Abstract
The estrogen receptor (ER) is a primary target for breast cancer (BC) treatment. As BC progresses to estrogen-independent growth, the IGF-1R and the ER interact in synergistic crosstalk mechanisms which results in enhanced activation of both receptors signaling cascades. Insulin-like growth factor 2 (IGF-2) is critical in BC progression and its actions are mediated by the IGF-1R. Our previous studies showed that IGF-2 regulates survival genes that protect the mitochondria and promote chemoresistance. In this study, we analyzed BC cells by subcellular fractionation, Western-Blot, qRT-PCR and siRNA analysis. Our results demonstrate that IGF-2 activates ER-α and ER-β and modulates their translocation to the nucleus, membrane organelles and the mitochondria. IGF-2 actions are mediated by the IGF-1R and the insulin receptor (IR). This novel mechanism of IGF-2 synergistic crosstalk signaling with ER-α and ER-β can promote estrogen-independent BC progression and provides new therapeutic targets for the treatment of breast cancer patients.
Keywords: IGF-2, estrogen receptors, IGF-1R, IR, breast cancer
Introduction
Estrogen signaling is mediated through two receptors, ERα and ERβ (Arpino, 2008; Kuiper, 1997; Webb, 1999) When the ER-β was discovered it was thought that it represented a splice variant of ERα, however current studies showed that ER-α and ER-β are different genes with distinct physiological functions (Flynn, 2008). While ERα mediates its effects in the nucleus, ER-β is predominantly localized in the mitochondria and regulates mitochondrial gene expression and membrane potential as well as ATP production (Chen, 2004; Demonacos, 1996; Nilsen, 2003; Richards, 1996; Sekeris, 1990; Wang, 2001; Yang, 2004). Activation of either ER-α or ER-β, promotes BC cell growth and survival, however, estrogen dependent tumors eventually develop mechanisms to activate ER mediated pathways without the requirement of estrogen (Szepeshazi, 1992).
The development of estrogen-independent BC involves cross-talk between several growth factors and the estrogen receptors (Gee, 2005; Shou, 2004). Insulin-like growth factor-2 (IGF-2) is a growth factor that plays a critical role in organ development, differentiation and metabolism by signaling via both the IGF-1R and the IR-A (Abbas, 2007). Signaling through both receptors also regulates the growth and differentiation of normal breast epithelial cells (Ishida, 1995; Rasmussen 1998; Singh, 2007; Vyas, 2005; Yballe, 1996).
The expression of the IGF-II gene is tightly regulated and inhibited by tumor suppressor genes such as p53, Pten, WT1, GC factor (GCF) and several other oncogenes (Zhang et al. 1996, Toretsky and Helman 1996, Kitadai et al. 1993). Mutations in tumor suppressors contribute to higher IGF-II expression not only for lack of suppression but also for gain of function as mutated p53 that stimulates IGF-II (Zhang et al., 1998). The hormonal regulation of IGF-II is positively regulated by E2, progesterone, prolactin and GH, all important hormones in normal breast development and in breast cancer progression (Brisken, et al 2002; Goldfine et. al., 1992; Hamelers et.al., 2003). Thus, IGF-II expression is important in normal breast development and increased IGF-II in the mammary gland contributes to tumor formation. In addition to hormonal stimulation and tumor suppressor inhibition of IGF-II, there are many other critical pathways that activate IGF-II that are also important in breast cancer development. The most significant include IGF-II stimulation by oxidative stress, the Wnt-pathway (disrupts eCadherin-βcatenin), integrins and mTOR (Erbay et al, 2003;Goel et.al., 2006, Morali et.al., 2001). Mutations in the IGF-II receptor (Byrd, et al., 1999) and pTEN also regulate IGF-II levels and IGF-II regulates pTEN (Sukmi Kang-Park et al, 2003, Perks et.al. 2007). The extensive regulatory mechanisms involved in IGF-2 expression also includes miRNAs, methylation, imprinting and other epigenetic alterations present in early development of cancer (Chao et al, 2008, Ge and Chen, 2011)
IGF-2 is highly expressed in breast cancer patients and plasma levels of free IGF-2 directly correlates to BC tumor size (Singer, 2004). Furthermore, transgenic animal models expressing high levels of IGF-2 develop aggressive breast cancer tumors (Pravtcheva, 1998; Pravtcheva, 2003; Bates, 1995).
We have shown that IGF-2 also promotes proliferation, inhibit apoptosis and stimulate the transformation of breast cancer cells (Singh, 2007; Singh, 2008). Our studies also showed that IGF-2 can activate estrogen-regulated genes like survivin, BCL-xl and BCL-2 independent of estrogen through IGF-1R and Insulin receptor. Thus, our particular interest in IGF-2 effects in breast cancer is based on our original observations that IGF-1 was not present in these breast cancer cells and that published studies about IGF-1 analysis by RIA reflected interference with IGFBPs and not IGF-I (De León et al, 1988,1989, 1992). Furthermore, IGF-1 is mainly regulated by GH while IGF-2, as discussed above, is tightly regulated by a wide range of tumor suppressors and hormones that are critical for breast cancer development. Thus, IGF-2 plays a critical role in normal breast differentiation and in the development and progression of breast cancer, demonstrating that IGF-2 have a broader range of biological functions at the cell or tumor level.
Since IGF-1 activation of the IGF-1R can cross-talk and activate the ER-α signaling pathway (Fagan and Yee, 2008) we propose that likewise, IGF-2 can activate the ER-α. We also propose that IGF-2 may activate ER-β signaling pathway via cross-talk with the IGF-1R. Moreover, since IGF-2 not IGF-1 binds the insulin receptor-A, also a member of the IGF-1R family, we thought that IGF-2 activation of the IGF-1R and IR-A signaling pathways results in the phosphorylation/activation of ER-α and ER-β in BC cells. Thus, the present study aims to elucidate if IGF-2 binding to both, IGF-1 receptor and the insulin receptor-A results in activation and translocation of the estrogen receptors.
Materials and Methods
Cell culture
CRL-2335, HS578T, and MCF-7 cell lines were obtained from the American Type Culture Collection (ATCC). The CRL-2335 cell line was derived from a 60-year old African-American (AA) woman and the HS578t cell line was derived from a 74-year old Caucasian (CA) woman and both cell lines are triple negative (ER-,PR-,Her2-). MCF-7 cells were derived from a pleural effusion of a CA woman (69 yo breast cancer patient) and it is used as a positive control for ER-α and ER-β expression. Cultures were maintained at 37C in a 5% CO2 incubator. CRL-2335 cell cultures were maintained in RPMI 1640 media (ATCC) supplemented with 10mL of penicillin/streptomycin (10,000 units/mL penicillin and 10,000 units/mL streptomycin sulfate, Cellgro), 3ug/ml B-amphotericin, and 10% fetal bovine serum (Hyclone). HS578T cell cultures were maintained in DMEM/F12 media (Cellgro) with 4mM L-glutamine, 1.5g/L sodium bicarbonate, 4.5g/L glucose, 0.01mg/ml bovine insulin (Sigma), 10mL of penicillin/streptomycin (10,000 units/mL penicillin and 10,000 units/mL streptomycin sulfate, Cellgro), and 10% fetal bovine serum (Hyclone). MCF-7 cells were maintained in DMEM/F12 media (Cellgro) supplemented with 10 ml of 5000 U penicillin/streptomycin (100 U/ml penicillin and 100 U/ml streptomycin sulfate, Cellgro), 4 mM L-glutamine (Cellgro), 3 µg/ml β-amphotericin, and 5% fetal bovine serum (Hyclone).
Quantitative Real Time PCR (qRT-PCR)
Total RNA was extracted using the Aurum™ Total RNA Mini Kit (BioRad). RNA integrity was evaluated by UV spectroscopy and RNA Quality Indicator (RQI) values were obtained using the Experion™ Automated Electrophoresis System (BioRad). Total RNA samples were stored at −80C until assayed. Quantitative reverse-transcription PCR (qRT-PCR) was performed to determine ER-α and ER-β mRNA expression in MCF-7, CRL-2335 and HS578T cells.
The primer pairs used for ER-α (SA Biosciences) are:
ER-α forward – 5’-ATCCTGATGATTGGTCTCGTCT-3’
ER-α reverse - 5’-TCTGGAAGAGAAGGAACCATATCC-3’
The primer pairs used for ER-β (SA Biosciences) are:
ER-β forward - 5’-GCTCATCTTTGCTCCAGATCTTG-3’
ER-β reverse - 5’-GATGCTTTGGTTTGGGTGATTG-3’
cDNA synthesis was performed using the iScript™ cDNA synthesis kit (Bio-Rad) with 400ng total RNA in a 20 µL reaction. The reaction protocol is as follows: 5 min 25C, 30 min 42C, and 5 min 85C. Each 50 µL qRT-PCR reaction contained 1 µl of cDNA product from the first-strand synthesis, 25 µl of iQ™ SYBR® Green Supermix (Bio-Rad:100mM KCL, 6 mM MgCl2, 40mM Tris-HCL, pH 8.4, 0.4mM of dATP, dCTP, dGTP and dTTP, iTaq DNA Polymerase 50 U/mL, SYBR Green I, 20mM Fluorescein), and 300nM of each forward and reverse primer. Before PCR amplification, iTaq DNA polymerase was activated at 95C for 10 minutes. This was followed by forty cycles of PCR amplification: 30 sec denaturation at 95C, 15 sec annealing at 57C, and 90 sec elongation at 72C. Fluorescence was detected at the end of every 72C extension phase. To confirm the presence of specific PCR products, melting curve analyses were performed on the PCR products after the cycling protocol.
