Abstract
Rasgrf1 is genomically imprinted; only the paternally-inherited allele is expressed in the neonatal mouse brain until weaning, at which time expression becomes biallelic. Whereas Rasgrf1 has been implicated in learning and memory via knockout studies in adult mice, the effect of its normal imprinted expression on these phenotypes has not yet been examined. Neonatal mice with experimentally manipulated patterns of imprinted Rasgrf1 expression were assessed on an associative olfactory task. Neonates lacking the normally-expressed wildtype paternal allele exhibited significant impairment in olfactory associative memory. Adult animals in which neonatal imprinting had been manipulated were also behaviorally assessed; while neonatal imprinting significantly affects body weight even into adulthood, no learning and memory phenotype attributable to imprinting was observed in adults. Additional analyses of neonates revealed imprinted Rasgrf1 transcript selective to olfactory bulb even in mice that were null for Rasgrf1 in the rest of the brain, and showed that Rasgrf1 affects Ras and Rac activation in the brain. Taken together, these results indicate that Rasgrf1 expression from the wildtype paternal allele contributes to learning and memory in neonatal mice.
Keywords: genomic imprinting, olfactory learning, development, memory consolidation, conflict hypothesis
INTRODUCTION
Rasgrf1 is an imprinted gene that is expressed solely from the paternally-inherited allele in neonatal mouse brain until weaning (postnatal day 21; P21), at which time its expression becomes biallelic (Drake et al. 2009; Plass et al. 1996). Rasgrf1 imprinting is controlled by a binary switch consisting of a differentially-methylated domain (DMD) and a series of repeats immediately 3’ of the DMD. These repeats direct the placement of methylation on the paternal DMD, which regulates gene transcription at the Rasgrf1 locus. In contrast, the maternal DMD is unmethylated, which permits CTCF binding and results in the inhibition of gene transcription from the maternal allele (Yoon et al. 2005; Yoon et al. 2002). Hence, only the paternal allele is normally expressed in neonates. Rasgrf1 is most highly expressed in central nervous system neurons (Sturani et al. 1997; Zippel et al. 2000), with lower expression levels in other somatic tissues (Font de Mora et al. 2003; Plass et al. 1996). Its product, RasGRF1 protein, is a guanine nucleotide exchange factor for Ras and Rac (Cen et al. 1993; Innocenti et al. 1999), activating these G-proteins in response to cellular calcium influx (Farnsworth et al. 1995) or serine phosphorylation (Mattingly et al. 1999; Yang et al. 2003) in pathways downstream of muscarinic receptor activity (Mattingly & Macara 1996), heterotrimeric G-protein subunit dissociation (Kiyono et al. 2000; Shou et al. 1995), and neurotrophin binding to TrkA, TrkB, and TrkC receptors (MacDonald et al. 1999; Robinson et al. 2005).
Rasgrf1 is involved in multiple neuronal learning and plasticity mechanisms, including Ras-MAPK-dependent memory consolidation and long-term plasticity in the amygdala (Brambilla et al. 1997) and Ras-ERK pathway activation (Fasano et al. 2009; Krapivinsky et al. 2003). Correspondingly, adult Rasgrf1 knockout mice are impaired in several learning and memory tasks (Brambilla et al. 1997; Fasano et al. 2009; Giese et al. 2001). These results suggested that the normal neonatal imprinting of Rasgrf1 expression may affect learning performance in neonatal mice – a prediction consistent with the conflict hypothesis of genomic imprinting (Moore & Haig 1991; Wilkins & Haig 2003). Here, we evaluated the effects of perturbation of Rasgrf1 imprinting on olfactory associative learning during the neonatal period when Rasgrf1 expression is normally imprinted. Specifically, we engineered two mutant Rasgrf1 alleles that, respectively, prevented expression of the paternally-inherited allele or forced expression of the maternally-inherited allele; with these we generated neonates expressing Rasgrf1 from either, neither, or both parentally-inherited alleles and tested them with an olfactory associative learning assay. Additionally, as Rasgrf1 imprinting in neonates affects postnatal growth well into adulthood (Drake et al. 2009), we also measured the effects of neonatal imprinting on a battery of behavioral assays in adults.
MATERIALS AND METHODS
Animals
Mice utilized for behavioral experiments carried various combinations of wildtype and two mutated alleles designed to alter the pattern of neonatal imprinting (allele-specific expression). One mutated allele, Rasgrf1tm1Pds, or tm1, prevents Rasgrf1 expression in neonates when paternally inherited (Yoon el al. 2002). The second mutated allele, Rasgrf1tm2Pds, or tm2, activates expression of Rasgrf1, including expression from the normally silent maternal allele. The Pgk enhancer enforces expression of the tm2 allele (Yoon et al. 2005). Mutations were prepared using J1 ES cells (129S4Jae background); mice bearing the mutations were backcrossed a minimum of 10 times onto the C57BL/6J background before the additional crosses described herein were performed. For imprinting tests, we crossed C57BL/6J mice with FVB/NJ mates.
Animals used in behavioral experiments were derived from two crosses. First, +/tm1 males were bred with tm2/+ females, which generated four genotypes and facilitated the use of littermate controls. Specifically, in addition to the wildtype genotype, in which neonatal Rasgrf1 expression is monoallelic and derived from the paternal allele (MP, +/+ or wt), mutant genotypes were generated that exhibited biallelic expression (B, tm2/+; maternal allele is listed first), were null for Rasgrf1 expression (N, +/tm1), or that exhibited monoallelic expression from the maternal allele (MM, tm2/tm1). Second, +/+ females were crossed with tm2/+ males, which generated a fifth genotype, +/tm2, as well as +/+ (MP) littermates to use as controls. The +/tm2 genotype was constructed to replicate wildtype (MP) imprinted expression of Rasgrf1 in behavioral experiments so as to control for any effects of the manipulations used to express the tm2 allele. Both tm1 and tm2 alleles were maintained on a C57BL/6J background. Mice used for RNA quantification were derived by crossing tm2/tm2, tm1/tm1, and +/+ homozygotes to generate the five genotypes (tm2/+, tm2/tm1, +/tm1, +/+, and +/tm2).
Mice were maintained on a regular 12:12-hour light/dark cycle and had ad libitum access to food and water except where specified in Materials and Methods. All experiments were carried out under a protocol approved by the Cornell University Institutional Animal Care and Use Committee in accordance with NIH guidelines.
Neonatal olfactory associative learning assay
Training
Mouse pups, eight days postnatal (P8, with P1 defined as the day of birth), were assessed for olfactory associative learning using established methods (Armstrong et al. 2006); except as noted, all procedures used were identical to those described therein. Briefly, two odor stimuli were prepared: 2-furyl methyl ketone (FMK) and n-hexyl acetate (HA), differentially diluted in mineral oil to concentrations theoretically emitting vapor-phase partial pressures of 5.0 Pa (Cleland et al. 2002); the corresponding liquid-phase (vol/vol) dilution ratios were 13.0 × 10−3 for FMK and 11.4 × 10−3 for HA. Mouse pups (individually identified on P2 using footpad tattoos) were separated from their dam by removing the dam from the home cage for 90 minutes. One of the diluted test odorants was then applied to all of the dam’s nipples, after which she was returned to the home cage so that the pups could suckle and thereby associate the experimental odorant with a milk reward. This training procedure was repeated daily from P3 through P8. Whereas each litter was presented with the same odor CS across the five training trials, odorants were counterbalanced across litters, such that half of the litters associated reward with FMK and the other half with HA.
Testing
After the final training session on P8, pups were allowed to suckle for 45 minutes and then were again separated from the dam for 120 minutes before being assessed for an associatively learned odor preference (Alleva & Calamandrei 1986; Armstrong et al. 2006). Briefly, pups were tested for place preference in a 32 × 19 cm plexiglass arena (13 cm wall height) with a wire mesh placed above two adjacent 12 × 19 × 7 cm deep compartments. One compartment contained a Kimwipe (Kimberly-Clark, Neenah, WI, USA) saturated with 500 µL of diluted FMK, the other the same but with diluted HA; note that for any given pup one of these odorants had been used for training whereas the other had never before been presented. The two compartments were separated by 0.7 cm (wall thickness) where they met under the center of the arena; the pup was placed on the mesh atop this wall, facing away from the experimenter, such that their right limbs were placed over one compartment and their left limbs were placed over the other compartment.
The exploratory behavior of each pup was recorded for 180 seconds (s). Each pup was scored as investigating a compartment whenever the pup moved its muzzle or limbs completely off the center wall and directly over a compartment. Pups were replaced upon the midline when the following criteria were met so that their limited mobility would not dominate the assessment of preference: if a pup fell over so as to be unable to regulate its movements, reached the external wall of the arena, froze for 3 s without head movements, or began grooming, it was replaced on the midline in the opposite orientation. The total accumulated time spent sniffing or moving over each of the two compartments was recorded by stopwatch. The orientation of the two compartments was varied with respect to both odor identity and odor contingency between test trials; furthermore, the spatial orientation of the scented compartments was also varied with respect to the room to control for differences in external cues. The dam and littermates of the pup being tested were removed to a distant location to avoid possible distraction via ultrasonic calls. The experimenter was blind to genotype during testing.
Immediately following testing, tail biopsies were taken for genotyping. The group sizes for neonatal behavioral testing were as follows: 68 wildtype/MP, 59 biallelic (B), 31 monoallelic maternal (MM), 46 null (N), and 29 +/tm2 mutants.
