or C-terminus to mCherry, a protein that is more than twice the size of the DUF583 domain. Fusion to a protein of this size, however, has a profound effect on the conformation and localization of the fibre bundles. Cells encoding BacM–mCherry alone or in addition to BacM have a crooked morphology and display bright fluorescent clusters or, after moderate overproduction, polar screws. In contrast, native BacM in wild-type cells does not form such clusters or screws.
Analysis of the N- and C-terminal regions of bactofilins from a variety of bacteria reveals the presence of putative transmembrane regions near the N-termini of bactofilins from a limited subset of gamma proteobacteria, which includes members of the genera Proteus, Yersinia, Sodalis, Photorhabdus, Anaeromonas and Wigglesworthia (Fig. S2). These regions might anchor these bactofilins to the cell membrane. The analysis furthermore shows a number of bactofilins, including M. xanthus BacM and BacP and C. crescentus BacA and BacB, in which one or both of these regions are rich in prolines (Table S1), suggesting their involvement in protein– protein interactions (Williamson, 1994). This observation supports the idea that these regions of bactofilins interact with PG modification enzymes to exert the bactofilins’ morphogenetic effects.
The importance of the N- and C-terminal regions is further highlighted by the fact that BacM and P. mirabilis CcmA both exist in the cell as a full-length and an N-terminally truncated form. The observation that native BacM fibres from M. xanthus contain predominantly the N-terminally truncated version supports the idea that the removal of the N-terminal tail occurs prior to or during fibre formation in vivo. The N-terminal truncation of BacM seems to be the result of a specific proteolytic cleavage, consistent with its absence in recombinant E. coli BL21 cells that produce the full-length version of BacM. In contrast, the generation of the truncated version of CcmA has previously been suggested to occur at the transcriptional level (Hay et al., 1999). However, a post-translational proteolytic scenario can currently not be ruled out. In such a case, the N-terminal TM helices of CcmA would be cleaved off, potentially resulting in a release of truncated CcmA from the membrane. The variability in composition and processing of the N- and C-terminal tails of bactofilins might be the basis for the variability in the morphogenetic functions of bactofilins. These tails might determine the bactofilins’ cellular localizations, their suprastructures and their interactions with specific PG synthesis/ modification enzymes resulting in the specific effects on cell morphology.
The importance of the tails is also evident from observations of mCherry fusions of BacM. While C-terminal fusions disturb the native arrangement of the protein in a dose-dependent manner, N-terminal fusion initially appeared to produce wild-type cell morphology. However, immunoblot analysis shows that the N-terminal fusion protein mCherry–BacM constitutes only a low proportion of total BacM in cells that encode BacM only as the fusion protein. Consequently, an N-terminal fusion is less disruptive than a C-terminal one. In line with a dose-dependent effect of the fusion protein, the presence of only a low proportion of mCherry–BacM in copolymers with native BacM enables the fibres to adapt more or less wild-type conformation and function. Copolymerization of fusion protein and native bactofilin is consistent with the observed colocalization of BacM and BacM–mCherry (Fig. 5). It is also in line with the previously reported finding that BacA-CFP adopts the filament-like distribution of overproduced BacB-Venus when these fusion proteins are coproduced in C. crescentus (Kühn et al., 2010).
A similar dose-dependent disturbance of protein supra-structure and cell shape has previously been reported for C. crescentus cytoskeletal protein crescentin (Ausmees et al., 2003). Replacement of CreS by a CreS–GFP fusion protein results in straightened cells in which the fusion protein forms a helix in the cell centre. By contrast, native CreS forms a membrane-associated ribbon. When producing CreS–GFP together with CreS, the crescent cell shape is restored. This is similar to the preservation of more or less wild-type morphology in cells containing only a low proportion of BacM in the form of mCherry–BacM. The consequences of fusing GFP to CreS resemble the drastic changes that occur when the C-terminal fusion protein BacM–mCherry is moderately overproduced instead of or in addition to BacM. In this case, prominent highly regular polar fluorescent screws are formed (Fig. 5 and Fig. S7), and the wild-type cell morphology is disturbed. Potentially, the modification of the C-terminus of BacM interferes with a function of BacM as a scaffold for PG modification enzymes resulting in morphogenetic defects. Moreover, as a consequence of the remarkably different behaviours of BacM–mCherry and native BacM, observations made with bactofilin fusion proteins should generally be interpreted with caution. Only a comparison with the location and function of the native protein can conclusively tell whether observations made with fusion proteins actually reflect the wild-type behaviour of the bactofilin.