Subcellular Fractionation
HS578T, CRL-2335 and MCF-7 cells were plated at a density of 1 × 106 cells/ml and grown to 80% confluency in 10mm culture plates. Cultures were treated with 100ng/ml proIGF-2 (GroPep) or mIGF-2 (PeproTech) in phenol-red free and serum free media for 20 minutes. MCF-7 cells were left untreated. The Proteo-Extract Subcellular Proteome Extraction Kit (Calbiochem) was used according to manufacturer’s protocol to obtain cytosolic, membrane/organelle (mitochondria) and nuclear fractions. All fractions were stored at −80C until assayed.
Western Blot Analysis
The Coomassie Plus Protein Assay Reagent (Pierce Biotechnology) was used to assess protein concentration. Forty micrograms of each protein sample were loaded into a 4–12% polyacrylamide-SDS gradient gel then transferred onto a PVDF membrane (Invitrogen) using an X-Cell SureLock electrophoretic transfer module (Invitrogen). Membranes were blocked for 1 hour at room temperature with 5% nonfat dry milk in 1X PBS/0.05% Tween. Phosphorylated ER-α was detected by incubating membranes at 4C overnight with either rabbit anti-human pER-α (Tyr 537) (Santa Cruz Biotechnology) diluted 1:500 in 3% w/v Bovine Serum Albumin (BSA) or mouse anti-human pERα (Ser 118) (Cell Signaling Technology) diluted 1:500 in 5% w/v nonfat dry milk in 1X PBS/0.05% Tween. Phosphorylated ER-β was detected by incubating membranes with rabbit anti-human pERβ (Ser 87, 1:1000, Santa Cruz Biotechnology). Unphosphorylated ERs were detected by incubating membranes at 4C overnight in 3% w/v non-fat dry milk in 1X PBS/0.05% Tween with either mouse anti-human ER-α (1:1000, Cell Signaling Technology) or mouse anti-human ER-β (1:1000, GeneTex). Proteins specific for the membrane/organelle (mitochondrial) and nuclear fractions were used to confirm the integrity of the subcellular compartments: membrane/organelle (mitochondrial) fraction - MnSOD (1:500, BD Biosciences) and nuclear fraction- LEDGF (1:1000, BD Biosciences). Membranes were then washed 3 times for 5 minutes in 10 ml of 1X PBS/0.05% Tween. The appropriate biotinylated secondary antibody was diluted in 1X PBS/0.05% Tween and incubated with each membrane for 1 hour (anti-rabbit IgG & anti-mouse IgG 1:5000, Amersham), followed by streptavidin-horseradish peroxidase (HRP, 1:2000, Amersham) for 1 hour. Each membrane was washed 4 times for 10 minutes in 10ml of 1X PBS/0.05% Tween. Protein visualization was achieved using enhanced chemiluminescence (ECL) (Pierce) followed by autoradiography with Hyperfilm ECL (Amersham). Band density obtained from WB analysis was quantified using the ChemiImager 4000 (Alpha Innotech Corporation).
siRNA Analysis
CRL-2335 and HS578T cells were plated in 6-well plates at a density of 1 × 105 cells/well and allowed to attach overnight in a 37C 5% CO2 incubator. Cells were grown to 60 −80% confluency then treated with 60 pmols of either IR (sc-29370, Santa Cruz Biotechnology), IGF-1R (sc-29358, Santa Cruz Biotechnology) or scrambled (sc-Santa Cruz Biotechnology) siRNA in the supplied transfection media and incubated for 9 hours at 37C in a 5% CO2 incubator. Complete media containing 20% FBS was added to the serum-free media so that the final serum concentration was 10% FBS. The cells were then incubated for 24 hours in a 37C 5% CO2 incubator. Media was replaced with fresh media followed by an additional 24-hour incubation period in a 37C 5% CO2 incubator. Afterwards, cells were incubated for 6 hours in serum-free, phenol-free media and then treated with either 100ng/ml of mIGF-2 or proIGF-2 for 20 minutes. Cells treated with scrambled siRNA did not receive treatment with IGF-2. Cellular fractions were prepared as previously described and stored at −20°C until assayed.
Statistical Analysis
The data for rtPCR was analyzed by One-way ANOVA with Tukey's Multiple Comparison Post Test using GraphPad Prism software version 5.02 for Windows (GraphPad Software, San Diego, CA). *** - p <0.001, ** - p <0.01.
Statistical analysis for the Western blots scans was performed using one-way ANOVA, SPSS 11.0 software (SPSS, Inc.). Values are expressed as the mean ± SEM of three or more replicate experiments performed in triplicate. Values of p < 0.05 were considered statistically significant.
Results
ER-α and ER-β mRNA in breast cancer cells
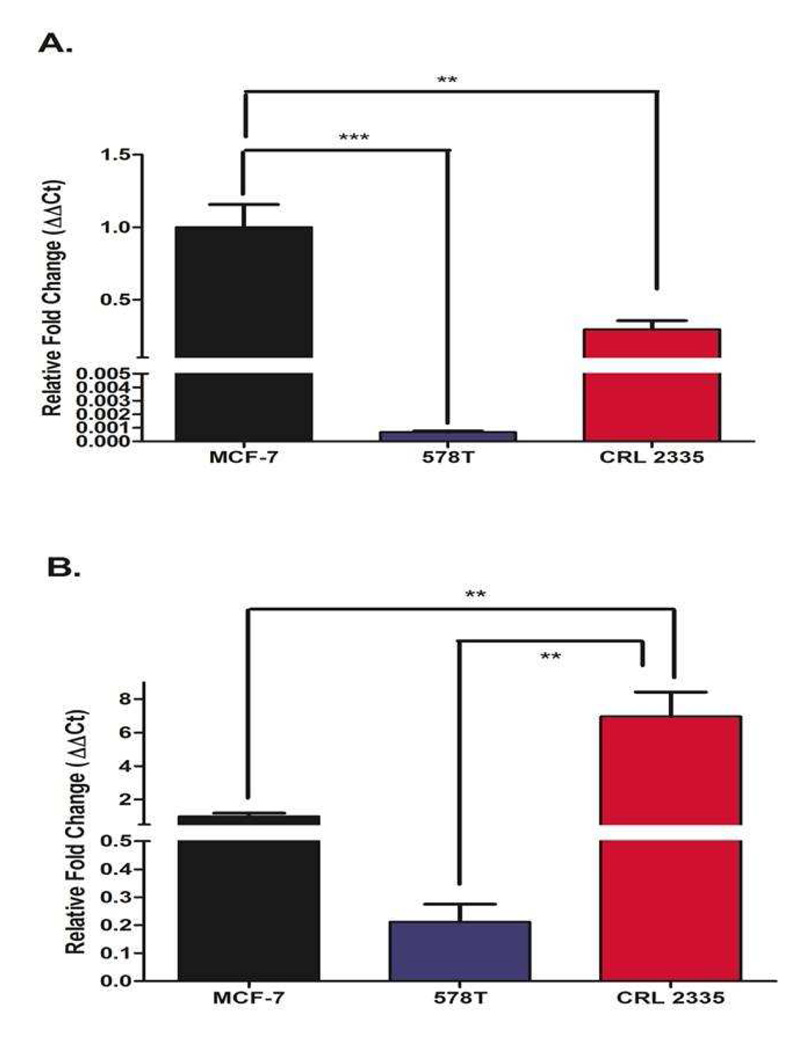
ER status is the golden test to determine whether a breast cancer tumor is sensitive to endocrine therapy. Tumors are designated as ER- when there is less than 10% staining for ER-α in the nuclei as assessed by immunohistochemistry (IHCC). Likewise, when cell lines are established from tumors their ER status is assigned by IHCC. Figure 1 depicts the expression of ER-α and ER-β mRNA in MCF-7 cells (positive control for ER-α and ER-β) and in the ER- cell lines CRL-2335 and Hs578T assessed by qRT-PCR. As expected, MCF-7 cells (ER+) expressed significantly higher levels of ER-α mRNA (Fig. 1A) compared to the ER- cell lines CRL-2335 and Hs578T. Of note, ER- CRL-2335 cells expressed 30% of the ER-α mRNA detected in ER+ MCF-7 cells. Fig.1 B shows the expression of ER-β mRNA. The highest levels of ER-β mRNA were detected in the CRL-2335 cells (6 fold higher than MCF-7 cells) while the ER- Hs578t cells expressed the lowest levels of ER-β mRNA amongst all 3 cell lines studied.
Figure 1.
ER-α and ER-β mRNA expressed as fold change of mRNA expression in MCF-7, CRL-2335 and Hs578T cells. Figure 1A depicts the expression of ER-α in MCF-7, CRL-2335 and Hs578T cell lines. Figure 1B shows the expression of ER-β in these same cell lines. Results are expressed as relative fold change (ΔΔCt). A p-value of less than 0.01 is represented by ** and a p value of less than 0.001 is represented by ***.