Rasgrf1 Quantification
For quantification of Rasgrf1 transcript levels, brains were collected from P8 neonates and the olfactory bulbs and hippocampi were dissected out. Rasgrf1 transcript levels were separately measured by quantitative PCR in olfactory bulb, hippocampus, and in the remainder of the brain (ROB, comprising the whole brain excepting olfactory bulbs, hippocampus, hypothalamus and pituitary gland) for each of the five genotypes. Specifically, RNA was extracted, reverse-transcribed, quantified in triplicate using an ABI Taqman© probe specific for Rasgrf1, and normalized to 18S rRNA levels. Alpha levels for statistical significance were Bonferroni-corrected for ten multiple comparisons such that p < 0.005 indicated significance.
Rasgrf1 Imprinted Expression
RNA was separately isolated from the olfactory bulbs and the rest of the brain (in this case including all structures other than the olfactory bulbs) of P8 neonates bred from reciprocal crosses between C57BL/6J and FVB/NJ males and females. RNA was reverse-transcribed into cDNA, and PCR amplified with the following primers and cycling conditions: (F) 5’-ggctcatgatgaatgccttt-3’ (R) 5’-tacagaagcttggcgttgtg-3’; 95 °C × 3 minutes, followed by 40 cycles of 95 °C × 20 seconds, 58 °C × 30 seconds, 72 °C × 50 seconds, followed by 72 °C × 5 minutes. PCR products were then digested with 10U AciI, which recognizes a restriction site (C’CGC) in exon 14 that distinguishes expression derived from either of the two parental strains (SNP ID rs29947965). C57BL/6J-derived expression was indicated by bands of 210/146 bp, and FVB/N-derived expression was indicated by bands at 226/130 bp.
Ras and Rac signaling assays
To determine whether Ras or Rac activation was influenced by mutations at Rasgrf1, we quantified the levels of the active forms of both G proteins in the olfactory bulb, hippocampus, and isocortex of P8 wildtype (MP) and null mutant (N) neonates. Similarly-sized tissue samples of olfactory bulb, hippocampus, and isocortex were removed from extracted brains and placed in Krebs-Ringer solution (11.1 mM glucose, 1.1 mM MgCl2, 1 mM Na2HPO4, 1.3 mM CaCl2, 25 mM NaHCO3, 120 mM NaCl, 4.7 mM KCl). To extract protein, tissues were homogenized in 0.5 ml of extraction buffer on ice using a Dounce 1-ml homogenizer with a tightly fitting pestle. The extraction buffer included 1 mM sodium orthovanadate (Na2VO4), 25 mM sodium fluoride (NaF), and EDTA-free protease inhibitor tablets (Roche) in magnesium-containing buffer supplied by Millipore (#20–168; 125 mM HEPES, pH 7.5, 750 mM NaCl, 5% IGEPAL CA-630, 50 mM MgCl2, 5 mM EDTA, and 10% glycerol). Extracts then were affinity purified using Ras and Rac activation assay kits (Millipore 17–218 and 17–283 respectively). Purified protein extracts then were SDS-PAGE electrophoresed, blotted, and probed using α-Ras (05–516) and α-Rac (05–389) antibodies supplied with the kits in conjunction with goat α-mouse HRP-conjugated IgG secondary antibody (Millipore 12–349). Blots were visualized using SuperSignal West Dura Substrate (Pierce 34075), captured using a LAS-4000 imager’s CCD camera and chemiluminescent detection function, and quantified using MultiGauge software. The amounts of precipitated (active) protein were normalized to the amounts of input (total) protein for each structure, and then normalized to wildtype levels; N=4 mice for all comparisons.
Adult behavioral assays
Adult mice (37–52 days old) of four genotypes (B, MM, MP, N) were tested on a battery of standard behavioral phenotyping tasks in order to assess whether a learning deficit phenotype such as that observed in neonates with abnormal Rasgrf1 imprinting persisted into adulthood (i.e., after the onset of biallelic Rasgrf1 expression in all genotypes). Such persistent effects of neonatal Rasgrf1 imprinting have been observed for body mass (Drake et al. 2009). Specifically, measurements of working memory, olfactory nonassociative memory, and short- and long-term fear memory tasks dependent on amygdala and hippocampus were performed; additionally, several control studies assessing motor function and basal activity and anxiety levels were performed to aid interpretation of the learning task results. The same mice were used for all studies except fear conditioning (inhibitory avoidance, cued and contextual conditioning), for which tests the mice were divided into groups such that each animal was subjected to only one shock. Moreover, in fear conditioning studies, mice were either assessed at 0.5 hours after training (short-term fear memory) or 24 hours (long-term fear memory), but not both.
Mice were assessed on all behavioral tasks in the following order across four consecutive days. Day 1: neurological screening, open field, balance beams, wire forelimb suspension, vertical pole, hanging wire grip test. Day 2: olfactory habituation, visual cliff, prehabituation to fear-conditioning test cage, spontaneous alternation, social recognition. Day 3: fear conditioning (either light/dark inhibitory avoidance or auditory tone-cued conditioning) and 0.5 hour assessment. Day 4: 24-hour assessment of fear conditioning. Detailed methods for behavioral tests other than learning and memory tests are provided in Supplemental Materials.
Learning and memory tests
A spontaneous alternation test was performed to assess working memory (King & Arendash 2002). Mice were placed in a radially symmetric plexiglass Y-maze, with arms 4 cm wide × 21 cm long and with 40 cm high walls, and allowed to explore for 300 seconds. The total number of arm entries was recorded as an additional measure of baseline activity. Spontaneous alternation was measured as the proportion of arm choices differing from the previous two choices; i.e., the number of such choices divided by the total number of opportunities to alternate (the total number of arm entries minus two). The score for spontaneous alteration reflects each animal’s memory of their exploration, since mice tend to avoid re-entering the arm that they explored most recently.
An olfactory habituation task was performed in a standard mouse housing box to assess nonassociative memory performance (Cleland et al. 2002). Presentation of an odorant elicits active investigation of the odor source; the extent of this investigation declines gradually over repeated presentations of the same odorant. Subsequent presentation of a different test odorant will elicit an increased investigative response depending upon the degree of similarity of the habituation and test odorants. In this study, acetic acid was used for three sequential habituation trials and the moderately similar odorant pentanoic acid for one subsequent test trial. Both odorants were diluted in mineral oil to theoretically emit vapor-phase partial pressures of 0.01 Pa, and were presented for 60 s per trial with 120 s intertrial intervals.
For light/dark inhibitory avoidance conditioning, mice were placed in the brightly-lit side of an automated, two-chamber light/dark shuttle cage (Coulbourn Instruments, Whitehall, PA, USA) to which they had been prehabituated on the previous day. After 10 seconds, the door between the two chambers was opened, and the latency for mice to enter the dark side was recorded. Once the mouse entered the dark side, the door was closed and a 2 s, 0.5 mA footshock was delivered. After an additional ten seconds, the mouse was returned to its home cage. Half of the mice were then re-tested 30 minutes later (short-term fear memory), while the other half were tested 24 hours later (long-term fear memory); the latency to enter the dark side of the shuttle cage was again recorded (up to a maximum of 180 s).
Finally, a joint cued/contextual learning paradigm was administered to mice that had not undergone inhibitory avoidance conditioning. Freezing behavior was assessed during each of four training/testing epochs, noted parenthetically below by name. To score freezing, mice were assessed every 5 seconds and scored as freezing or non-freezing; scoring was based on the average of these assessments. Mice were first placed into a square enclosure to which they had been previously habituated and given 120 s to explore (Pre). An 80 dB white noise auditory cue was then delivered for 30 s; during the last 2 s of cue presentation a 0.5 mA footshock was delivered. After an additional 60 s in the training enclosure, the mouse was removed to its home cage. Half of the mice began testing 30 minutes later (short-term memory) while the other half began testing 24 hours later (long-term memory). For testing at either latency, mice were again placed in the training enclosure for 120 s during which freezing was measured (Context), and then returned to the home cage. One hour later, mice were placed in an octagonal enclosure that was dissimilar from the training enclosure in floor texture, shape, wall design, lighting, and odor. After measuring freezing for 120 s (Switch), another 80 dB white noise cue was delivered for 30 s (with no shock), after which freezing was scored for an additional 120 s (Cue).
Statistical analysis
Neonatal behavior and most adult strength/motor and exploratory/sensory tests were analyzed with nonparametric tests (Mann-Whitney U for neonates, Kruskal-Wallis H for adults) because of the imposed maximum times or arbitrary scoring methods used. The normal approximation was used for the Mann-Whitney U-test (n > 20 in all cases), hence z-scores rather than the U statistic are reported. In nearly all cases (except where noted in Results), initial testing determined that sex was not a significant factor and the sexes were grouped together for analysis. The social recognition, spontaneous alternation, and inhibitory avoidance tests were analyzed by parametric ANOVA. Olfactory habituation and cued/contextual conditioning tests were analyzed using repeated-measures ANOVA (Wilks’ lambda criterion). Sex was included as a factor in parametric ANOVA designs but was never significant. Post hoc testing for ANOVA was performed using Tukey’s honestly significant difference (HSD) criterion. Body weight trajectories were analyzed using repeated-measures ANOVA. Expression assays were analyzed using Student’s t-test; the alpha criterion for the QPCR analyses was Bonferroni-corrected to account for multiple comparisons.