In summary, bactofilins are fibre-forming proteins characterized by the DUF583 domain, which by itself is sufficient for fibre integrity. In the organisms studied so far, bactofilins exist either only as a full-length, or additionally as a shorter processed version. Outside of the conserved DUF583 polymerization domain, the various N- and C-terminal tails of bactofilins show considerable variability with respect to their length and amino acid composition, and might be implicated in membrane attachment and/or protein–protein interactions. The only consistent function observed for bactofilins to date is their contribution to cell morphogenesis. Their absence, overproduction or modification can cause cells to curve, lose their helical shape, produce kinks in the cells, or reduce the length of stalks. So far, native bactofilins have been found to form a variety of suprastructures in vivo: small, sheet-like clusters in C. crescentus underneath the cell membrane close to the stalked pole, cytoplasmic cables that span the entire cell in a helix-like fashion, and rod-like structures typically located laterally near one of the cell poles. It is currently unclear why some bacterial species have multiple bactofilin homologues. In C. crescentus the two bactofilins appear to function together in the same morphogenetic process. This appears not to be true for BacM and BacN-P in M. xanthus. Finally, it is not well understood how bactofilins exert their morphogenetic effect, although the direct interaction with PG synthesis/modification enzymes appears to emerge as a common theme. Future studies might reveal whether bactofilins have additional functions other than their involvement in cellular morphogenesis, whether they exist in even more supramolecular arrangements, and how the different forms and suprastructures of bactofilins exert their effect on cell morphology.
Experimental procedures
Bacterial strains and growth conditions
Table 1 lists the strains used in this study. The construction of plasmids and strains is described in Table S2. M. xanthus and S. aurantiaca strains were cultured in CTT medium (Kaiser, 1979) or on CTT agar plates. E. coli and P. mirabilis strains were cultured in LB medium or on LB agar plates. When appropriate, antibiotics were added at the following concentrations: kanamycin, 40 µg ml−1; ampicillin, 100 µg ml−1; tetracycline, 10 µg ml−1.
Table 1.
Bacterial strains used in this study.
| Strain | Relevant descriptiona | Source or reference |
|---|---|---|
| M. xanthus | ||
| DK1622 | Wild-type (wt) | Kaiser (1979) |
| DK8615 | ΔpilQ | Wall et al. (1999) |
| DK10410 | ΔpilA | Wu et al. (1997) |
| EH301 | [DK1622] ΔbacM. Complete coding region removed from wt locus | This study |
| EH302 | [DK8615] ΔbacM. Complete coding region removed from wt locus | This study |
| EH309 | [EH301] bacM. Complementation at the wt bacM locus | This study |
| EH310 | [EH302] bacM. Complementation at the wt bacM locus | This study |
| EH332 | [DK1622] MXAN7474∷kan. Coding region disrupted by plasmid insertion. KanR | This study |
| EH341 | [EH301] ΔbacN-P. Complete coding regions replaced by 10 codons comprising the start of bacN (MXAN4637) and the end of bacP (MXAN4635) |
This study |
| EH342 | [DK1622] ΔbacN-P. Complete coding regions replaced by 10 codons comprising the start of bacN and the end of bacP |
This study |
| EH344 | [EH301] Poar-bacM. Complementation at the chromosomal attB site under control of the oar promoter. TetR |
This study |
| EH345 | [DK1622] PpilA-bacM. Integration of bacM under control of the pilA promoter at the chromosomal attB site. TetR |
This study |
| EH351 | [DK1622] bacM–mCherry. Integration of the gene fusion into the wt bacM locus. KanR | This study |
| EH358 | [EH301] bacM–mCherry. Integration of the gene fusion into the wt bacM locus at which endogenous bacM is absent. KanR |
This study |
| EH362 | [DK1622] Poar-bacM–mCherry. Integration of the gene fusion at the chromosomal attB site under control of the oar promoter. TetR |
This study |
| EH364 | [EH301] Poar-bacM–mCherry. Integration of the gene fusion at the chromosomal attB site under control of the oar promoter. TetR |
This study |
| EH366 | [EH302] bacM–mCherry. Integration of the gene fusion into the wt bacM locus at which endogenous bacM is absent. KanR |
This study |
| MT295 | [DK1622] ΔbacN-P | Kühn et al. (2010) |
| MT299 | [DK1622] ΔbacM mCherry–bacM | Kühn et al. (2010) |
| MT300 | [DK1622] ΔbacM | Kühn et al. (2010) |
| JK328 | [MT295] ΔbacN-P DbacM | Kühn et al. (2010) |
| E. coli | ||
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(araleu)7697 galU galK rpsL (StrR) endA1 nupG. E. coli host for cloning. |
Invitrogen |
| BL21 Star (DE3) | F−ompT hsdSB(rB−, mB−) gal dcm rne131 (DE3). E. coli host for overexpression. | Invitrogen |
| DH10B | F−mcrA Δ(mrr, hsdRMS-mcrBC) F80lacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara leu)7697 galU galK rpsL nupG λ−E. coli host for cloning. |
Invitrogen |
| BL21-Tag-7475TR | [BL21] his6-V5-TEV∷bacM(codons 28–151). Contains plasmid encoding a fusion of hexahistidine tag, V5 epitope and TEV site to bacM lacking the first 27 codons, all preceded by an inducible lac promoter. |
This study |
| BL21-7475FL | [BL21] bacM. Contains plasmid encoding bacM, under control of an inducible lac promoter. | This study |
| BL21-7475TR | [BL21] bacM(codons28–151). Contains plasmid encoding bacM lacking the first 27 condons, under control of an inducible lac promoter. |
This study |
| Others: | ||
| DW4/3–1 | Stigmatella aurantiaca wild-type | Qualls et al. (1978) |
| Pm-AAG1 |
Proteus mirabilis wild-type. Isolate from Aedes aegypti gut.Verified via 16S rRNA gene sequencing. |
Jose Luis Ramirez (unpublished) |
Strain names in brackets indicate the parental strain and its genotype.