Phosphorylation and subcellular translocation of ER-α and ER-β
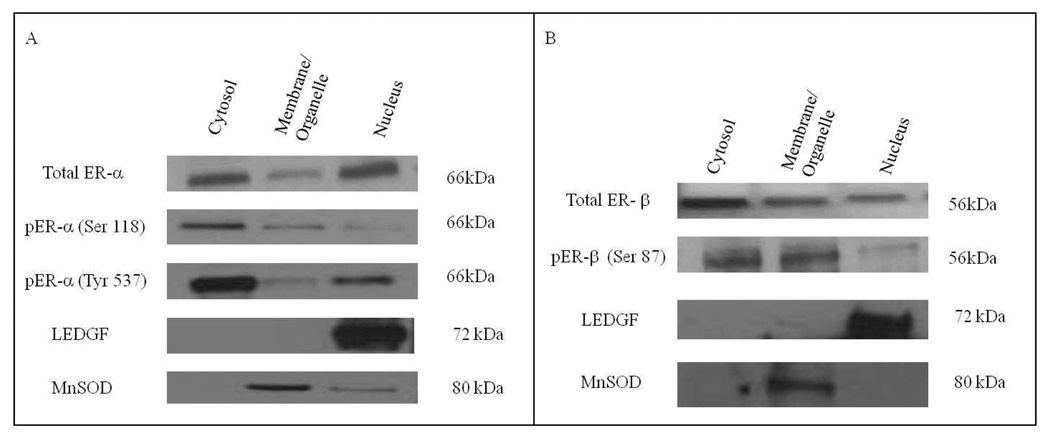
Since IGF-2 stimulates estrogen regulated genes, we chose two ER- cell lines, CRL-2335 and Hs578T to determine whether IGF-2 treatment regulates the expression or localization of ER-α and ER-β in the absence of estrogen. Figure 2 depicts the basal levels of ER-α and ER-β in MCF-7 cells. As expected, there is significant expression of ER-α and ER-β protein in the nuclear compartment of MCF-7 ER+ cells. MCF-7 cells were grown in serum-free, phenol-free media without estrogen, yet, ER-α and ER-β are present in cytosolic, membrane/organelle (mitochondrial) and nuclear fractions. Thus, in MCF-7 cells we can detect activated nuclear and organelle ER-α and ER-β receptors without the requirement of estrogen.
Figure 2.
Western Blot of Subcellular localization of Total and Phosphorylated ER-α and ER-β in MCF-7 cells. The basal expression of total and phosphorylated ER-α and ER-β was detected in MCF-7 cells. In order to verify separation and quatify loading of the subcellular compartments LEDGF and MnSOD were used as controls. Western blot is representative of three independent experiments in triplicate.
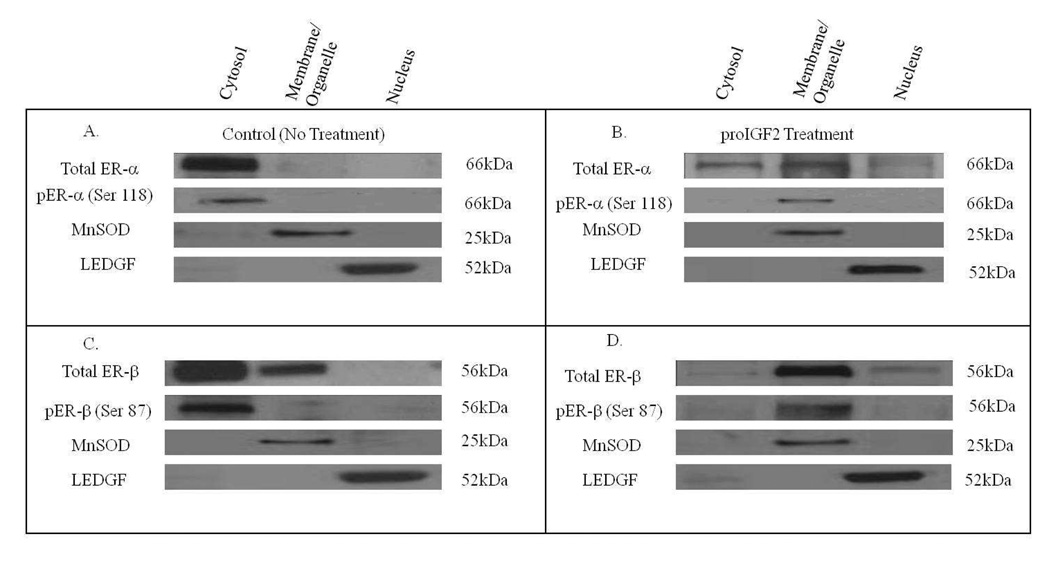
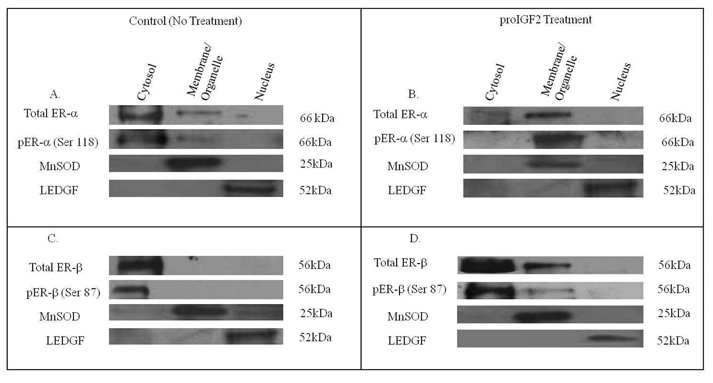
Likewise, the expression of ER-α and ER-β in Hs578T and CRL-2335 cell lines was evaluated by Western blot analysis (WB). Fig. 3A and Fig. 3C shows the levels of total and phosphorylated ER-α (Fig. 3A) and ER-β (Fig. 3C) in the cell compartment of ER- Hs578t cells. The basal expression of total ER-α and ER-α (Ser 118) in ER- Hs578t cells is localized in the cytosol (Fig. 3A). In contrast, ER-β is also detected in the membrane/organelle (mitochondrial) fraction in ER- Hs578t cells. No pER-β (Ser 87) was detected in the membrane/organelle, suggesting that the ER-β present in the organelle/mitochondrial fraction is not phosphorylated. Interestingly, when ER- Hs578t cells were treated with proIGF-2, ER-α translocates from the cytosol to membrane/organelle (mitochondrial) fraction and to the nucleus. (Fig. 3B). Of great significance, proIGF-II treatment to ER- Hs578t cells stimulates the translocation of ER-β from the cytosol to the membrane/organelle (mitochondria fraction and the nucleus (Fig. 3D). proIGF-2 also stimulated the phosphorylation of the ER-β receptor present in the membrane/organelle (mitochondria) fraction pER-β (Ser 87). Thus, in ER- Hs578t cells, the basal expression of total and phosphorylated ER-α (Ser 118) is localized in the cytosol. When ER- Hs578t cells are treated with proIGF-II, ER-α translocates to the membrane/organelle (mitochondria) fraction and nucleus while pER-α (Ser 118) is localized in the mitochondrial/membrane fraction. Of note, treatment with mIGF-2 did not stimulate any changes in ER-α or ER-β subcellular localization in CRL-2335 and Hs578T BC cells (Data not shown).
Figure 3.
Western Blot of Subcellular localization of Total and Phosphorylated ER-α and ER-β in Hs578T cells in response to proIGF2 (100ng/mL) treatment. Figure 3A and 3C shows the basal expression of total and phosphorylated ER-α and ER-β. Figure 3B and 3D shows the subcellular translocation of total and phosphorylated ER-α and ER-β in cells treated with proIGF2. Western blot is representative of three independent experiments in triplicate.
Figure 4 shows a western blot of the subcellular localization of total and phosphorylated ER-α and ER-β in ER- CRL-2335 cells. Figs. 4A and 4C shows that in basal conditions, total and phosphorylated ER-α and ER-β are localized in the cytosol, while ER-α is also localized in the membrane/organelle (mitochondria) fraction. Treatment with proIGF-2 stimulated cytosolic ER-α translocation to the membrane/organelle (mitochondria) fraction (Fig. 4B) where all of the pER-α (Ser 118) was localized. In contrast, treatment with proIGF-II in CRL-2335 cells only partially stimulated ER-β and pER-β translocation to the membrane/organelle (mitochondria) fraction (Fig. 4D). Thus, both cell lines designated as estrogen negative cells, CRL-2335 and Hs578T, express phosphorylated ER-α and ER-β and treatment with proIGF-2 stimulates translocation of both receptors. These data suggests that ER-α and ER-β play an important extranuclear function in estrogen negative breast cancer cells and also shows that both receptors can be regulated by proIGF-2.
Figure 4.
Western Blot of Subcellular localization of Total and Phosphorylated ER-α and ER-β in CRL-2335 cells in response to proIGF2 (100ng/mL) treatment. Figure 4A and 4C shows the basal expression of total and phosphorylated ER-α and ER-β. Figure 4B and 4D shows the subcellular translocation of total and phosphorylated ER-α and ER-β in cells treated with proIGF2. WB is representative of at least three separate experiments.