RESULTS
Rasgrf1 imprinting affects olfactory associative learning in neonates
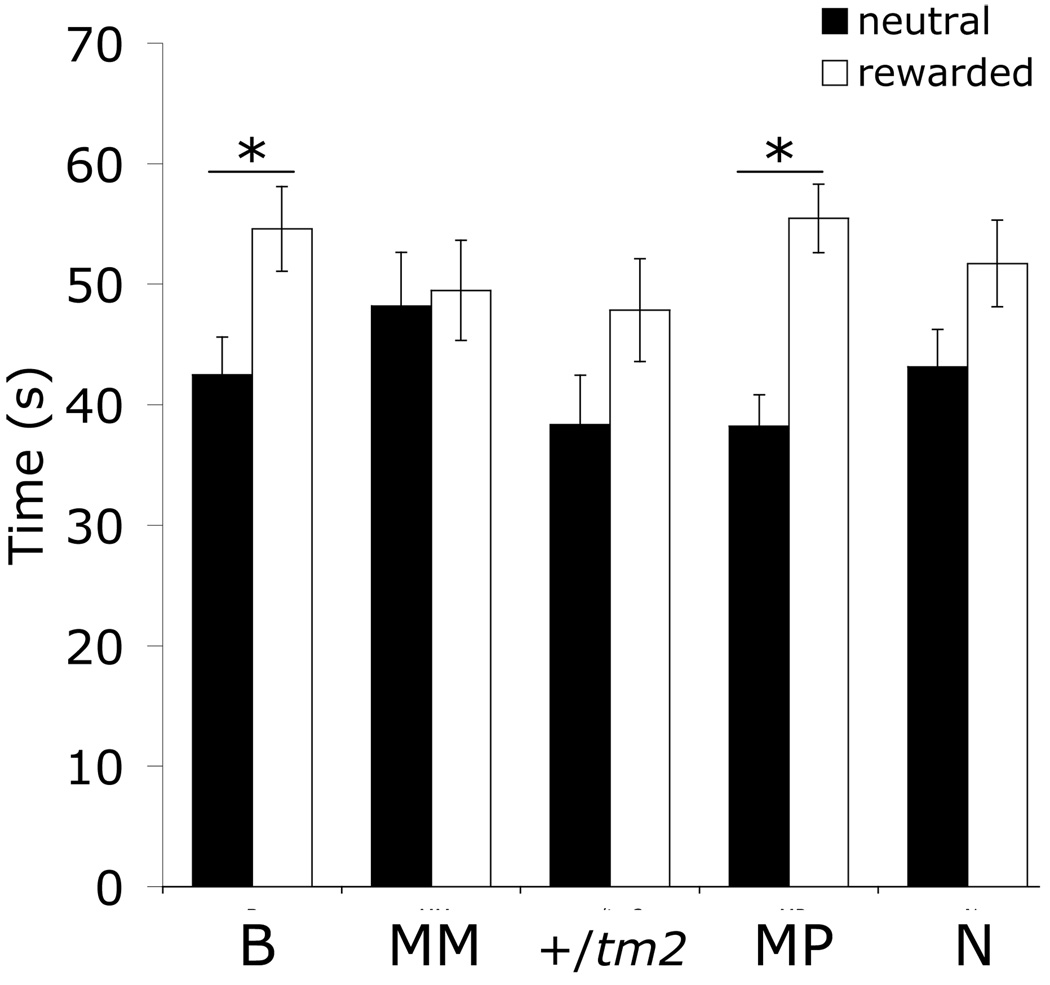
In wildtype mouse neonates, Rasgrf1 expression is monoallelic, expressed only from the paternally-inherited allele. In addition to wildtypes (MP), four mutant imprinting genotypes were generated that, as neonates, exhibited biallelic expression (B), were nominally null for Rasgrf1 (N), that expressed Rasgrf1 solely from the maternal allele (MM), or that monoallelically expressed the paternal allele under the control of the Pgk enhancer used to drive maternal allele expression in B and MM mutants (+/tm2) (Fig. S1). We asked whether these alterations in Rasgrf1 imprinting influenced learning by employing an olfactory associative learning paradigm suitable for use in neonatal mice (Alleva & Calamandrei 1986; Armstrong et al. 2006). Among the five Rasgrf1 imprinting genotypes studied, both genotypes with paternally-inherited wildtype alleles, the biallelics (B, z = 2.44, n = 59, p = 0.015) and the wildtypes (MP, z = 4.02, n = 68, p < 0.0001; Fig. 1), demonstrated a significant preference for the positively conditioned odor over the neutral odor. The other three genotypes (MM, +/tm2, N) did not demonstrate a significant preference for the rewarded odor (MM, z = 0.34, n = 31, p = 0.735; +/tm2, z = 1.35, n = 29, p = 0.176; N, z = 1.62, n = 46, p = 0.104), suggesting an impairment in olfactory associative learning associated with reduced or abnormal Rasgrf1 expression. Simple anosmia was ruled out as an alternative hypothesis because anosmic and hyposmic neonatal mice are suckling-impaired and often starve to death unaided (Turgeon & Meloche 2009); our neonates exhibited no such difficulties. However, hyposmia could be a contributing factor to the observed impairments, particularly given that reducing the perceived intensity of odorants reduces associative learning in adult mice (Cleland et al. 2009). The fact that the +/tm2 genotype did not fully recapitulate wildtype (MP) performance further suggested that paternally-inherited expression of the tm2 allele did not fully restore wildtype expression.
Figure 1. Associative olfactory learning in neonatal mice.
Neonatal mice were tested for their ability to learn and remember an introduced maternal odor. Mice were tested for place preference by measuring time spent over either the neutral or rewarded odor during a 120-second trial period (see Methods). B, biallelic, N=59; MM, monallelic maternal, N=31; +/tm2, N=29; MP, monoallelic paternal (wildtype), N=68; N, null mutant, N=44. Asterisks indicate p < 0.01. Error bars indicate SEM.
Quantification of Rasgrf1 transcript
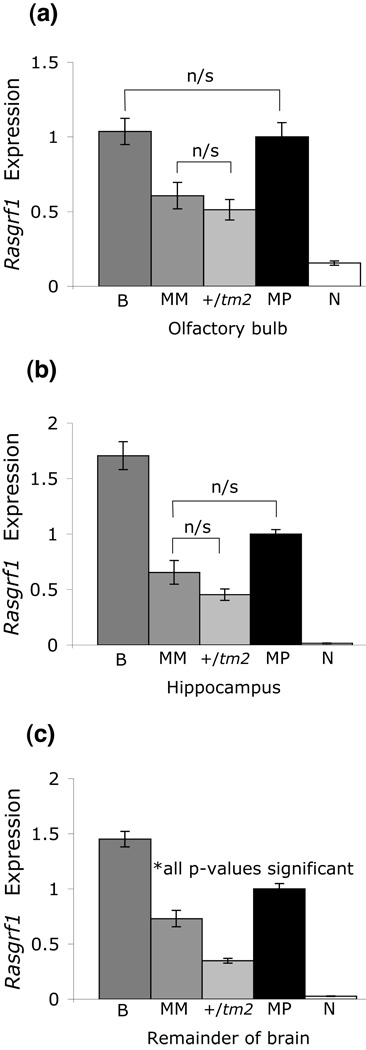
The tm2 allele essentially functioned as a null allele in the neonatal associative learning task, as neither the +/tm2 nor MM (tm2/tm1) genotypes differed from null mutants (+/tm1) in their performance and biallelic animals (tm2/+) did not differ from wildtypes. We therefore asked whether the level of tm2-derived Rasgrf1 expression differed from wildtype allele expression. Using quantitative PCR (Q-PCR), Rasgrf1 transcript levels were quantified for each of the five genotypes in three regions of the P8 neonatal brain: olfactory bulb, hippocampus, and the remainder of the brain.
The neural plasticity underlying odor preference learning in P8 neonatal rats is largely limited to the olfactory bulb (Moriceau & Sullivan 2004; Sullivan 2001; Sullivan & Leon 1987). Biallelic (B) and wildtype (MP) mice expressed significantly elevated Rasgrf1 transcript levels in olfactory bulb compared to the other three genotypes (Fig. 2a; p < 0.005 for all pairwise comparisons; see Table S1 for complete statistics), correlating with the significance of olfactory preference learning in the B and MP genotypes (Fig. 1). Transcript levels in B and MP mice were similar (Table S1; p > 0.05). Interestingly, Rasgrf1 expression levels in MM and +/tm2 animals – both of which have one tm2 allele and one inactive allele – were similar to each other (Table S1; p > 0.05) and intermediate between those of the B/MP animals and the nulls (Table S1; p < 0.005 in comparison to each), indicating that the tm2 allele successfully produces Rasgrf1 transcript in the olfactory bulb, but not to an extent comparable to wildtype expression. Moreover, uniquely among the brain structures tested, Rasgrf1 transcript was detected in the olfactory bulbs of null mice.
Figure 2. Rasgrf1 transcript quantification in P8 brain regions.
Rasgrf1 transcript from P8 neonates was quantified in (a) olfactory bulb, (b) hippocampus, and (c) the remainder of the brain (ROB; see Methods for details). Data were normalized to 18S rRNA levels and then further normalized to the Rasgrf1 expression level of the wildtype (MP) genotype. Except for those pairs depicted as nonsignificant (n/s), all pairwise comparisons were significantly different (p < 0.005, Bonferroni-corrected for ten multiple comparisons; Tables S1–S3). Note that higher Rasgrf1 expression levels tend to correspond to genotypes that are the best performers in the learned odor preference test, and that olfactory bulb expression patterns exhibit the best correlation with behavioral performance. Error bars indicate standard deviations.