Protein expression and purification
Recombinant BacM proteins were expressed in E. coli BL21 from genes cloned into the pET151/D-TOPO vector (Invitrogen). Cells were induced with 1 mM IPTG (isopropyl β-d-1-thiogalactopyranoside) at mid-log phase, then grown overnight at 30°C for protein expression. The cells were then harvested and lysed by sonication. The resulting lysate was centrifuged at 40 000 g for 30 min to remove cellular debris, and the supernatant was applied to a 5 ml HisTrap column (Invitrogen) pre-equilibrated with loading buffer (50 mM Tris pH 7.5, 100 mM NaCl, 0.5% Triton X-100, 0.1 mM PMSF and 1 mM DTT). The column was washed and eluted with imidazole according to the manufacturer’s instructions. The 250 mM imidazole elution fraction containing the purified protein was dialysed into Tris buffer (50 mM Tris, 100 mM NaCl, pH 7.5) and concentrated to 0.5 mg ml−1 using an Amicon 3 kDa molecular weight cut-off centrifugal filter. Protein concentrations were determined using a modified Bradford assay (Bio-Rad Protein Assay). Untagged BacM proteins were generated by growing cells after induction for 3 h at 32°C. After harvesting in microfuge tubes at 12 000 g for 2 min, the cell pellets were treated for immunoblot analysis as described.
Antibody generation and purification
Purified tagged N-terminally truncated BacM protein was used by Cocalico Biologicals (Reamstown, PA, USA) to immunize a rabbit. Initial immunoblot analysis of the generated anti-BacM antiserum revealed some unspecific binding in addition to a strong reaction to BacM.
To purify the antiserum as well as the pre-immune serum, in each case cells from 4 l of EH302 culture (ΔpilQ ΔbacM) were harvested at an OD600 of 0.7. Cell pellets (approximately 12 g) were resuspended in 24 ml PM-buffer, mixed with 12 ml 16% paraformaldehyde, fixed at 4°C for 14 h, washed twice with 30 ml (same volume in all subsequent steps) PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na-phosphate, pH 7.4), and permeabilized for 15 min with 0.2% Triton X-100 in PBS. Next, the cells were washed twice with PBS and once with PBST (PBS with 0.05% Tween 20), and then incubated rocking for 20 min in PBST/2% BSA. Anti-BacM and pre-immune sera were each diluted 1:25 in PBST/2% BSA, and 30 ml of each were incubated separately with the pretreated EH302 cells for 1 h at 25°C on a rocking shaker to allow for unspecific binding of serum components to the cells. After removal of the cells via centrifugation for 15 min at 26 000 g, no unspecific binding was detected when using these purified sera in immunoblots or immunofluorescence experiments (Fig. 3E and Fig. S13).
Immunoblot analysis
For immunoblot analysis of total cellular protein, cells were grown to mid-logarithmic phase, harvested by centrifugation, and resuspended in TPM buffer (10 mM Tris-HCl, 8 mM MgSO4, 1 mM KH2PO4, pH 7.6) to an OD600 of 1.0. One millilitre of cell suspension, each, was centrifuged and the pellets were immediately frozen in liquid nitrogen. When testing BacM stability against degradation, freezing was performed after incubation of the cell pellet at 25°C for 8 h, or after sonication of the cells in a cup sonicator and incubation of the lysate at 25°C for 1.5 or 8 h. Subsequently, the frozen cells were boiled in Laemmli buffer and equal amounts of total cellular protein (30 mg) were loaded onto a 4–20% Tris-Glycine Gel (Invitrogen). SeeBlue (Invitrogen) was used as MW standard. The proteins were separated using SDS-PAGE and transferred to nitrocellulose membrane. After 30 min blocking with 3% non-fat dry milk in TBS (20 mM Tris-HCl, 137 mM NaCl, pH 7.6), the blots were incubated for 1 h at 25°C with purified anti-BacM serum diluted 1:500 in TBS containing 1% BSA and 0.05% Tween 20. When testing whether the anti-MreB serum (Jones et al., 2001) can detect MreB in M. xanthus lysate, the serum was used at a dilution of 1:8000. As the next step, blots were washed 3× with TBS and incubated for 1 h at 25°C with ImmunoPure peroxidase-conjugated goat anti-rabbit IgG secondary antibody (Thermo Scientific) diluted 1:10 000 in TBS containing 1% BSA and 0.05% Tween 20. After washing three times with TBS, blots were developed with SuperSignal West Pico chemiluminescent substrate (Pierce), X-ray film and a Mini-Medical film processor (AFP imaging).