IGF-2 stimulates the translocation of the ERs through IGF-1R and IR
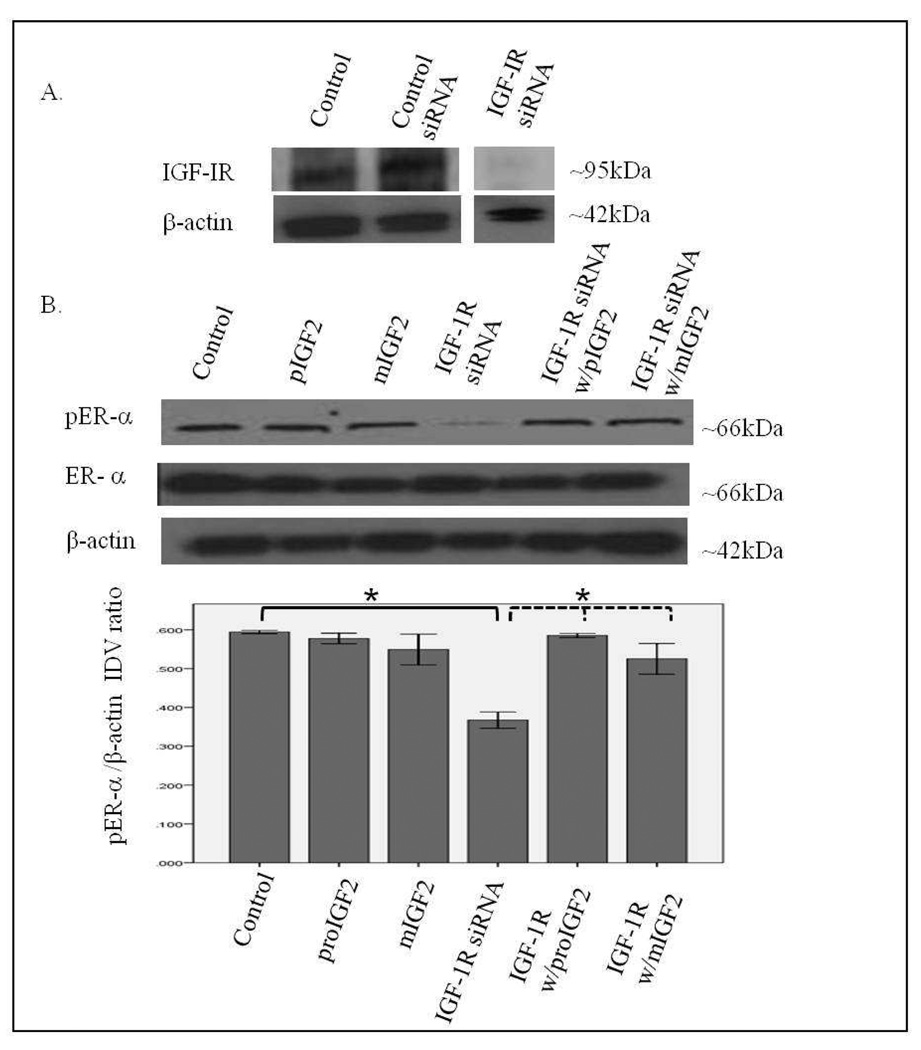
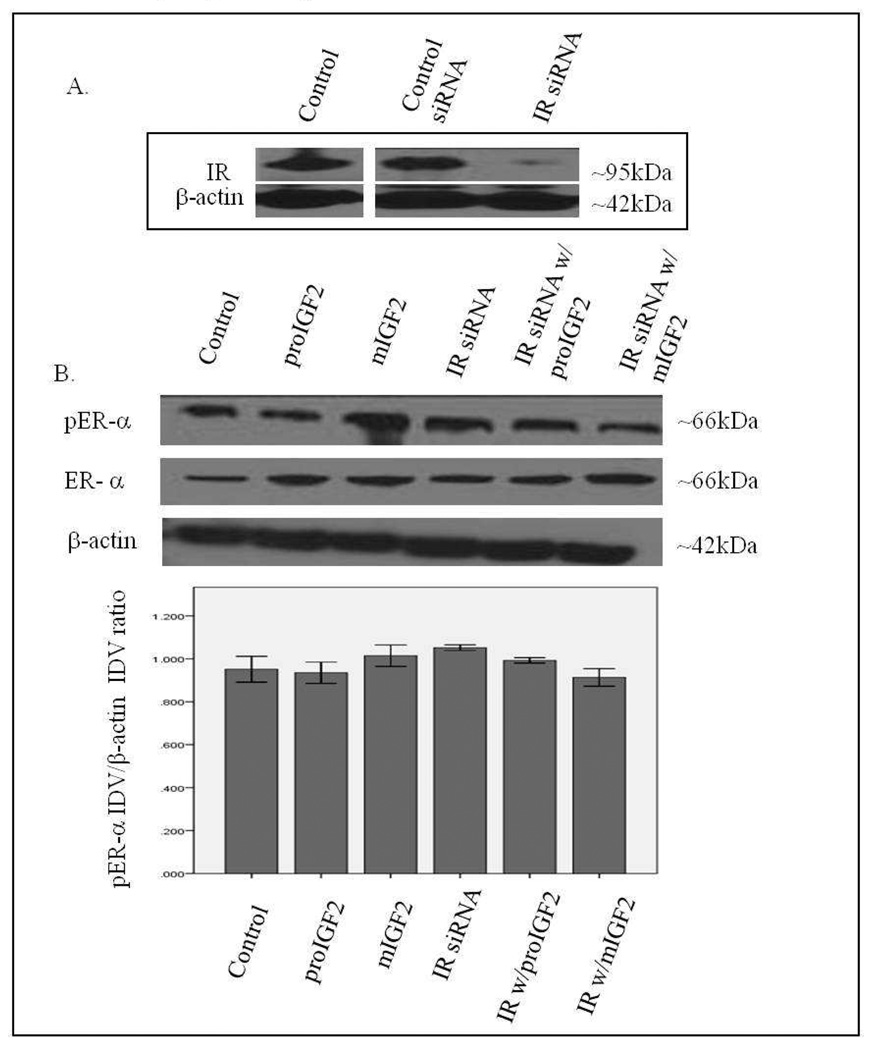
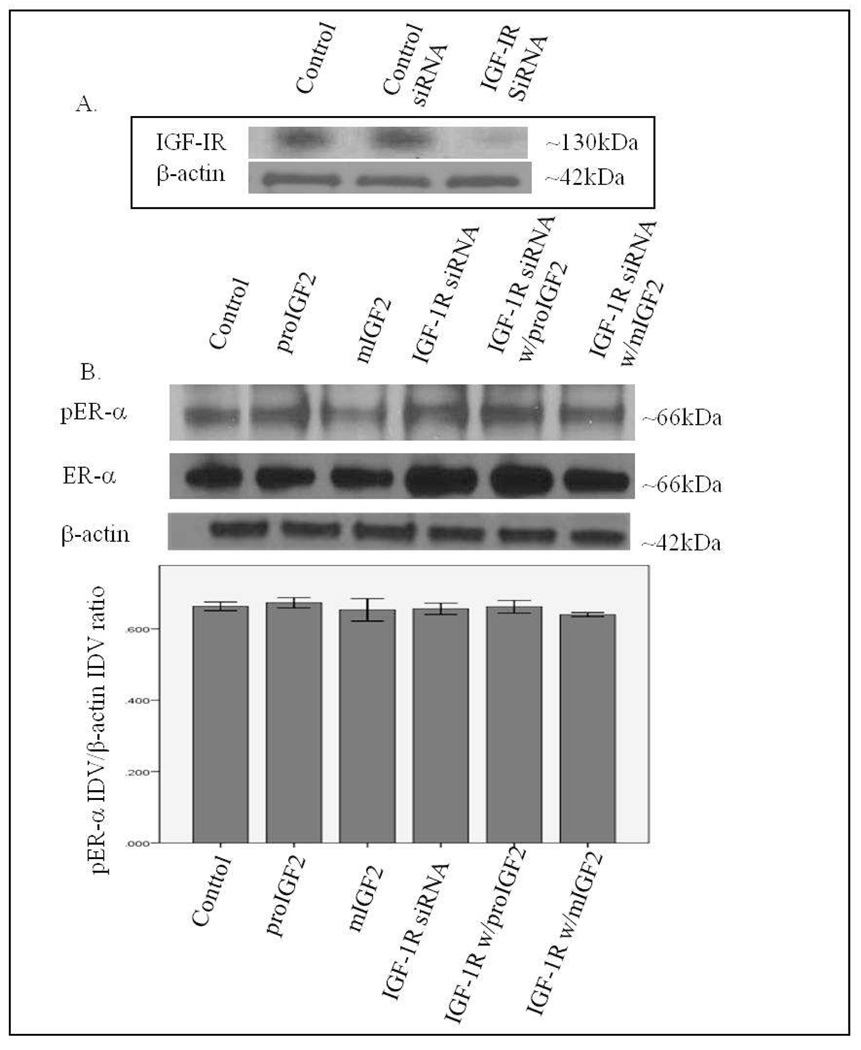
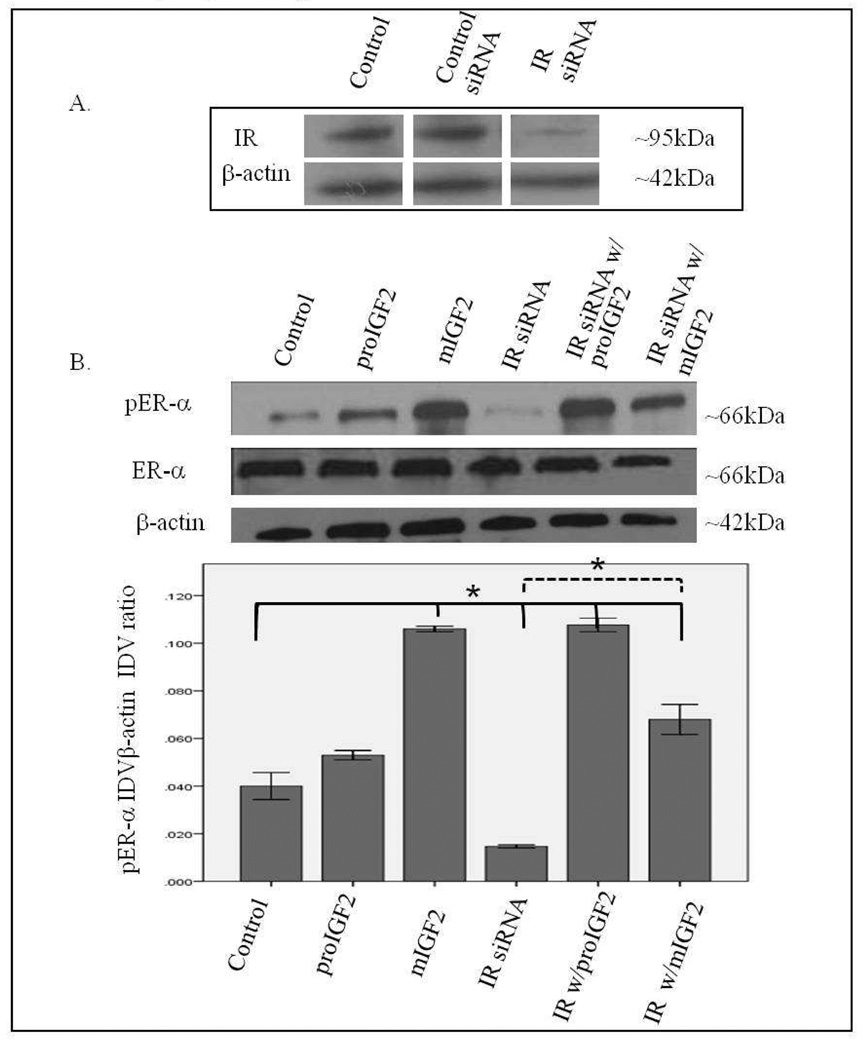
Since IGF-2 actions are mediated by binding to the insulin-like growth factor-1 receptor (IGF-1R) and the insulin receptor-A (IR-A) we assessed whether proIGF-2 mediates the activation and translocation of the estrogen receptors through IGF-1R, the IR-A or both. Hs578T and CRL-2335 cell lines were treated with IGF-1R and IR-A siRNA to determine which of these receptors were mediating IGF-2 actions. Figure 5A shows a Western Blot of Hs578t cells treated with IGF-1R siRNA, scrambled siRNA and control and demonstrates that siRNA successfully blocked the expression of the IGF-1R. Knock-down of the IGF-1R (Fig. 5B) in the Hs578T cell line significantly (p<.05) reduced the phosphorylation of both pERα and pERβ (Fig. 5B & 5C). Interestingly, when siRNA transfected cells were treated with IGF-2 the levels of pERα and pERβ were restored comparable to the control cells (Fig. 5B & C). Thus, IGF-2 rescued the phosphorylation of pERα and pERβ when IGF-1R was reduced or knocked-down, suggesting that another receptor(s) were mediating IGF-2 signaling in the absence of the IGF-1R. In contrast, no effect in the phosphorylation of ERα or ERβ was observed when the IR was successfully knocked-down in these same cells (Fig. 6A–C). These results suggest that IGF-2 mediated phosphorylation and translocation of ERα and ERβ is dependent on activation of the IGF-1R but in its absence IGF-2 can activate the IR or other receptor to restore ER-α and ER-β phosphorylation.
Figure 5.
A & B. Western Blot of Total and Phosphorylated expression of ER-α in Hs578T cells treated with IGF-1R siRNA. Figure 5A shows a WB of Hs578T cells untreated (Control), treated with “scrambled” siRNA (siRNA control) and IGF-1R siRNA. Figure 5B shows WB of ER-α expression following proIGF2 and mIGF2 treatment of IGF-1R siRNA treated Hs578T cells. The bar graphs show the results of the densitometry analysis of the WBs phosphorylated ER-α normalized to β-actin and represent the mean +/− SE of three separate experiments. Solid brackets and * represents values significantly different from control (*p<0.05). Dashed brackets and * represents values significantly different from values between IGF-1R siRNA only treated cells and IGF-IR siRNA with proIGF2 and/ or IGF1R siRNA with mIGF2 treated cells.