The hippocampus is not thought to underlie learning in rodents until after weaning (Sullivan 2001). Interestingly, the effect of imprinting genotype on Rasgrf1 expression levels in hippocampus differed from the pattern observed in olfactory bulb, and consequently did not correlate well with neonatal behavioral data. Specifically, biallelic neonates expressed significantly more Rasgrf1 transcript than any other genotype (Fig. 2b; p < 0.005 for all pairwise comparisons; see Table S2 for complete statistics), and transcript levels between MP (wildtype) and MM neonates were similar (Table S2; p > 0.05). However, like the olfactory bulb pattern, the MM and +/tm2 genotypes expressed similar levels of Rasgrf1 (Table S2; p > 0.05), and null (N) animals expressed significantly lower Rasgrf1 transcript levels than did any other genotype (Table S2; p < 0.005 for all pairwise comparisons).
In the remainder of the brain, Rasgrf1 expression patterns were significantly different in all ten pairwise comparisons among genotypes (Fig. 2c; p < 0.005 for all pairwise comparisons; see Table S3 for complete statistics). Specifically, as observed in hippocampus, biallelic mice overexpressed, and null mice underexpressed, Rasgrf1 transcript relative to wildtype (MP; Table S3; p < 0.005 for both comparisons). Rasgrf1 expression levels in MM and +/tm2 animals were intermediate between the MP animals and nulls, and significantly different from both; moreover, Rasgrf1 expression was significantly greater in MM than in +/tm2 neonates (Table S3; p < 0.005 for all comparisons). Overall, these results support previous findings that early olfactory learning is dependent upon olfactory bulb.
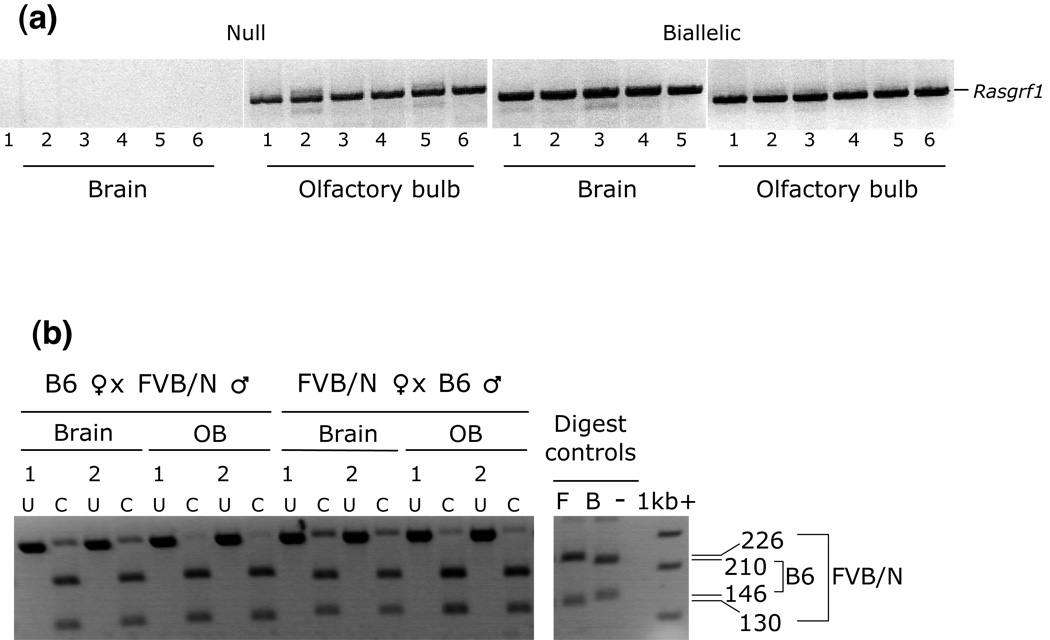
Rasgrf1 expression and imprinting in null olfactory bulb
Because Rasgrf1 expression has never before been detected in any brain structures in null mice, we performed two tests to verify the Rasgrf1 expression in the olfactory bulbs of null mice observed in our Q-PCR studies. First, gel analysis of the PCR products confirmed the presence of Rasgrf1 bands of identical size to those detected in mice carrying fully expressed alleles, indicating that the Q-PCR assays detected bona fide Rasgrf1 transcript. (Fig. 3a). In a second test, we asked whether the Rasgrf1 transcript detected in olfactory bulb was imprinted, or if imprinting mechanisms in this structure were behaving differently than in the remainder of the brain (see Methods). Reciprocal crosses between wildtype C57BL/6J and FVB/NJ animals were set up. Olfactory bulbs from P8 pups derived from these crosses were isolated, cDNAs were generated, and Rasgrf1 transcript was assayed for parent-specific expression. The assay relied on an AciI restriction site that produces a unique digestion pattern for each of the two strains. Progeny of C57BL/6J mothers and FVB/NJ fathers displayed 226/130 bp bands diagnostic of expression from the paternal FVB/NJ allele. From the reciprocal cross, progeny of FVB/NJ mothers and C57BL/6J fathers displayed the 210/146 bands diagnostic of expression from the paternal C57BL/6J allele (Fig. 3b). The restriction patterns verify amplification of Rasgrf1 and demonstrate that expression in olfactory bulb is imprinted, with expression coming from the paternal allele, as is the case in the rest of the brain. Because the above crosses demonstrate that the maternal Rasgrf1 allele is silent in olfactory bulb, it is likely that a level of expression from the paternal tm1 allele is permitted in olfactory bulb and that olfactory bulb regulates Rasgrf1 differently than other tissues.
Figure 3. Rasgrf1 is expressed and imprinted in neonatal null olfactory bulb.
(a) Rasgrf1 transcript was amplified by RT-PCR in the olfactory bulbs and the remainder of the brain in null and biallelic animals using RT-PCR. Six mice were used for each assay. (b) Imprinted expression was assayed by RT-PCR followed by AciI digestion of the PCR products generated from tissues of progeny from reciprocal crosses between C57BL/6J (B6) and FVB/NJ (FVB) parents. Maternal strain is shown first. FVB expression generates 226bp and 130bp bands; B6 expression generates 210bp and 146bp bands. Digestion of amplicons (lanes labeled “C”) produced exclusively paternal banding patterns. Two to four mice were used for each test (lane pairs labeled “1” and “2” contain results from two different mice for each tissue type from each cross). C: cut/digested; U: undigested PCR products. Digest control lanes show transcript from each of the parent strains (F: FVB; B: B6).
Rasgrf1 mutation dysregulates Ras and Rac activation in neonates
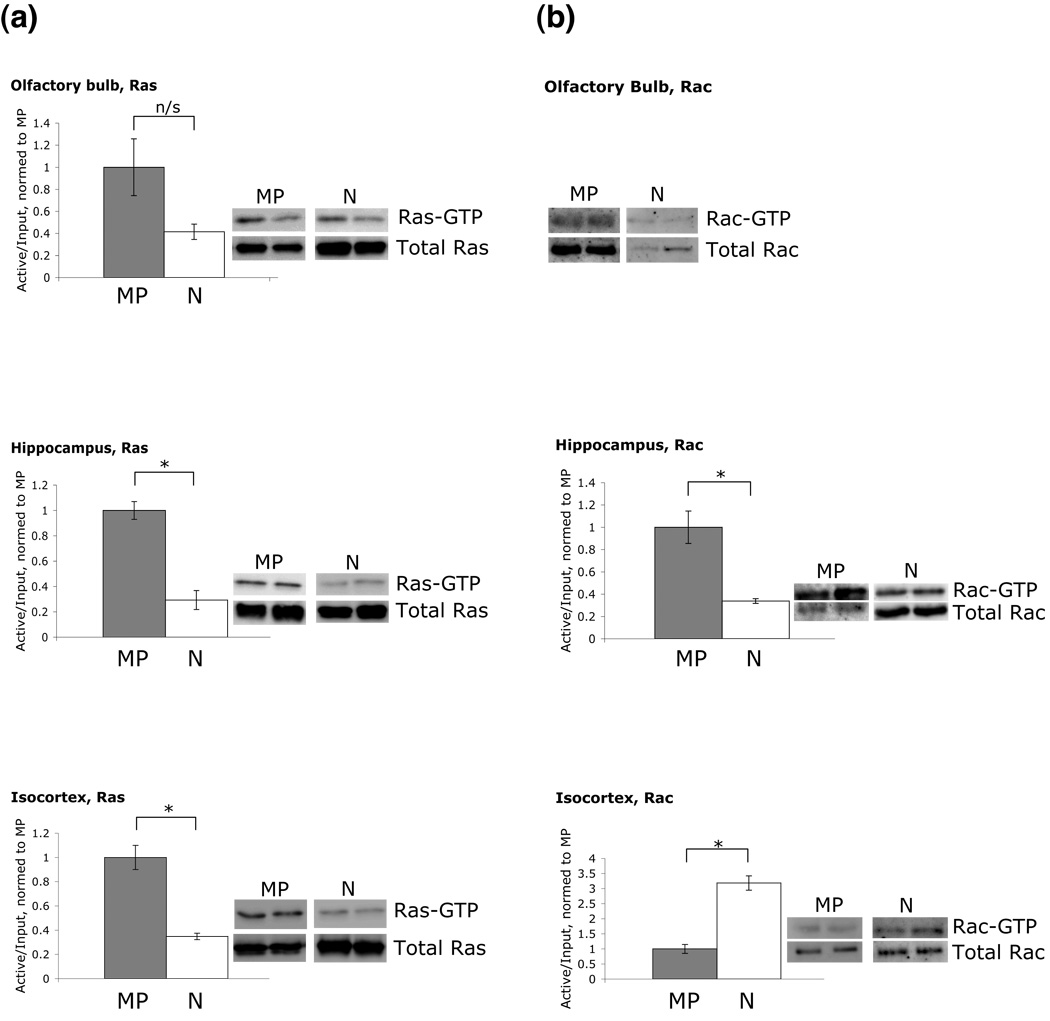
RasGRF1 acts as a guanine-nucleotide exchange factor for both Ras and Rac proteins. Although prior work has indicated that RasGRF1 is not an active signaling intermediate in neonatal mouse cortical neurons (Tian et al. 2004), the differences in Rasgrf1 expression that we observed among neonatal genotypes and brain structures, along with the differential effects of genotype on neonatal associative learning, suggest that this finding may not be general and that impaired expression of Rasgrf1 could in fact affect Ras or Rac activation in neonates. To determine whether Ras or Rac activation was influenced by mutations at Rasgrf1, we quantified the levels of the active forms of both proteins in tissue samples from the olfactory bulb, hippocampus, and isocortex of P8 wildtype (MP) and null mutant (N) neonates.