Light and fluorescence microscopy
Microscopic images were acquired with a Nikon Plan Apo 100x/NA 1.4 phase-contrast oil immersion objective on a Nikon Eclipse 90i microscope. For fluorescence microscopy a DAPI-, a FITC-, or a TRITC-filter cube were applied. Volocity software was used to acquire images with an ORCA ER CCD camera (Hamamatsu), and for subsequent image processing. The original image dimension of 1344 × 1024 pixels resulted in coverage of 65 nm distance on the slide per pixel at 100× magnification. All images were further processed with Adobe Photoshop software. Brightness and contrast were adjusted so that the features visible in the unprocessed images were optimally represented.
To document cell shapes, cells were grown to mid-logarithmic or stationary phase, pelleted, resuspended in CTT medium, applied to a glass slide, and observed under phase contrast.
To investigate M. xanthus or S. aurantiaca cells via immunofluorescence microscopy, cells were grown to mid-logarithmic phase unless stated otherwise. Cells were harvested, resuspended in PM buffer (20 mM Na-phosphate, 1 mM MgSO4, pH 7.4) to a density of 8 × 108 cells per ml, fixed for 40 min in 4% paraformaldehyde in PM buffer, and 15 µl aliquots per well were applied to 10-well HTC Super Cured slides (Thermo Scientific, Cel-Line). Cells attached to the slides were permeabilized by incubation in 0.2% Triton X-100 in PBS for 10 min, followed by washing with PBS and GTE buffer (50 mM glucose, 20 mM Tris/HCl, 10 mM EDTA, pH 7.5), and finally by incubation in 1 mg ml−1 lysozyme in GTE buffer for 30 min at 37°C. After washing with PBS and blocking with PBST/2% BSA (PBS supplemented with 0.05% Tween 20 and 2% BSA), cells were incubated for 1 h at 25°C in PBST/2% BSA with 1:10-diluted purified anti-BacM serum, 1:10-diluted purified pre-immune serum (as negative control), or 1:4000-diluted anti-MreB antiserum (Jones et al., 2001). Alexa Fluor 594- or Alexa Fluor 488-conjugated goat anti-rabbit IgG antibodies (Invitrogen) diluted 1:1000 in PBST/ 2%BSA were used as secondary antibodies (1 h at 25°C). After washing with PBS, cells were optionally incubated with 1 mg ml−1 DAPI in PBS for 10 min, and finally mounted with ProLong Gold antifade reagent (Molecular Probes), optionally containing 1 µg ml−1 DAPI.
To visualize the BacM–mCherry fusion protein in suspended cells, the cells were collected from liquid cultures at different growth phases or from 1.5% agar plates containing CTT. After resuspension, the cells were optionally fixed and applied to glass slides, and red fluorescence was observed using a FITC filter cube.
To observe BacM–mCherry screws in motile, growing cells, the cells were collected after 3 days of growth on CTT agar and resuspended in CTT to an OD600 of approximately 0.2. Then 1 µl cells was spotted onto a 0.7 mm thick patch of 1.5% agar in TPM buffer, and covered with a coverslip. The agar patch optionally contained CTT medium plus 100 µM cephalexin. To avoid dehydration, all edges of the coverslip were carefully sealed with a molten mixture of paraffin/ vaseline (2:1 w/w). Under these conditions the cells maintained growth and motility for several days.
To visualize DNA in live unfixed cells, cells from mid-logarithmic CTT cultures were used, optionally after additional growth in CTT with 100 µM cephalexin for 6–13 h to an OD600 of 0.6–0.8. To 100 ml of these cultures DAPI was added to a final concentration of 1 µg ml−1. After 10 min incubation, 1.5 µl of each mixture was applied to a glass slide, and blue fluorescence was recorded using a DAPI filter cube.