Western Blot of Total and Phosphorylated expression of ER-β in Hs578T cells treated with IGF-1R siRNA. Figure 5C shows WB of ER-β expression following proIGF2 and mIGF2 treatment of IGF-1R siRNA treated Hs578T cells. The bar graphs show the results of the densitometry analysis of the WBs phosphorylated ER-β normalized to β-actin and represent the mean +/− SE of three separate experiments. Solid brackets and * represents values significantly different from control (*p<0.05). Dashed brackets and * represents values significantly different from values between IGF-1R siRNA only treated cells and IGF-IR siRNA with proIGF2 and/ or IGF1R siRNA with mIGF2 treated cells.
Figure 6.
Western Blot of Total and Phosphorylated expression of ER-α in Hs578T cells treated with IR siRNA. Figure 6A shows a WB of Hs578T cells untreated (Control), treated with “scrambled” siRNA (Control siRNA) and IR siRNA. Figure 6B shows a WB of ER-α following proIGF2 and mIGF2 treatment of IR siRNA transfected Hs578T cells. The bar graphs show the results of the densitometry analysis of the WBs of phosphorylated ER-α normalized to β-actin and represent the mean +/− SE of three separate experiments.
Western Blot of Total and Phosphorylated expression of ER-β in Hs578T cells treated with IR siRNA. Figure 6C shows a WB of ER-β following proIGF2 and mIGF2 treatment of IR siRNA transfected Hs578T cells. The bar graphs show the results of the densitometry analysis of the WBs of phosphorylated ER-β normalized to β-actin and represent the mean +/− SE of three separate experiments.
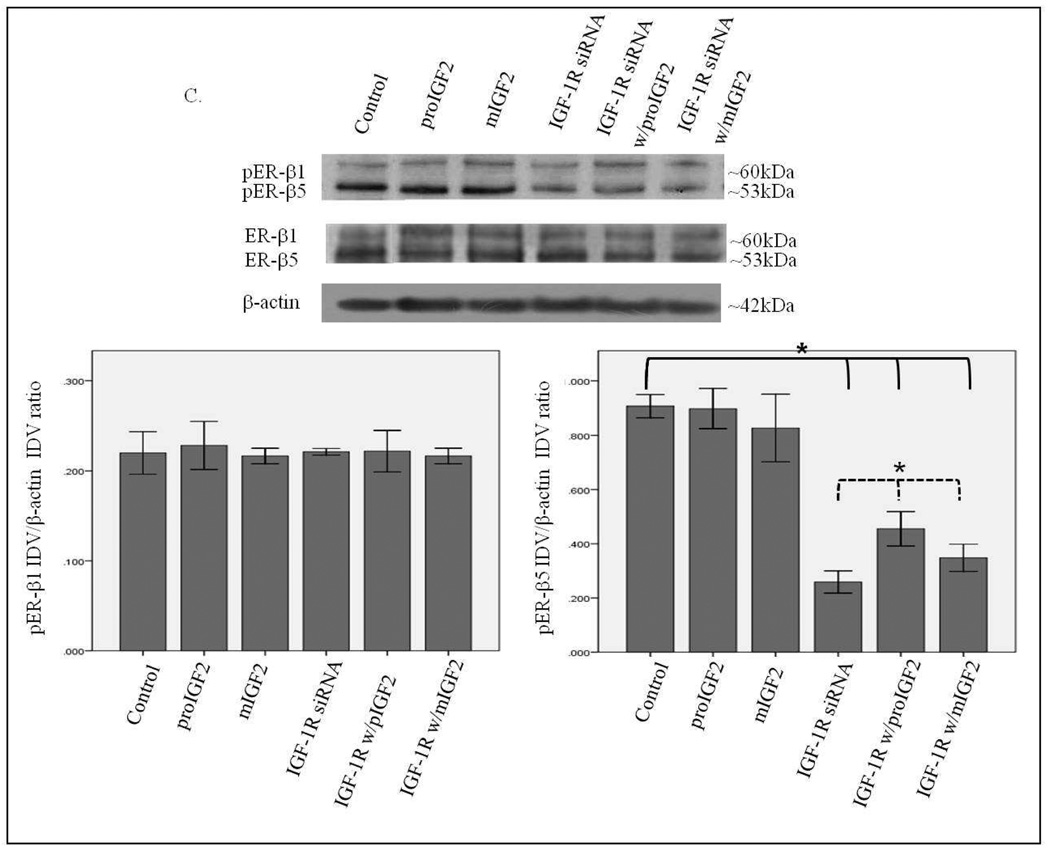
Figure 7 A–C shows that successful knockdown of IGF-1R in the CRL-2335 breast cancer cells had no effect on the phosphorylation of ER-α and ER-β1. In contrast, a significant reduction in the phosphorylation of ER-β5 (53kDa) variant (Fig. 7C) was observed when the IGF-1R was blocked with siRNA. The phosphorylation of the ER-β5 variant was not restored with IGF-2 treatment suggesting that it requires the activation of the IGF-1R signaling. This is significant because ER-β5 is expressed in breast tumors from African-American women and CRL-2335 cells were established from an AA breast cancer patient.