Western blot analyses indicated significant differences between wildtype and null animals in the proportion of Ras and Rac protein that was activated (Fig. 4a,b). Among the three brain structures, extracts from olfactory bulb contained the lowest levels of activated Ras and Rac proteins, such that longer exposures were needed for their detection than that required for the other tissues. There was no significant difference in the amount of activated Ras protein between genotypes in olfactory bulb extracts (t(6) = 1.09, p = 0.315). Rac levels in olfactory bulb of null mice were too low to reliably measure the level of activation, so no ratio was calculated. In hippocampus, in contrast, extracts from null mice contained significantly lower proportions of activated Ras and Rac proteins than did wildtype animals (Ras: t(6) = 3.44, p = 0.014; Rac: t(6) = 2.96, p = 0.0251). Finally, in isocortex, null animals produced a significantly lower proportion of activated Ras (t(6) = 3.18, p = 0.0191) but a significantly higher proportion of activated Rac (t(6) = 3.89, p = 0.0081) than did wildtypes.
Figure 4. Relative expression levels of activated Ras and Rac proteins in null and wildtype brains regions depend on brain region.
Levels of active and inactive Ras protein (a) and Rac protein (b) in the olfactory bulb, hippocampus, and isocortex of wildtype (MP) and null mutant (N) neonates at P8. Levels of Rac in olfactory bulb were too low to quantify. The amounts of precipitated (active) protein were normalized to the amounts of input (total) protein for each structure, and then normalized to wildtype levels. Representative blots are shown next to each graph. Three to four animals of each genotype were used in each test. Error bars indicate SEM.
Together, these data indicate that Rasgrf1 is expressed and functional in the neonatal brain, that it affects Ras and Rac activation in neonatal mice, and that there are multiple, region-specific differences in the activity of Rasgrf1 signaling proteins. These effects of Rasgrf1, presumably in olfactory bulb, include the regulation of normal olfactory associative learning in neonates.
Body mass assays
The effects of Rasgrf1 imprinting on body mass persist into adulthood, well after expression becomes biallelic at approximately P21 (Drake et al. 2009). Body mass was measured in wildtype and +/tm2 mice from P8 through P63 for comparison with these published data, which do not include the +/tm2 genotype. No significant differences in body mass were observed between wildtype and +/tm2 males or females at any age measured, indicating that, similar to maternally-derived tm2 expression, paternally-derived tm2 expression is sufficient to produce a wildtype size phenotype (Fig. S2a,b). In contrast, mice that were null for Rasgrf1 as neonates (+/tm1) remained significantly smaller than wildtypes even into adulthood (measured up to P63), whereas imprinted biallelics (tm2/+) were significantly heavier (Drake et al. 2009).
Adult behavioral assays
Because the effects of Rasgrf1 imprinting on body mass persist into adulthood, (Drake et al. 2009), we asked whether the same was true of its effect on associative learning. Accordingly, we performed a battery of standard behavioral phenotyping tests on cohorts of adult mice drawn from the B, MM, MP (wildtype), and N genotypes. As mice of all genotypes exhibit biallelic expression of Rasgrf1 as adults; the sole difference among the adult cohorts tested was a history of differential Rasgrf1 expression profiles as neonates.
Strength and motor coordination
Adult mice first were tested on a series of strength and motor coordination tasks. First, escape latency from 11 mm and 5 mm balance beams was tested over three sequential trials per beam. Because there was a significant effect of sex on latency to escape for both beam diameters (Kruskal-Wallis test; 11 mm, H(1) = 4.510, p = 0.034; 5 mm, H(1) = 13.551, p < 0.001), the sexes were analyzed separately for this task. There were no significant differences among the four genotypes on escape latency from either diameter of beam for either sex (11 mm, females, H(3) = 4.650, p = 0.199; males, H(3) = 3.587, p = 0.310; 5 mm, females, H(3) = 4.888, p = 0.180; males, H(3) = 4.157, p = 0.245) (Fig. S3a). In other tests, there was no significant effect of sex (forelimb suspension latency H(1) = 0.136, p = 0.713, hand-scoring H(1) = 0.007, p = 0.935; vertical pole H(1) = 0.048, p = 0.826; hanging-wire grip test H(1) = 0.384, p = 0.535), so the sexes were combined for analysis. There was no effect of genotype on the latency to fall from a single wire forelimb suspension (H(3) = 5.287, p = 0.152), a vertical pole (H(3) = 0.403, p = 0.940), or a hanging-wire grip test using a modified cage lid (H(3) = 2.929, p = 0.403). Hand-scoring of the single-wire forelimb suspension test (see Methods) also revealed no significant differences among genotypes (H(3) = 2.819, p = 0.420) (Fig. S3b).
Exploratory behaviors
Mice were then tested to assess their relative mobility and exploratory tendencies. In a small open field test (18" square, 4×4 grid, 300 s trial period), there was no significant effect of genotype on either the number of line crossings (mobility; H(3) = 0.723, p = 0.868) or the proportion of time spent in the 12 edge squares of the open field (thigmotaxis; H(3) = 3.151, p = 0.369) (Fig. S3c). In the step-down visual cliff apparatus, nearly all (87%) of the mice across both sexes and all genotypes stepped down onto the opaque side. There was no effect of genotype on the latency to step down, irrespective of whether analysis included both sides (H(3) = 1.309, p = 0.727) or only the opaque side (H(3) = 1.669, p = 0.644) (Fig. S3d). The sexes were pooled for analysis in these exploratory tests because pretests revealed no significant effects attributable to sex (open field mobility, H(1) = 3.408, p = 0.065; thigmotaxis, H(1) = 0.242, p = 0.622; step-down latency, H(1) = 1.406, p = 0.236, opaque side only, H(1) = 0.721, p = 0.396). In the social recognition task, the time spent investigating a newly introduced mouse was significantly greater than that spent investigating a cagemate, irrespective of sex or genotype (three-factor ANOVA; main effect of intruder familiarity, F(1,84) = 25.864, p < 0.001; for main effects of sex and genotype and all interactions, p > 0.05) (Fig. S3e). A discrimination index also was calculated for each subject as the difference between the two investigation times divided by their sum, such that an index of 0 indicated no distinction between cagemates and newly introduced mice, −1 indicated investigation only of the cagemate, and +1 indicated investigation only of the newly introduced mouse. Neither genotype nor sex, nor their interaction, significantly affected the discrimination index (two-factor ANOVA; main effect of genotype, F(3,42) = 2.428, p = 0.079; main effect of sex, F(1,42) = 0.006, p = 0.938; interaction, F(3,42) = 0.988, p = 0.408).
Learning and memory
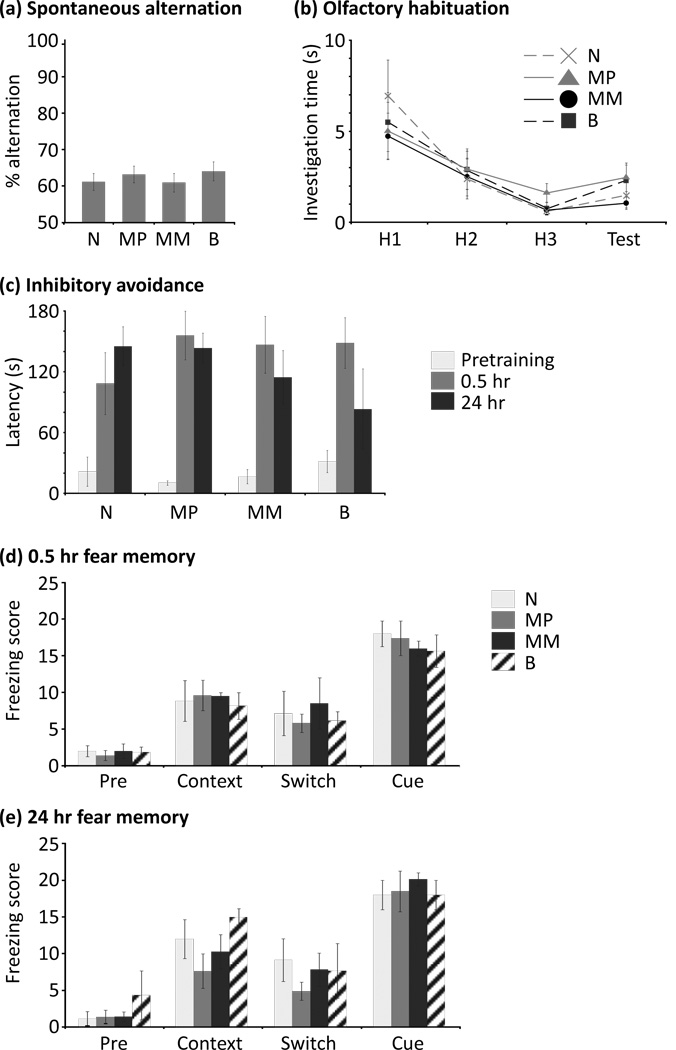
There was no effect of genotype on the total number of arm entries (H(3) = 2.493, p = 0.477) or the proportion of alternations (H(3) = 1.214, p = 0.750) in a five-minute spontaneous alternation task (Fig. 5a). The sexes were pooled for analysis in this test because pretests revealed no significant effects attributable to sex (number of entries, H(1) = 0.306, p = 0.580; proportion of alternations, H(1) = 1.050, p = 0.305). Olfactory habituation significantly affected investigation times in all animals (Wilks’ lambda; F(3,41) = 9.072, p < 0.001), but was not affected by genotype, sex, or their interaction (Wilks’ lambda, interaction of habituation×genotype; F(9,99.9) = 0.383, p = 0.941; habituation × sex; F(3,41) = 0.684, p = 0.567; habituation × genotype × sex; F(9,99.9) = 1.054, p = 0.403) (Fig. 5b). That is, neither habituation to the first odor nor the degree of cross-habituation to the second, similar odor was significantly affected by genotype or sex.