Electron microscopy
For analysis by scanning electron microscopy (SEM), M. xanthus cells grown in CTT to an OD600 of 0.7 were attached to a 0.01% poly-l-lysine-treated glass slide and fixed for 1.5 h at 25°C in 2.5% glutaraldehyde EM grade in PM buffer. Subsequently, cells were washed 3× with PM buffer, post-fixed with 1% osmium tetroxide in PM buffer, washed twice with water, incubated in aqueous 2% uranyl acetate for 1 h at 25°C, and washed 2× with water. Cells were dehydrated overnight through a graded series of ethanol, 5 min each in 50%, 70% and 90% ethanol, then three times 5 min in 100% ethanol, and finally 10 min in hexamethyldisilazane (Ted Pella). After draining the liquid, the slides were desiccated overnight, attached to aluminum stubs, and coated with 20 nm of Au/Pd with a Desk III Sputter Coater (Denton Vacuum). After installing the stubs, digital images of the cells were captured by a Leo 1530 field emission scanning electron microscope operating at 1 kV.
For examination by transmission electron microscopy, isolated and purified BacM fibres were negatively stained on glow discharge-treated 400 mesh carbon-coated copper grids (Electron Microscopy Sciences) using 2% (w/v) uranyl acetate. For specimen observation, a Philips CM120 was used at an acceleration voltage of 100 kV. Images were recorded on Kodak ISO 165 black and white film at nominal magnifications ranging from 10 000 to 52 000×. Aliquots of the isolated purified BacM fibre samples were analysed using SDS-PAGE to biochemically confirm the absence of any contaminating proteins.
Isolation and purification of native BacM fibres from M. xanthus cells
M. xanthus wild-type cells (DK1622) were either grown in liquid CTT medium to a density of approximately 5 × 109 cells ml−1 or for 24 h on CTT agar trays (1.5% agar, 30 × 50 cm commercially available aluminum cookie sheets). About 30 g of cells was harvested and re-suspended in 120 ml of 10 mM Tris buffer pH 8.0 and subjected to blending in a household mixer (3 min highest speed setting). Unbroken cells were pelleted by centrifugation at 12 000 g and membranes and large cell debris were removed by a second centrifugation for 15 min at 47 800 g. 2% PEG 8000 and 100 mM NaCl were added to the supernatant and the suspension was recentrifuged for 15 min at 47 800 g. The pelleted fibres, membrane fragments and cellular proteins were dissolved in about 10 ml Tris buffer containing 0.5% dodecylmaltoside. After addition of CsCl to 0.3 g ml−1 the mixture was transferred to clear ultracentrifuge tubes and centrifuged overnight at 368 000 g in an SW55 rotor. Isolated fibres formed a small whitish pellet at the bottom of the tubes. After careful removal of the supernatant, the pellet was resuspended in a small amount of 10 mM Tris buffer pH 8.0 and examined under the electron microscope to confirm the presence of the fibres.
Protein analysis
For N-terminal sequencing, purified fibres were separated on SDS-PAGE, blotted onto PVDF membrane (Millipore) and stained with Coomassie Blue G250. Appropriate bands were cut out and subjected to Edman degradation at the Macromolecular Structure Facility, Michigan State University. MALDI-MS/MS analysis of in-gel trypsin-digested proteins was performed according to standard protocols (Shevchenko et al., 1996) with some modifications as described in the Supporting Information.
S-motility assays
The ability of strains to swarm on soft agar plates using S-motility was investigated essentially as previously described (Jelsbak and Kaiser, 2005). In brief, after inoculation of CTT medium with pre-cultures, strains were grown to an OD600 of 1.0 (5 × 108 cells per ml), pelleted and resuspended in CTT medium to an OD of 10. Then 2 × 107 cells were spotted on 50% CTT, 0.4% agar plates and incubated at 32°C. Pictures of the plates were taken after 24 and 48 h with an Epson Perfection 636U scanner in transparent mode and processed with Adobe Photoshop software.
Immunoprecipitation
For immunoprecipitation of BacM, 14 ml of a culture of strain DK1622 at an OD600 of 0.8 was centrifuged, and the pelleted cells were resuspended in 1.5 ml lysis buffer (PBS with 5 mM EDTA, 0.5% Triton X-100, and 1 mM PMSF) and lysed by sonication on ice. After clearing by centrifugation, 1.4 ml of the lysate was added to 20 µl protein A sepharose beads (Sigma-Aldrich) and incubated at 4°C for 1 h on a rocking shaker to remove non-specifically bound proteins. After pelleting of the beads, 1.25 ml of the pre-incubated lysate was removed and added to 250 µl purified anti-BacM antiserum. Twenty-five-microlitre fresh protein A sepharose beads were incubated with this mixture for 4 h at 4°C. After centrifugation, the supernatant was removed, the pelleted beads were washed twice with 1 ml PBS, each, and finally mixed with 25 µl 2× Laemmli buffer and boiled for 10 min to elute bound antibodies together with antigen. Cells of strain EH301 were treated in the same way and 15 µl of each eluate was subjected to SDS-PAGE. Distinct bands at 13 and 11 kDa apparent molecular weight were present only after incubation with DK1622 lysate and were excised and analysed using mass spectrometry.