Figure 7.
Western Blot of Total and Phosphorylated expression of ER-α in CRL-2335 cells treated with IGF-1R siRNA. Figure 7A shows a WB of CRL-2335 cells untreated (Control), transfected with “scrambled” siRNA (Control siRNA) and IGF-1R siRNA. Figure 7B shows a WB of ER-α in CRL-2335 cells treated with IGF-1R siRNA. The bar graphs show phosphorylated ER-α normalized to β-actin and represent the mean +/− SE of three separate experiments.
Western Blot of Total and Phosphorylated expression of ER-β1 and ER-β5 in CRL-2335 cells treated with IGF-1R siRNA. Figure 7C shows a WB of ER-β1 and ER-β5 in CRL-2335 cells treated with IGF-1R siRNA. The bar graphs show phosphorylated ER-β1 and ER-β5 normalized to β-actin and represent the mean +/− SE of three separate experiments. Bar graphs with solid brackets and * represents values significantly different from control (*p<0.05). Bar graphs with dashed brackets and * show significantly different (*p<0.05) values between IGF-1R siRNA only treated cells and IGF-IR siRNA with proIGF2 and/ or IGF1R siRNA with mIGF2 treated cells.
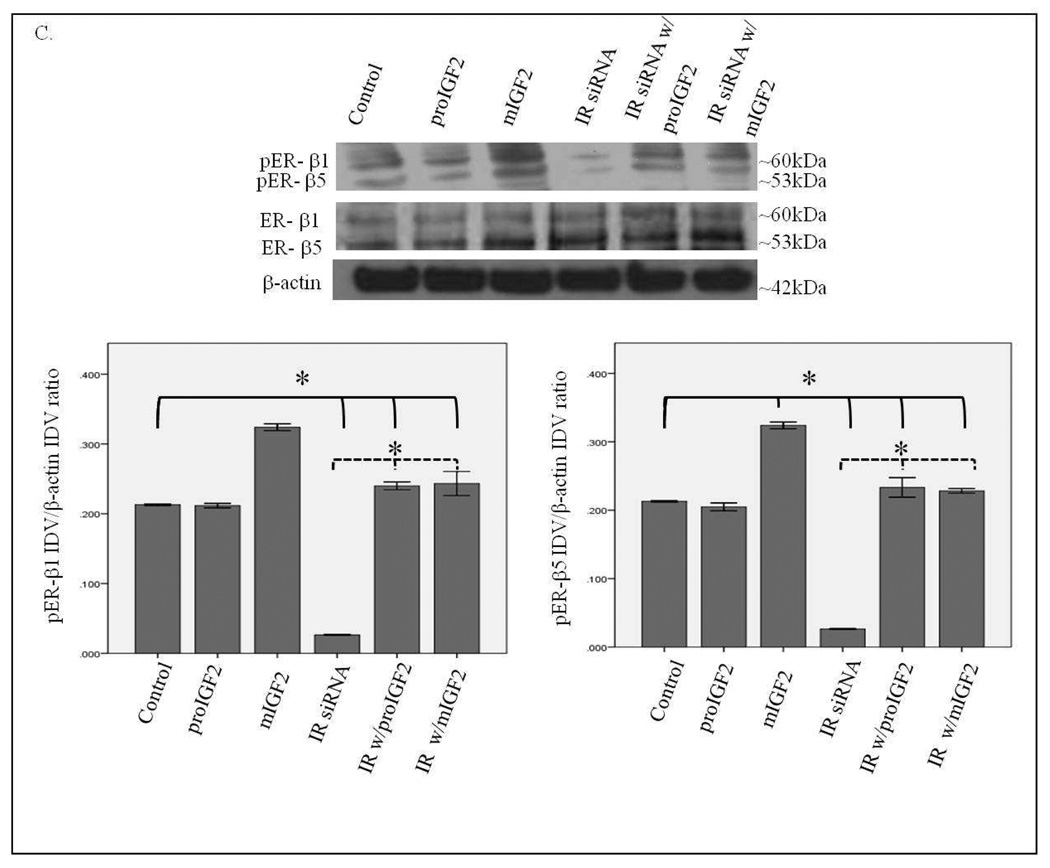
Notably, when CRL-2335 cells were treated with IR siRNA a significant reduction in the phosphorylation of ER-α, ER-β1 and ER-β5 (Fig. 8 A–C) was observed. In contrast to the Hs578t BC cells, phosphorylation of ER-α and ER-β in the CRL-2335 cells is more dependent on the activation of the IR signaling. Also distinct in the CRL-2335 cells is the expression of the ER-β5 variant which is not detected in the Hs578t BC cells. Treatment with IGF-2 restored phosphorylation of ER-α, ER-β1 and ER-β5 in the CRL-2335 BC cells (Fig. 8 B&C). Simultaneous knockdown of IR and IGF-1R was lethal (Data not shown).
Figure 8.
Western Blot of Total and Phosphorylated expression of ER-α in CRL-2335 cells treated with IR siRNA. Figure 8A shows a WB of CRL-2335 cells untreated (Control), transfected with “scrambled” siRNA (Control siRNA) and IR siRNA. Figure 8B shows a WB of total and phosphorylated ER-α following proIGF2 and mIGF2 treatment and/or IR siRNA treatment in CRL-2335 cells. The bar graphs show phosphorylated ER-α normalized to β-actin and represent the mean +/− SE of three separate experiments. Bar graphs with solid brackets and * represents values significantly different from control (*p<0.05). Bar graphs with dashed brackets and * significantly different (*p<0.05) values between IR siRNA only treated cells and IR siRNA with proIGF2 and/ or IGF1R siRNA with mIGF2 treated cells.
Western Blot of Total and Phosphorylated expression of ER-β1 and ER-β5 in CRL-2335 cells treated with IR siRNA. Figure 8C shows a WB of total and phosphorylated ER-β1 and ER-β5 following proIGF2 and mIGF2 treatment and/or IR siRNA treatment in CRL-2335 cells. The bar graphs show phosphorylated ER-β1 and ER-β5 normalized to β-actin and represent the mean +/− SE of three separate experiments. Bar graphs with solid brackets and * represents values significantly different from control (*p<0.05). Bar graphs with dashed brackets and * significantly different (*p<0.05) values between IR siRNA only treated cells and IR siRNA with proIGF2 and/ or IGF1R siRNA with mIGF2 treated cells.
Discussion
The acquired ability of hormone refractory breast cancer cells to avoid cell death in the presence of anti-estrogen therapy means that the cells have developed the ability to maintain cell survival signaling pathways without the requirement of estrogen. The mechanism(s) used by breast cancer to acquire this ability are not well-defined. A general consensus in the field is that “cross-talk” mechanisms between growth factor receptors (IGF-1R and EGFR) and estrogen receptors (ER-α) facilitate the progression of breast cancer tumors that become hormone insensitive and refractory (Schiff, 2004). In fact, activation of the IGF-1R (Kato, 1995; Lannigan, 2003) can lead to the activation of ER-α through activation of the mitogen-activated protein (MAP) kinase pathway resulting in the phosphorylation of ER-α at Ser 118. Since IGF-2, not IGF-I, is the growth factor expressed in breast cancer cells (Pezzino, 1996), we propose that IGF-2 can maintain survival signals in an autocrine fashion by binding to the IGF-1R and possibly to the IR (LeRoith, 1995; Sciacca, 1999). Furthermore, since many breast cancers express both ER-α and ER-β, we deduced that IGF-2 can bind and activate the IGF-1R and IR leading to the activation of both ER-α and ER-β in BC cells. This IGF-1R and IR/ER cross-talk allows the cells to activate/phosphorylate the ERs without the need of estrogen. Thus, current anti-estrogen therapies would not be able to effectively prevent ER activation in these BC cells.