Figure 5. Learning and memory performance in adult mice.
Adult mice all express Rasgrf1 biallelically; genotype designations apply to each animal’s history of neonatally imprinted expression. (a) Assessment of working memory in the spontaneous alternation task. The proportion of alternations was not affected by genotype. (b) Assessment of nonassociative olfactory learning by habituation to odors. Neither habituation nor cross-habituation to a moderately different test odorant were affected by genotype. (c) Light-dark inhibitory avoidance. All genotypes learned to associate dark entry with shock in one trial; learning and memory performance did not depend on genotype at either 0.5 hours (short-term memory) or 24 hours (long-term memory). (d) Short-term memory assessed by one-trial cued/contextual fear conditioning. There were no effects of genotype on the levels of freezing behavior scored. Pre, freezing in the training cage prior to tone-shock pairing; Context, freezing after being replaced in the training cage 0.5 hr after tone-shock pairing (contextual response); Switch, freezing after being placed in a novel test cage (60 min later; baseline for cued response); Cue, freezing in the novel test cage after presentation of tone CS (cued response). (e) Long-term memory assessed by one-trial cued/contextual fear conditioning. There were no effects of genotype on the levels of freezing behavior scored. Testing was performed identically to the short-term memory task. Separate cohorts were tested at 0.5 hr and 24 hr latencies. Error bars indicate SEM.
In a light-dark inhibitory avoidance task, all animals exhibited strong one-trial fear learning (three-factor ANOVA; main effect of latency, F(2,60) = 39.076, p < 0.001) with latency to enter the dark side differing significantly between pretraining and both posttraining latencies (Tukey’s HSD; p < 0.001 for both comparisons) but not between the 0.05 and 24 hour posttraining latencies (p = 0.478). Genotype was not a significant main effect (F(3,60) = 0.298, p = 0.826); it did not affect the latency to enter the dark compartment either before shock, 30 minutes after delivery of a shock in the dark compartment (short-term memory), or 24 hours after shock delivery (consolidated long-term memory (simple effects analysis; p > 0.05 in all cases; Fig. 5c). Sex also was not a significant main effect (F(1,60) = 0.015, p = 0.904), nor were any interactions significant (p > 0.05 in all cases).
Finally, we ran an auditory cued/contextual conditioning task, in which an 80 dB white noise cue was paired with shock in a single conditioning trial, and freezing behavior was measured before training (Pre), when animals were placed back in the original context (contextual conditioning; Context), when animals were subsequently placed into a novel context (prior to testing cued conditioning; Switch) and finally in response to the auditory cue in this novel context (cued conditioning; Cue). There were no significant effects of genotype on freezing behavior either when tested 0.5 hours later (short-term fear memory; Wilks’ lambda; F(9,29.4) = 0.894; p = 0.542) or 24 hours later (long-term fear memory; F(9,39.1) = 0.543; p = 0.834) (Fig. 5d,e). Sex also was not a significant main effect at either latency (0.5 hr, F(3,12) = 2.742, p = 0.089; 24 hr, F(3,16) = 0.788, p = 0.518), nor were any interactions significant (p > 0.05 in all cases).
DISCUSSION
Abnormal imprinted expression of Rasgrf1 produces learning deficits in neonates
Several learning and neuronal phenotypes have been ascribed to G-protein signaling effects influenced by RasGRF1 (Brambilla et al. 1997; Farnsworth et al. 1995; Fasano et al. 2009; Giese et al. 2001; Kesavapany et al. 2006; Krapivinsky et al. 2003; Tonini et al. 2001; Yang & Mattingly 2006). We here demonstrate that neonatal learning and memory are dependent upon Rasgrf1 expression levels in neonatal brain. Impaired Rasgrf1 imprinting produced learning deficits in neonates, though this effect did not persist into adulthood, when Rasgrf1 expression becomes uniformly biallelic. This contrasts with the body mass growth phenotypes associated with the manipulation of neonatal Rasgrf1 expression, which persist beyond the age when expression becomes biallelic (Drake et al. 2009). Interestingly, in olfactory bulb, the location of neonatal olfactory preference learning (Sullivan 2001), the biallelic Rasgrf1 genotype did not yield increased gene expression over wildtype (Fig. 2a), in contrast to its effect in other brain regions (Fig. 2b,c). That is, Rasgrf1 transcript levels in olfactory bulb correlated directly with behavioral performance on the olfactory associative learning task, whereas other, non-olfactory phenotypes presumably depend on potentially dissimilar Rasgrf1 expression patterns in other tissues. These results demonstrate tissue-specific regulation underlying a multiplicity of functions for Rasgrf1.
The tm2 allele essentially behaved like a null allele in terms of its contribution to neonatal learning and memory performance, irrespective of whether it was maternally or paternally inherited; neither MM, +/tm2, or N animals demonstrated a learned preference for the rewarded odorant. Although these impairments were expected in the null animals based on previous assessments of Rasgrf1 −/− knockout mice (Brambilla et al. 1997; Giese et al. 2001), it was a surprising result in the MM and +/tm2 genotypes, which are phenotypically indistinguishable from MP animals in terms of body size and growth (Drake et al. 2009) (Fig. S2). No significant differences in weight were observed between wildtype and +/tm2 males or females, indicating that, similar to maternally-derived tm2 expression, paternally-derived tm2 expression is sufficient to produce a wildtype size phenotype, but insufficient to restore wildtype learning and memory performance, as assessed by the olfactory odor learning paradigm. However, the persistence of Rasgrf1 expression observed in the olfactory bulbs of null mutant animals may mitigate their olfactory learning deficit and conceal a greater dependence of olfactory learning on Rasgrf1 than was observed; notably, while MM and +/tm2 mice expressed significantly more transcript than N mice, even in olfactory bulb, the nonzero expression level in N mice implies that we were unable to observe a neonatal olfactory learning phenotype for a mouse with zero Rasgrf1 expression.
Rasgrf1-related learning and memory phenotypes in adult mice
Manipulation of Rasgrf1 imprinting during the neonatal period had no persistent effect on learning and memory performance in adult mice. In contrast, adult Rasgrf1 knockout animals exhibit clear learning and memory deficits, although the nature of these deficits is uncertain. Brambilla et al. (1997) found that mice lacking RasGRF1 were impaired in amygdala-dependent memory consolidation; retention of fear conditioning in inhibitory avoidance, cued conditioning, and contextual conditioning tests was normal when tested 0.5 hours after conditioning but severely impaired after 24 hours. In contrast, their performance on two hippocampus-dependent tasks (hidden-platform water maze, eight-arm radial maze) was unaffected across multiple days. Substantially different results were obtained by Giese et al. (2001), who found that RasGRF1-deficient mice were impaired on multiple hippocampus-dependent learning tasks (contextual discrimination, water maze with hidden platform, and social transmission of food preference) but not on inhibitory avoidance or contextual conditioning. All testing was performed at 21–24 hour post-conditioning latencies, except that the social transmission of food preference was also tested immediately after training, and RasGRF1-deficient mice were impaired at that latency as well. Finally, Fasano et al. (2009) replicated two of the results from Brambilla et al. (1997), showing that mice lacking RasGRF1 had impaired memory for inhibitory avoidance conditioning after 24 hours but not after 1 hour, and that they performed normally when tested in the water maze. The differences between these groups’ findings presumably result from the different derivations of the mutant strains used, illustrating that genetic manipulations are not necessarily uniform in their phenotypic effects.
Possible molecular mechanisms underlying Rasgrf1-dependent olfactory learning
Early olfactory associative learning involves NMDA-receptor dependent processes (Lincoln et al. 1988; Weldon et al. 1997), and is mediated by CREB phosphorylation (Cui et al. 2007; McLean et al. 1999; Raineki et al. 2009; Yuan et al. 2003). In adults, NMDA receptors mediate olfactory learning at multiple levels in diverse species, including Drosophila (Hudson & Distel 1986), honeybees (Si et al. 2004), rats (Tronel & Sara 2003), and mice (Brennan 1994). RasGRF1 is known to directly associate with NMDA receptors via the NR2B subunit (Krapivinsky et al. 2003), which is striking given that certain olfactory memory paradigms are specifically NR2B-dependent (White & Youngentob 2004).