Paper disc diffusion assay
Cells of different strains were grown in CTT medium to an OD600 of 0.25. Eight hundred microlitres of cell suspension (1 × 108 cells) was mixed with 8 ml CTT overlay agar (0.75%, 55°C) and poured onto 50 ml CTT agar (1.5%) in 15 cm diameter culture plates. The suspension of the cells in agar prevented them from moving across the plates during the assay. Solutions of different antibiotics were applied to 6 mm diameter paper discs (Whatman 3MM chromatography paper) as previously described (Ericsson, 1960). This resulted in defined amounts of antibiotic per disk ranging from 50 to 600 µg, depending on the antibiotic. After drying, the discs were placed on top of the overlay agar and the plates were sealed with parafilm and incubated at 32°C for 100 h. Finally, the plates were scanned to record the sizes of growth inhibition zones.
Test of susceptibility to antibiotics in a 96-well microtitre plate format
Cells of different strains were grown in CTT medium, harvested at exponential growth phase, and diluted to an OD600 of 0.002 (1 × 106 cells per ml). Freshly prepared sterile 10 mg ml−1 stock solutions of amoxicillin, mezlocillin and ampicillin were diluted to 400 µg ml−1 in CTT medium, and serial twofold dilutions were produced. The 300 µl wells of 96-well plastic trays were then each filled with 60 µl of antibiotic solution. Subsequent mixing with 60 µl of cell suspension resulted in the final antibiotic concentrations. The number of cells per well was 6 × 104, corresponding to a concentration of 5 × 105 cells per ml, a common concentration in such assays (Wiegand et al., 2008). All wells were then covered with UV-sterilized adhesive plastic foil to prevent evaporation and the plates were incubated at 32°C while shaking at 300 r.p.m. After 72 h, the contents of each well were mixed by pipetting them up and down five times, and absorption at 595 nm was determined with a plate reader. Subsequently, the plates were scanned to obtain a visual record of the cell densities in the wells. MICs were determined as the concentrations at which no cell growth was detectable in the wells.
Supplementary Material
Acknowledgements
We would like to thank Jeff Errington for providing the anti-MreB antibody, Mike Delannoy from the Microscope Facility at the Johns Hopkins School of Medicine for his kind help with the sample preparation and acquisition of the SEM pictures, and Tatjana Boronina from the Johns Hopkins University Mass Spectrometry and Proteomics Facility for her excellent help with the acquisition of the mass spectrometry data. Furthermore, we thank Martin Thanbichler for kindly providing us with M. xanthus strains MT295, MT299, MT300 and JK328, Susanne Müller for sending us the S. aurantiaca strain DW4/3–1, Jose Luis Ramirez for providing us with the P. mirabilis wild-type strain, and the members of the Hoiczyk laboratory for helpful discussions and comments on the manuscript. The research in our laboratory is funded by a grant from the National Institutes of Health (GM85024) and a Faculty Innovation Fund from the Bloomberg School of Public Health to E.H., an NIEHS Training Grant (ES07141) to C.A. M., and a fellowship from the Jane Russell Fund for Innovative Research in Infectious Diseases to M.K.K.
Footnotes
Supporting information
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Adams DW, Errington J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol. 2009;7:642–653. doi: 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- Ausmees N, Kuhn JR, Jacobs-Wagner C. The bacterial cytoskeleton: an intermediate filament-like function. Cell. 2003;115:705–713. doi: 10.1016/s0092-8674(03)00935-8. [DOI] [PubMed] [Google Scholar]
- Bi E, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Cabeen MT, Charbon G, Vollmer W, Born P, Ausmees N, Weibel DB, Jacobs-Wagner C. Bacterial cell curvature through mechanical control of cell growth. EMBO J. 2009;28:1208–1219. doi: 10.1038/emboj.2009.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballido-López R. The bacterial actin-like cytoskeleton. Microbiol Mol Biol Rev. 2006;70:888–909. doi: 10.1128/MMBR.00014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballido-López R, Errington J. The bacterial cytoskeleton: in vivo dynamics of the actin-like protein Mbl of Bacillus subtilis. Dev Cell. 2003;4:19–28. doi: 10.1016/s1534-5807(02)00403-3. [DOI] [PubMed] [Google Scholar]