Binding of IGF-2 to the IGF-1R and IR activates different signaling pathways (Valentinis, 2001; Chen, 2009). Our study suggests that the activation of these different signaling pathways give each receptor a unique role in the phosphorylation of ER-α and ER-β. In ER- Hs578T cells, IGF-1R knockdown decreased the phosphorylation of both ER-α and ER-β. However, treatment with IGF-2 was able to increase the phosphorylation of both, ER-α and ER-β, back to control levels in IGF-1R siRNA transfected Hs578T cells. Thus, IGF-1R appears to be important in the phosphorylation of ER-α and ER-β in Hs578T cells, however IGF-2 treatment was able to restore ER phosphorylation presumably by acting through the IR-A. Indeed, Hs578t was the first breast cancer cell line shown to express high levels of IR-A (Sciacca, 1999). In contrast, knocked down expression of the IGF-1R in the ER- CRL-2335 cells had no effect on the phosphorylation of ER-α or ER-β1 but it decreased the activation of the ER-β5 variant. The ER-β5 variant is overexpressed in African-American (AA) women with aggressive BC (Poola, 2005) and this study shows that ER-β5 variant is expressed in the ER-CRL-2335 cells derived from an AA breast cancer patient (Gazdar, 1998). Treatment with IGF-2 was unable to restore ER-β5 phosphorylation in the IGF-1R siRNA transfected ER-CRL-2335 cells. These results suggest that IGF-1R expression is required for the phosphorylation of the ER-β5 variant. In contrast, ER-β5 variant is not expressed in the Hs578T cell line derived from a Caucasian breast cancer patient (Hackett, 1977).
IR knockdown further demonstrated the different roles of IGF-1R and IR in the phosphorylation of ER-α and ER-β. In contrast to the inhibition of the IGF-1R, IR knockdown in Hs578T cells had no effect on the phosphorylation of either estrogen receptor. Of note, IR siRNA transfected CRL-2335 cells showed a decrease in the phosphorylation of ER-α, ER-β and ER-β5. IGF-2 treatment restored the phosphorylation of all, ER-α, ER-β and ER-β5, possibly thorough the IGF-1R. These findings are very significant because they show the unique ability of IGF-2 to activate both the IGF-1R and IR to enhance ER activation and signaling in BC cells. Thus, both, IGF-1 and IR are important targets in the treatment of estrogen independent breast cancers.
Our study also shows that IGF-2 treatment not only phosphorylated ER-α and ER-β but it also stimulated the sub-cellular translocation of both estrogen receptors from the cytosol to the organelles/membrane (mitochondria) fraction. The organelles/membrane (mitochondria) fraction contains the plasma membrane where the estrogen receptors activate a unique signaling cascade that results in rapid changes that do not require estrogen receptor translocation to the nucleus (Bjornstrom, 2005). The translocation of the extranuclear ER to the plasma membrane allows for cross-talk between the growth factor receptors and ER which then promotes the activation of cell survival pathways such as the MAP kinase pathway (Kahlert, 2000; Klotz, 2002).
These data also demonstrates that IGF-2 stimulated extranuclear ER translocation to the mitochondria where it can regulate mitochondrial function (Bjornstrom, 2005; Pietras, 2007; Singh, 2007; Song, 2006; Xu, 2004). We have previously shown that IGF-2 regulation of the mitochondria allows breast cancer cells to prevent apoptosis and provide energy for cell growth, thus, promoting cell survival (Singh, 2007).
This paper shows that IGF-2 is able to promote breast cancer cell survival by activating the IGF-1R and the IR-A which promotes the activation of cell survival pathways. We also demonstrate that IGF-2 is able to regulate the translocation of ER-α and ER-β to the mitochondria and plasma membrane in order to promote cell survival (Anders, 2008). Our study presents compelling evidence that IGF-2 plays an important role in promoting growth factor and estrogen receptor cross-talk mechanisms thus, activating estrogen signaling pathways without the requirement of estrogen.
In summary, IGF-2 promotes the activation of estrogen cell survival pathways without the requirement for estrogen. The novel proposed mechanism is that IGF-2 is able to bind to the IGF-1R and IR-A to promote ER-α and ER-β phosphorylation and translocation to the nucleus, plasma membrane and mitochondria leading to the activation of cell survival pathways. Breast cancers that grow independent of estrogen currently have no targeted treatments due to a gap in knowledge of cell survival mechanisms in these cells. The mechanism proposed in this paper may lead to the development of targeted treatments for these breast cancers.
Acknowledgments
This research was supported by 5P20 MD001632, and NIGMS 5R25GM060507.
Footnotes
Declaration of Interest
No conflicts of interest are declared by the authors.
References
- Abbas A, Yakar S, LeRoith D, Brodt P. The Role of the IGF System in Cancer Growth and Metastasis: Overview and Recent Insights. Endocrinology. 2007;28(1):20–47. doi: 10.1210/er.2006-0001. [DOI] [PubMed] [Google Scholar]
- Anders C, Carey LA. Understanding and treating triple-negative breast cancer. Oncology (Williston Park) 2008;22(11):1233–1239. discussion 1239–1240, 1243. [PMC free article] [PubMed] [Google Scholar]
- Arpino G, Wiechmann L, Osborne CK, Schiff R. Crosstalk between the estrogen receptor and the HER tyrosine kinase receptor family: molecular mechanism and clinical implications for endocrine therapy resistance. Endocr Rev. 2008;29(2):217–233. doi: 10.1210/er.2006-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates P, Fisher R, Ward A, Richardson L, Hill DJ, Graham CF. Mammary cancer in transgenic mice expressing insulin-like growth factor II (IGF-II) Br. J. Cancer. 1995;72(5):1189–1193. doi: 10.1038/bjc.1995.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd JC, Devi GR, de Souza AT, Jirtle RL, MacDonald RG. Disruption of ligand binding to the insulin-like growth factor II/mannose 6-phosphate receptor by cancer-associated missense mutations. J. Biol. Chem. 1999;274:24408–24416. doi: 10.1074/jbc.274.34.24408. [DOI] [PubMed] [Google Scholar]
- Bjornstrom L, Sjoberg M. Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol Endocrinol. 2005;19(4):833–842. doi: 10.1210/me.2004-0486. [DOI] [PubMed] [Google Scholar]
- Brisken C, Ayyannan A, Nguyen C, Heineman A, Reinhardt F, Tan J, Dey SK, Dotto GP, Weinberg RA. IGF-2 is a mediator of prolactin-induced morphogenesis in the breast. Dev Cell. 2002 Dec;3(6):877–887. doi: 10.1016/s1534-5807(02)00365-9. [DOI] [PubMed] [Google Scholar]
- Chao, Wendy, Patricia D'Amore IGF2:Epigenetic regulation and role in development and disease. Cytokine & Growth Factors Reviews. 2008;19:111–120. doi: 10.1016/j.cytogfr.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JQ, Delannoy M, Cooke C, Yager JD. Mitochondrial localization of ERalpha and ERbeta in human MCF7 cells. Am J Physiol Endocrinol Metab. 2004;286(6):E1011–E1022. doi: 10.1152/ajpendo.00508.2003. [DOI] [PubMed] [Google Scholar]
- Chen Y, Boyartchuk V, Lewis BC. Differential Roles of Insulin-like Growth Factor Receptor- and Insulin Receptor-mediated Signaling in the Pheonotypes of Hepatocellular Carcinoma Cells. Neoplasia. 2009;11(9):835–845. doi: 10.1593/neo.09476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demonacos CV, Karayanni N, Hatzoglou E, Tsiriyiotis C, Spandidos DA, Sekeris CE. Mitochondrial genes as sites of primary action of steroid hormones. Steroids. 1996;61(4):226–232. doi: 10.1016/0039-128x(96)00019-0. [DOI] [PubMed] [Google Scholar]
- De León DD, Bakker B, Wilson DM, Hintz RL, Rosenfeld R. Demonstration of Insulin-like growth factor (IGF-I and -II) receptors and binding protein in human breast cancer cells lines. Biochem and Biophys. Res. Commun. 1988;152:398–405. doi: 10.1016/s0006-291x(88)80727-7. [DOI] [PubMed] [Google Scholar]
- De León DD, Wilson DM, Bakker B, Lamsom G, Hintz RL, Rosenfeld R. Characterization of insulin-like growth factor (IGF) binding proteins from human breast cancer cells. Mol. Endocrinol. 1989;3:567–574. doi: 10.1210/mend-3-3-567. [DOI] [PubMed] [Google Scholar]
- De León DD, Powers M, Wilson DM, Rosenfeld RG. Effects of insulin-like growth factors (IGFs) and IGF receptor antibodies in human breast cancer cell proliferation. Growth factors. 1992;6:327–336. doi: 10.3109/08977199209021544. [DOI] [PubMed] [Google Scholar]