In the olfactory bulb, noradrenergic inputs from the locus coeruleus play a role in olfactory learning in neonatal rodent pups (Christie-Fougere et al. 2009; Moriceau & Sullivan 2004); specifically, activation of noradrenergic inputs to neonatal olfactory bulb during the presentation of an odor stimulus increases cAMP levels in mitral cells. Higher cAMP levels increase CREB phosphorylation, which mediates the formation of long-term odor preference memory (Cui et al. 2007; McLean et al. 1999). Rasgrf1 has been shown to transduce signals arriving at the NMDA receptor and to activate the ERK/MAPK pathway in response (Krapivinsky et al. 2003; Tian et al. 2004), which lies upstream of CREB phosphorylation. While we did not identify a direct link between Rasgrf1 and activation of the Ras or Rac pathways in olfactory bulb, we demonstrated that there are differences in the amounts of activated protein between wildtype and null mutant mice in other structures.
Our results also indicated that Rasgrf1 expression in olfactory bulb may be subject to different epigenetic regulatory mechanisms than in other tissues. The paternally inherited null tm1 allele reduced Rasgrf1 expression to 1–2% of wild type levels in hippocampus and whole brain; expression in olfactory bulb was also reduced, but only to 20% of wildtype levels in P8 mice. Normal Rasgrf1 expression in olfactory bulb at this age is exclusively paternally-derived, which indicates that the tm1 repeat deletion is either failing to produce relevant hypomethylation, or that the transcription regulatory mechanisms controlling imprinted expression in the olfactory bulb differ from those used in other tissues, where imprinting also occurs.
Genomic Imprinting and the Conflict Hypothesis
It has been established that Rasgrf1 expression influences postnatal growth, as Rasgrf1 knock-out mice are smaller than their wild-type littermates (Clapcott et al. 2003; Itier et al. 1998). Moreover, proper imprinting in neonates is critical for maintaining normal growth into adulthood (Drake et al. 2009), an experiment made possible by the tm1 and tm2 alleles. Here, we demonstrated that growth phenotypes are not influenced by the parental allele per se from which Rasgrf1 expression is derived, suggesting instead that growth is influenced by the overall level of Rasgrf1 expression in the governing tissue. This result is consistent with the central but untested assumption of the “conflict hypothesis” describing the evolution of genomic imprinting (Moore & Haig 1991; Wilkins & Haig 2003). This hypothesis describes the evolution of genomic imprinting in mammals as a battle between the two parental genomes over optimum expression levels at imprinted loci, particularly those governing growth and resource consumption (Moore & Haig 1991). Previously untested was the assumption that equivalent amounts of expression derived from either parental allele would produce an equivalent phenotype, as we observed via the size phenotype at the ages assayed (Drake et al. 2009), (Fig. S2). However, the different patterns of imprinted Rasgrf1 regulation observed in different tissues – even among three different regions of telencephalic cortex – indicate that such questions cannot be definitively answered without identifying the tissue or tissues within which Rasgrf1 levels govern somatic growth, and suggest a yet-unappreciated complexity in the selective mechanisms underlying genomic imprinting.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the technical assistance of Jim Putnam for maintenance of the breeding colony. We also thank Karim Boudadi for assistance with behavioral training and Yoon Jung Park for helpful suggestions. This research was supported by NIH grant CA098597 to PDS and by the Cornell Center for Vertebrate Genomics. NMD was supported by NIH Training Grant T32DK007158.
REFERENCES
- Alleva E, Calamandrei G. Odor-aversion learning and retention span in neonatal mouse pups. Behav Neural Biol. 1986;46:348–357. doi: 10.1016/s0163-1047(86)90317-1. [DOI] [PubMed] [Google Scholar]
- Armstrong CM, DeVito LM, Cleland TA. One-trial associative odor learning in neonatal mice. Chem Senses. 2006;31:343–349. doi: 10.1093/chemse/bjj038. [DOI] [PubMed] [Google Scholar]
- Brambilla R, Gnesutta N, Minichiello L, White G, Roylance AJ, Herron CE, Ramsey M, Wolfer DP, Cestari V, Rossi-Arnaud C, Grant SG, Chapman PF, Lipp HP, Sturani E, Klein R. A role for the Ras signalling pathway in synaptic transmission and long-term memory. Nature. 1997;390:281–286. doi: 10.1038/36849. [DOI] [PubMed] [Google Scholar]
- Brennan PA. The effects of local inhibition of N-methyl-D-aspartate and AMPA/kainate receptors in the accessory olfactory bulb on the formation of an olfactory memory in mice. Neuroscience. 1994;60:701–708. doi: 10.1016/0306-4522(94)90498-7. [DOI] [PubMed] [Google Scholar]
- Cen H, Papageorge AG, Vass WC, Zhang KE, Lowy DR. Regulated and constitutive activity by CDC25Mm (GRF), a Ras-specific exchange factor. Mol Cell Biol. 1993;13:7718–7724. doi: 10.1128/mcb.13.12.7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie-Fougere MM, Darby-King A, Harley CW, McLean JH. Calcineurin inhibition eliminates the normal inverted U curve, enhances acquisition and prolongs memory in a mammalian 3'-5'-cyclic AMP-dependent learning paradigm. Neuroscience. 2009;158:1277–1283. doi: 10.1016/j.neuroscience.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Clapcott SJ, Peters J, Orban PC, Brambilla R, Graham CF. Two ENU-induced mutations in Rasgrf1 and early mouse growth retardation. Mamm Genome. 2003;14:495–505. doi: 10.1007/s00335-002-2258-4. [DOI] [PubMed] [Google Scholar]
- Cleland TA, Morse A, Yue EL, Linster C. Behavioral models of odor similarity. Behav Neurosci. 2002;116:222–231. doi: 10.1037//0735-7044.116.2.222. [DOI] [PubMed] [Google Scholar]
- Cleland TA, Narla VA, Boudadi K. Multiple learning parameters differentially regulate olfactory generalization. Behav Neurosci. 2009;123:26–35. doi: 10.1037/a0013991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley JN. What's wrong with my mouse: behavioral phenotyping of transgenic and knockout mice. 2nd ed. Hoboken, NJ: Wiley; 2007. [Google Scholar]
- Cui W, Smith A, Darby-King A, Harley CW, McLean JH. A temporal-specific and transient cAMP increase characterizes odorant classical conditioning. Learn Mem. 2007;14:126–133. doi: 10.1101/lm.496007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake NM, Park YJ, Shirali AS, Cleland TA, Soloway PD. Imprint switch mutations at Rasgrf1 support conflict hypothesis of imprinting and define a growth control mechanism upstream of IGF1. Mamm Genome. 2009;20:654–663. doi: 10.1007/s00335-009-9192-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnsworth CL, Freshney NW, Rosen LB, Ghosh A, Greenberg ME, Feig LA. Calcium activation of Ras mediated by neuronal exchange factor Ras-GRF. Nature. 1995;376:524–527. doi: 10.1038/376524a0. [DOI] [PubMed] [Google Scholar]
- Fasano S, D'Antoni A, Orban PC, Valjent E, Putignano E, Vara H, Pizzorusso T, Giustetto M, Yoon B, Soloway P, Maldonado R, Caboche J, Brambilla R. Ras-guanine nucleotide-releasing factor 1 (Ras-GRF1) controls activation of extracellular signal-regulated kinase (ERK) signaling in the striatum and long-term behavioral responses to cocaine. Biol Psychiatry. 2009;66:758–768. doi: 10.1016/j.biopsych.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Font de Mora J, Esteban LM, Burks DJ, Nunez A, Garces C, Garcia-Barrado MJ, Iglesias-Osma MC, Moratinos J, Ward JM, Santos E. Ras-GRF1 signaling is required for normal beta-cell development and glucose homeostasis. Embo J. 2003;22:3039–3049. doi: 10.1093/emboj/cdg280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese KP, Friedman E, Telliez JB, Fedorov NB, Wines M, Feig LA, Silva AJ. Hippocampus-dependent learning and memory is impaired in mice lacking the Ras-guanine-nucleotide releasing factor 1 (Ras-GRF1) Neuropharmacology. 2001;41:791–800. doi: 10.1016/s0028-3908(01)00096-x. [DOI] [PubMed] [Google Scholar]
- Hudson R, Distel H. Pheromonal release of suckling in rabbits does not depend on the vomeronasal organ. Physiol Behav. 1986;37:123–128. doi: 10.1016/0031-9384(86)90394-x. [DOI] [PubMed] [Google Scholar]
- Innocenti M, Zippel R, Brambilla R, Sturani E. CDC25(Mm)/Ras-GRF1 regulates both Ras and Rac signaling pathways. FEBS Lett. 1999;460:357–362. doi: 10.1016/s0014-5793(99)01374-5. [DOI] [PubMed] [Google Scholar]