- Carballido-López R, Formstone A, Li Y, Ehrlich SD, Noirot P, Errington J. Actin homolog MreBH governs cell morphogenesis by localization of the cell wall hydrolase LytE. Dev Cell. 2006;11:399–409. doi: 10.1016/j.devcel.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Charbon G, Cabeen MT, Jacobs-Wagner C. Bacterial intermediate filaments: in vivo assembly, organization, and dynamics of crescentin. Genes Dev. 2009;23:1131–1144. doi: 10.1101/gad.1795509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsson H. The paper disc method for determination of bacterial sensitivity to antibiotics. Studies on the accuracy of the technique. Scand J Clin Lab Invest. 1960;12:408–413. doi: 10.3109/00365516009065405. [DOI] [PubMed] [Google Scholar]
- Gayda RC, Henk MC, Leong D. C-shaped cells caused by expression of an ftsA mutation in Escherichia coli. J Bacteriol. 1992;174:5362–5370. doi: 10.1128/jb.174.16.5362-5370.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitai Z. The new bacterial cell biology: moving parts and subcellular architecture. Cell. 2005;120:577–586. doi: 10.1016/j.cell.2005.02.026. [DOI] [PubMed] [Google Scholar]
- Gitai Z. New fluorescence microscopy methods for microbiology: sharper, faster, and quantitative. Curr Opin Microbiol. 2009;12:341–346. doi: 10.1016/j.mib.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring NW, Beckwith J. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr Biol. 2005;15:R514–R526. doi: 10.1016/j.cub.2005.06.038. [DOI] [PubMed] [Google Scholar]
- Goldman BS, Nierman WC, Kaiser D, Slater SC, Durkin AS, Eisen J, et al. Evolution of sensory complexity recorded in a myxobacterial genome. Proc Natl Acad Sci USA. 2006;103:15200–15205. doi: 10.1073/pnas.0607335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann PL. Cytoskeletal elements in bacteria. Annu Rev Microbiol. 2007;61:589–618. doi: 10.1146/annurev.micro.61.080706.093236. [DOI] [PubMed] [Google Scholar]
- Graumann PL, Knust T. Dynamics of the bacterial SMC complex and SMC-like proteins involved in DNA repair. Chromosome Res. 2009;17:265–275. doi: 10.1007/s10577-008-9014-x. [DOI] [PubMed] [Google Scholar]
- Hay NA, Tipper DJ, Gygi D, Hughes C. A novel membrane protein influencing cell shape and multi-cellular swarming of Proteus mirabilis. J Bacteriol. 1999;181:2008–2016. doi: 10.1128/jb.181.7.2008-2016.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen GR, Wireman JW, Dworkin M. Effect of temperature on the growth of Myxococcus xanthus. J Bacteriol. 1977;130:561–562. doi: 10.1128/jb.130.1.561-562.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelsbak L, Kaiser D. Regulating pilin expression reveals a threshold for S motility in Myxococcus xanthus. J Bacteriol. 2005;187:2105–2112. doi: 10.1128/JB.187.6.2105-2112.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LJF, Carballido-López R, Errington J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–922. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- Kaiser D. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc Natl Acad Sci USA. 1979;76:5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai Y, Daniel RA, Errington J. Regulation of cell wall morphogenesis in Bacillus subtilis by recruitment of PBP1 to the MreB helix. Mol Microbiol. 2009;71:1131–1144. doi: 10.1111/j.1365-2958.2009.06601.x. [DOI] [PubMed] [Google Scholar]
- Kruse T, Moller-Jensen J, Lobner-Olesen A, Gerdes K. Dysfunctional MreB inhibits chromosome segregation in Escherichia coli. EMBO J. 2003;22:5283–5292. doi: 10.1093/emboj/cdg504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse T, Bork-Jensen J, Gerdes K. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol Microbiol. 2005;55:78–89. doi: 10.1111/j.1365-2958.2004.04367.x. [DOI] [PubMed] [Google Scholar]
- Kühn J, Briegel A, Morschel E, Kahnt J, Leser K, Wick S, et al. Bactofilins, a ubiquitous class of cytoskeletal proteins mediating polar localization of a cell wall synthase in Caulobacter crescentus. EMBO J. 2010;29:327–339. doi: 10.1038/emboj.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard TA, Møller-Jensen J, Löwe J. Towards understanding the molecular basis of bacterial DNA segregation. Philos Trans R Soc Lond B Biol Sci. 2005;360:523–535. doi: 10.1098/rstb.2004.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löwe J, Amos LA. Evolution of cytomotive filaments: the cytoskeleton from prokaryotes to eukaryotes. Int J Biochem Cell Biol. 2009;41:323–329. doi: 10.1016/j.biocel.2008.08.010. [DOI] [PubMed] [Google Scholar]