- Itier JM, Tremp GL, Leonard JF, Multon MC, Ret G, Schweighoffer F, Tocque B, Bluet-Pajot MT, Cormier V, Dautry F. Imprinted gene in postnatal growth role. Nature. 1998;393:125–126. doi: 10.1038/30120. [DOI] [PubMed] [Google Scholar]
- Kesavapany S, Pareek TK, Zheng YL, Amin N, Gutkind JS, Ma W, Kulkarni AB, Grant P, Pant HC. Neuronal nuclear organization is controlled by cyclin-dependent kinase 5 phosphorylation of Ras Guanine nucleotide releasing factor-1. Neurosignals. 2006;15:157–173. doi: 10.1159/000095130. [DOI] [PubMed] [Google Scholar]
- King DL, Arendash GW. Behavioral characterization of the Tg2576 transgenic model of Alzheimer's disease through 19 months. Physiol Behav. 2002;75:627–642. doi: 10.1016/s0031-9384(02)00639-x. [DOI] [PubMed] [Google Scholar]
- Kiyono M, Kaziro Y, Satoh T. Induction of rac-guanine nucleotide exchange activity of Ras-GRF1/CDC25(Mm) following phosphorylation by the nonreceptor tyrosine kinase Src. J Biol Chem. 2000;275:5441–5446. doi: 10.1074/jbc.275.8.5441. [DOI] [PubMed] [Google Scholar]
- Krapivinsky G, Krapivinsky L, Manasian Y, Ivanov A, Tyzio R, Pellegrino C, Ben-Ari Y, Clapham DE, Medina I. The NMDA receptor is coupled to the ERK pathway by a direct interaction between NR2B and RasGRF1. Neuron. 2003;40:775–784. doi: 10.1016/s0896-6273(03)00645-7. [DOI] [PubMed] [Google Scholar]
- Lincoln J, Coopersmith R, Harris EW, Cotman CW, Leon M. NMDA receptor activation and early olfactory learning. Brain Res. 1988;467:309–312. doi: 10.1016/0165-3806(88)90036-3. [DOI] [PubMed] [Google Scholar]
- MacDonald JI, Verdi JM, Meakin SO. Activity-dependent interaction of the intracellular domain of rat trkA with intermediate filament proteins, the beta-6 proteasomal subunit, Ras-GRF1, and the p162 subunit of eIF3. J Mol Neurosci. 1999;13:141–158. doi: 10.1385/JMN:13:1-2:141. [DOI] [PubMed] [Google Scholar]
- Mattingly RR, Macara IG. Phosphorylation-dependent activation of the Ras-GRF/CDC25Mm exchange factor by muscarinic receptors and G-protein beta gamma subunits. Nature. 1996;382:268–272. doi: 10.1038/382268a0. [DOI] [PubMed] [Google Scholar]
- Mattingly RR, Saini V, Macara IG. Activation of the Ras-GRF/CDC25Mm exchange factor by lysophosphatidic acid. Cell Signal. 1999;11:603–610. doi: 10.1016/s0898-6568(99)00034-0. [DOI] [PubMed] [Google Scholar]
- McLean JH, Harley CW, Darby-King A, Yuan Q. pCREB in the neonate rat olfactory bulb is selectively and transiently increased by odor preference-conditioned training. Learn Mem. 1999;6:608–618. doi: 10.1101/lm.6.6.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore T, Haig D. Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet. 1991;7:45–49. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- Moriceau S, Sullivan RM. Unique neural circuitry for neonatal olfactory learning. J Neurosci. 2004;24:1182–1189. doi: 10.1523/JNEUROSCI.4578-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plass C, Shibata H, Kalcheva I, Mullins L, Kotelevtseva N, Mullins J, Kato R, Sasaki H, Hirotsune S, Okazaki Y, Held WA, Hayashizaki Y, Chapman VM. Identification of Grf1 on mouse chromosome 9 as an imprinted gene by RLGS-M. Nat Genet. 1996;14:106–109. doi: 10.1038/ng0996-106. [DOI] [PubMed] [Google Scholar]
- Raineki C, De Souza MA, Szawka RE, Lutz ML, De Vasconcellos LF, Sanvitto GL, Izquierdo I, Bevilaqua LR, Cammarota M, Lucion AB. Neonatal handling and the maternal odor preference in rat pups: involvement of monoamines and cyclic AMP response element-binding protein pathway in the olfactory bulb. Neuroscience. 2009;159:31–38. doi: 10.1016/j.neuroscience.2008.12.012. [DOI] [PubMed] [Google Scholar]
- Robinson KN, Manto K, Buchsbaum RJ, MacDonald JI, Meakin SO. Neurotrophin-dependent tyrosine phosphorylation of Ras guanine-releasing factor 1 and associated neurite outgrowth is dependent on the HIKE domain of TrkA. J Biol Chem. 2005;280:225–235. doi: 10.1074/jbc.M410454200. [DOI] [PubMed] [Google Scholar]
- Shou C, Wurmser A, Suen KL, Barbacid M, Feig LA, Ling K. Differential response of the Ras exchange factor, Ras-GRF to tyrosine kinase and G protein mediated signals. Oncogene. 1995;10:1887–1893. [PubMed] [Google Scholar]
- Si A, Helliwell P, Maleszka R. Effects of NMDA receptor antagonists on olfactory learning and memory in the honeybee (Apis mellifera) Pharmacol Biochem Behav. 2004;77:191–197. doi: 10.1016/j.pbb.2003.09.023. [DOI] [PubMed] [Google Scholar]
- Sturani E, Abbondio A, Branduardi P, Ferrari C, Zippel R, Martegani E, Vanoni M, Denis-Donini S. The Ras Guanine nucleotide Exchange Factor CDC25Mm is present at the synaptic junction. Exp Cell Res. 1997;235:117–123. doi: 10.1006/excr.1997.3660. [DOI] [PubMed] [Google Scholar]
- Sullivan RM. Unique characteristics of neonatal classical conditioning: the role of the amygdala and locus coeruleus. Integr Physiol Behav Sci. 2001;36:293–307. doi: 10.1007/bf02688797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan RM, Leon M. One-trial olfactory learning enhances olfactory bulb responses to an appetitive conditioned odor in 7-day-old rats. Brain Res. 1987;432:307–311. doi: 10.1016/0165-3806(87)90056-3. [DOI] [PubMed] [Google Scholar]
- Tian X, Gotoh T, Tsuji K, Lo EH, Huang S, Feig LA. Developmentally regulated role for Ras-GRFs in coupling NMDA glutamate receptors to Ras, Erk and CREB. Embo J. 2004;23:1567–1575. doi: 10.1038/sj.emboj.7600151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonini R, Franceschetti S, Parolaro D, Sala M, Mancinelli E, Tininini S, Brusetti R, Sancini G, Brambilla R, Martegani E, Sturani E, Zippel R. Involvement of CDC25Mm/Ras-GRF1-dependent signaling in the control of neuronal excitability. Mol Cell Neurosci. 2001;18:691–701. doi: 10.1006/mcne.2001.1050. [DOI] [PubMed] [Google Scholar]
- Tronel S, Sara SJ. Blockade of NMDA receptors in prelimbic cortex induces an enduring amnesia for odor-reward associative learning. J Neurosci. 2003;23:5472–5476. doi: 10.1523/JNEUROSCI.23-13-05472.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgeon B, Meloche S. Interpreting neonatal lethal phenotypes in mouse mutants: insights into gene function and human diseases. Physiol Rev. 2009;89:1–26. doi: 10.1152/physrev.00040.2007. [DOI] [PubMed] [Google Scholar]
- Weldon DA, Fedorcik GG, LoRusso CM, Tiburzi MJ, Lenoci JM. Olfactory conditioning impairment following posttraining NMDA receptor blockade in neonatal rats. Neurobiol Learn Mem. 1997;67:34–42. doi: 10.1006/nlme.1996.3744. [DOI] [PubMed] [Google Scholar]
- White TL, Youngentob SL. The effect of NMDA-NR2B receptor subunit over-expression on olfactory memory task performance in the mouse. Brain Res. 2004;1021:1–7. doi: 10.1016/j.brainres.2004.05.114. [DOI] [PubMed] [Google Scholar]
- Wilkins JF, Haig D. What good is genomic imprinting: the function of parent-specific gene expression. Nat Rev Genet. 2003;4:359–368. doi: 10.1038/nrg1062. [DOI] [PubMed] [Google Scholar]
- Yang H, Cooley D, Legakis JE, Ge Q, Andrade R, Mattingly RR. Phosphorylation of the Ras-GRF1 exchange factor at Ser916/898 reveals activation of Ras signaling in the cerebral cortex. J Biol Chem. 2003;278:13278–13285. doi: 10.1074/jbc.M209805200. [DOI] [PubMed] [Google Scholar]
- Yang H, Mattingly RR. The Ras-GRF1 exchange factor coordinates activation of H-Ras and Rac1 to control neuronal morphology. Mol Biol Cell. 2006;17:2177–2189. doi: 10.1091/mbc.E05-10-0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon B, Herman H, Hu B, Park YJ, Lindroth A, Bell A, West AG, Chang Y, Stablewski A, Piel JC, Loukinov DI, Lobanenkov VV, Soloway PD. Rasgrf1 imprinting is regulated by a CTCF-dependent methylation-sensitive enhancer blocker. Mol Cell Biol. 2005;25:11184–11190. doi: 10.1128/MCB.25.24.11184-11190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon BJ, Herman H, Sikora A, Smith LT, Plass C, Soloway PD. Regulation of DNA methylation of Rasgrf1. Nat Genet. 2002;30:92–96. doi: 10.1038/ng795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q, Harley CW, Darby-King A, Neve RL, McLean JH. Early odor preference learning in the rat: bidirectional effects of cAMP response element-binding protein (CREB) and mutant CREB support a causal role for phosphorylated CREB. J Neurosci. 2003;23:4760–4765. doi: 10.1523/JNEUROSCI.23-11-04760.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zippel R, Balestrini M, Lomazzi M, Sturani E. Calcium and calmodulin are essential for Ras-GRF1-mediated activation of the Ras pathway by lysophosphatidic acid. Exp Cell Res. 2000;258:403–408. doi: 10.1006/excr.2000.4937. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.