- Lu C, Stricker J, Erickson HP. FtsZ from Escherichia coli, Azotobacter vinelandii, and Thermotoga maritime - quantitation, GTP hydrolysis, and assembly. Cell Motil Cytoskeleton. 1998;40:71–86. doi: 10.1002/(SICI)1097-0169(1998)40:1<71::AID-CM7>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:205–210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin W. FtsZ and the division of prokaryotic cells and organelles. Nat Rev Mol Cell Biol. 2005;6:862–871. doi: 10.1038/nrm1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Canamero M, Munoz-Dorado J, Farez-Vidal E, Inouye M, Inouye S. Oar, a 115-kilodalton membrane protein required for development of Myxococcus xanthus. J Bacteriol. 1993;175:4756–4763. doi: 10.1128/jb.175.15.4756-4763.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauriello EMF, Mouhamar F, Nan B, Ducret A, Dai D, Zusman DR, Mignot T. Bacterial motility complexes require the actin-like protein, MreB and the Ras homologue, MglA. EMBO J. 2010;29:315–326. doi: 10.1038/emboj.2009.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michie KA, Löwe J. Dynamic filaments of the bacterial cytoskeleton. Annu Rev Biochem. 2006;75:467–492. doi: 10.1146/annurev.biochem.75.103004.142452. [DOI] [PubMed] [Google Scholar]
- Morris DM, Jensen GJ. Toward a biomechanical understanding of whole bacterial cells. Annu Rev Biochem. 2008;77:583–613. doi: 10.1146/annurev.biochem.77.061206.173846. [DOI] [PubMed] [Google Scholar]
- Qualls GT, Stephens K, White D. Morphogenetic movements and multicellular development in fruiting myxobacterium Stigmatella aurantiaca. Dev Biol. 1978;66:270–274. doi: 10.1016/0012-1606(78)90291-9. [DOI] [PubMed] [Google Scholar]
- Rajagopala SV, Titz B, Goll J, Parrish JR, Wohlbold K, McKevitt MT, et al. The protein network of bacterial motility. Mol Syst Biol. 2007;3:128. doi: 10.1038/msb4100166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev. 2008;32 doi: 10.1111/j.1574-6976.2008.00105.x. 556-556. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Shih YL, Rothfield L. The bacterial cytoskeleton. Microbiol Mol Biol Rev. 2006;70:729–754. doi: 10.1128/MMBR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J, Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975;254:34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- Sycuro LK, Pincus Z, Gutierrez KD, Biboy J, Stern CA, Vollmer W, Salama NR. Peptidoglycan crosslinking relaxation promotes Helicobacter pylori’s helical shape and stomach colonization. Cell. 2010;141:822–833. doi: 10.1016/j.cell.2010.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takacs CN, Poggio S, Charbon G, Pucheault M, Vollmer W, Jacobs-Wagner C. MreB drives de novo rod morphogenesis in Caulobacter crescentus via remodeling of the cell wall. J Bacteriol. 2010;192:1671–1684. doi: 10.1128/JB.01311-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanbichler M, Shapiro L. Getting organized - how bacterial cells move proteins and DNA. Nat Rev Microbiol. 2008;6:28–40. doi: 10.1038/nrmicro1795. [DOI] [PubMed] [Google Scholar]
- Tsang PH, Li GL, Brun YV, Ben Freund L, Tang JX. Adhesion of single bacterial cells in the micronewton range. Proc Natl Acad Sci USA. 2006;103:5764–5768. doi: 10.1073/pnas.0601705103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanEngelenburg SB, Palmer AE. Fluorescent biosensors of protein function. Curr Opin Chem Biol. 2008;12:60–65. doi: 10.1016/j.cbpa.2008.01.020. [DOI] [PubMed] [Google Scholar]
- Wall D, Kolenbrander PE, Kaiser D. The Myxococcus xanthus pilQ (sglA) gene encodes a secretin homolog required for type IV pilus biogenesis, social motility, and development. J Bacteriol. 1999;181:24–33. doi: 10.1128/jb.181.1.24-33.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CL, Kitich A, Gober JW. Positioning cell wall synthetic complexes by the bacterial morphogenetic proteins MreB and MreD. Mol Microbiol. 2010;76:616–633. doi: 10.1111/j.1365-2958.2010.07108.x. [DOI] [PubMed] [Google Scholar]
- Wiegand I, Hilpert K, Hancock REW. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008;3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- Williamson MP. The structure and function of proline-rich regions in proteins. Biochem J. 1994;297:249–260. doi: 10.1042/bj2970249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SS, Kaiser D. Genetic and functional evidence that type IV pili are required for social gliding motility in Myxococcus xanthus. Mol Microbiol. 1995;18:547–558. doi: 10.1111/j.1365-2958.1995.mmi_18030547.x. [DOI] [PubMed] [Google Scholar]
- Wu SS, Wu J, Kaiser D. The Myxococcus xanthus pilT locus is required for social gliding motility although pili are still produced. Mol Microbiol. 1997;23:109–121. doi: 10.1046/j.1365-2958.1997.1791550.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.