- Erbay E, Park IH, Nuzzi PD, Schoenherr CJ, Chen J. IGF-II transcription in skeletal myogenesis is controlled by mTOR and nutrients. J Cell Biol. 2003 Dec 8;163(5):931–936. doi: 10.1083/jcb.200307158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan D, Yee D. Crosstalk between IGF1R and Estrogen Receptor Signaling in Breast Cancer. J Mammary Gland Biol Neoplasia. 13:423–429. doi: 10.1007/s10911-008-9098-0. [DOI] [PubMed] [Google Scholar]
- Flynn JM, Dimitrijevich SD, Younes M, Skliris G, Murphy LC, Cammarata PR. Role of wild-type estrogen receptor-beta in mitochondrial cytoprotection of cultured normal male and female human lens epithelial cells. Am J Physiol Endocrinol Metab. 2008;295(3):E637–E647. doi: 10.1152/ajpendo.90407.2008. [DOI] [PubMed] [Google Scholar]
- Gazdar AF, et al. Characterization of paired tumor and non-tumor cell lines established from patients with breast cancer. Int. J. Cancer. 1998;78:766–774. doi: 10.1002/(sici)1097-0215(19981209)78:6<766::aid-ijc15>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Ge Y, et al. IGF-II is regulated by microRNA-125b in skeletal myogenesis. Journal of Cell Biology. 2011 doi: 10.1083/jcb.201007165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee JM, Robertson JF, Gutteridge E, Ellis IO, Pinder SE, Rubini M, et al. Epidermal growth factor receptor/HER2/insulin-like growth factor receptor signalling and oestrogen receptor activity in clinical breast cancer. Endocr Relat Cancer. 2005;12 Suppl 1:S99–S111. doi: 10.1677/erc.1.01005. [DOI] [PubMed] [Google Scholar]
- Goel HL, Loredana Moro, Michael King, et al. β1 Integrins Modulate Cell Adhesion by regulating Insulin-Like Growth Factor-II Levels in the Microenvironment. Cancer Res 2006. 2006;66:331–342. doi: 10.1158/0008-5472.CAN-05-2588. [DOI] [PubMed] [Google Scholar]
- Goldfine ID, Papa V, Vigneri R, Siiteri P, Rosenthal S. Progestin regulation of insulin and insulin-like growth factor I receptors in cultured human breast cancer cells. Breast Cancer Res Treat. 1992;22(1):69–79. doi: 10.1007/BF01833335. [DOI] [PubMed] [Google Scholar]
- Hackett AJ, et al. Two syngeneic cell lines from human breast tissue: the aneuploid mammary epithelial (Hs 578T) and the diploid myoepithelial (Hs 578Bst) cell lines. J. Natl. Cancer Inst. 1977;58:1795–1806. doi: 10.1093/jnci/58.6.1795. [DOI] [PubMed] [Google Scholar]
- Hamelers IHL, Steenbergh PH. Interactions between estrogen and insulin-like growth factor signaling pathways in human breast tumor cells. Endocrine-Related Cancer. 2003;10:331–345. doi: 10.1677/erc.0.0100331. [DOI] [PubMed] [Google Scholar]
- Ishida S, Noda M, Kuzuya N, Kubo F, Yamada S, Yamanaka T, et al. Big insulin-like growth factor II-producing hepatocellular carcinoma associated with hypoglycemia. Intern Med. 1995;34(12):1201–1206. doi: 10.2169/internalmedicine.34.1201. [DOI] [PubMed] [Google Scholar]
- Kahlert S, Nuedling S, van Eickels M, Vetter H, Meyer R, Grohe C. Estrogen receptor alpha rapidly activates the IGF-1 receptor pathway. J Biol Chem. 2000;275(24):18447–18453. doi: 10.1074/jbc.M910345199. [DOI] [PubMed] [Google Scholar]
- Kato S, Endoh H, Masuhiro Y, Kitamoto T, Uchiyama S, Sasaki H, et al. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science. 1995;270(5241):1491–1494. doi: 10.1126/science.270.5241.1491. [DOI] [PubMed] [Google Scholar]
- Kitadai Y, Yamazaki H, Yasui W, Kyo E, Yokozaki H, Kajiyama G, Johnson AC, Pastan I, Tahara E. GC factor represses transcription of several growth factor/receptor genes and causes growth inhibition of human gastric carcinoma cell lines. Cell Growth Differ. 1993;4(4):291–296. [PubMed] [Google Scholar]
- Klotz DM, Hewitt SC, Ciana P, Raviscioni M, Lindzey JK, Foley J, et al. Requirement of estrogen receptor-alpha in insulin-like growth factor-1 (IGF-1)-induced uterine responses and in vivo evidence for IGF-1/estrogen receptor cross-talk. J Biol Chem. 2002;277(10):8531–8537. doi: 10.1074/jbc.M109592200. [DOI] [PubMed] [Google Scholar]
- Kuiper GG, Carlsson B, Grandien K, Enmark E, Haggblad J, Nilsson S, et al. Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors alpha and beta. Endocrinology. 1997;138(3):863–870. doi: 10.1210/endo.138.3.4979. [DOI] [PubMed] [Google Scholar]
- Lannigan DA. Estrogen receptor phosphorylation. Steroids. 2003;68(1):1. doi: 10.1016/s0039-128x(02)00110-1. [DOI] [PubMed] [Google Scholar]
- LeRoith D, Baserga R, Helman L, Roberts CT., Jr Insulin-like growth factors and cancer. Ann Intern Med. 1995;122(1):54–59. doi: 10.7326/0003-4819-122-1-199501010-00009. [DOI] [PubMed] [Google Scholar]
- Morali OG, Delmas V, Moore R, Jeanney C, Thiery JP, Larue L. IGF-II induces rapid beta-catenin relocation to the nucleus during epithelium to mesenchyme transition. Oncogene. 2001;20:4942–4950. doi: 10.1038/sj.onc.1204660. [DOI] [PubMed] [Google Scholar]
- Morali OG, Delmas V, Moore R, Jeanney C, Thiery JP, Larue L. IGF-II induces rapid beta-catenin relocation to the nucleus during epithelium to mesenchyme transition. Oncogene. 2001;20:4942–4950. doi: 10.1038/sj.onc.1204660. [DOI] [PubMed] [Google Scholar]
- Nilsen J, Diaz Brinton R. Mechanism of estrogen-mediated neuroprotection: regulation of mitochondrial calcium and Bcl-2 expression. Proc Natl Acad Sci U S A. 2003;100(5):2842–2847. doi: 10.1073/pnas.0438041100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzino V, Papa V, Milazzo B, et al. Insulin-like Growth Factor-I Receptors in Breast Cancer. Annuals of the NY Academy of Sciences. 1996;784(1):189–201. doi: 10.1111/j.1749-6632.1996.tb16236.x. [DOI] [PubMed] [Google Scholar]
- Perks CM, Vernon EG, Rosendahl AH, Tonge D, Holly JM. IGF-II and IGFBP-2 differentially regulate PTEN in human breast cancer cells. Oncogene. 2007 Aug 30;26(40):5966–5972. doi: 10.1038/sj.onc.1210397. [DOI] [PubMed] [Google Scholar]
- Pietras RJ, Marquez-Garban DC. Membrane-associated estrogen receptor signaling pathways in human cancers. Clin Cancer Res. 2007;13(16):4672–4676. doi: 10.1158/1078-0432.CCR-07-1373. [DOI] [PubMed] [Google Scholar]
- Poola I, Fuqua SA, De Witty RL, Abraham J, Marshallack JJ, Liu A. Estrogen receptor alpha-negative breast cancer tissues express significant levels of estrogen-independent transcription factors, ERbeta1 and ERbeta5: potential molecular targets for chemoprevention. Clin Cancer Res. 2005;11(20):7579–7585. doi: 10.1158/1078-0432.CCR-05-0728. [DOI] [PubMed] [Google Scholar]
- Pravtcheva DD, Wise TL. Metasizing mammary carcinomas in H19 enhancers-Igf2 transgenic mice. J. Exp. Zool. 1998;281:43–57. [PubMed] [Google Scholar]
- Pravtcheva DD, Wise TL. Transgene instability in mice injected with an in vitro methylated Igf2 gene. Mutation Res. 2003;529:35–50. doi: 10.1016/s0027-5107(03)00110-6. [DOI] [PubMed] [Google Scholar]
- Rasmussen AA, Cullen KJ. Paracrine/autocrine regulation of breast cancer by the insulin-like growth factors. Breast Cancer Res Treat. 1998;47(3):219–233. doi: 10.1023/a:1005903000777. [DOI] [PubMed] [Google Scholar]
- Richards RG, DiAugustine RP, Petrusz P, Clark GC, Sebastian J. Estradiol stimulates tyrosine phosphorylation of the insulin-like growth factor-1 receptor and insulin receptor substrate-1 in the uterus. Proc Natl Acad Sci U S A. 1996;93(21):12002–12007. doi: 10.1073/pnas.93.21.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiff R, Massarweh SA, Shou J, Bharwani L, Mohsin SK, Osborne CK. Cross-talk between estrogen receptor and growth factor pathways as a molecular target for overcoming endocrine resistance. Clin Cancer Res. 2004;10(1 Pt 2):331S–336S. doi: 10.1158/1078-0432.ccr-031212. [DOI] [PubMed] [Google Scholar]
- Sciacca L, Costantino A, Pandini G, Mineo R, Frasca F, Scalia P, et al. Insulin receptor activation by IGF-II in breast cancers: evidence for a new autocrine/paracrine mechanism. Oncogene. 1999;18(15):2471–2479. doi: 10.1038/sj.onc.1202600. [DOI] [PubMed] [Google Scholar]
- Sekeris CE. The mitochondrial genome: a possible primary site of action of steroid hormones. In Vivo. 1990;4(5):317–320. [PubMed] [Google Scholar]
- Shou J, Massarweh S, Osborne CK, Wakeling AE, Ali S, Weiss H, et al. Mechanisms of tamoxifen resistance: increased estrogen receptor-HER2/neu cross-talk in ER/HER2-positive breast cancer. J Natl Cancer Inst. 2004;96(12):926–935. doi: 10.1093/jnci/djh166. [DOI] [PubMed] [Google Scholar]
- Singer C, Mogg M, Koestler W, Pacher M, Marton E, Kubista E, et al. Insulin like Growth Factor (IGF)- I and IGF-II Serum Concentrations in Patients with Benign and Malignant Breast Lesions. Clinical Cancer Research. 2004;10:4003–4009. doi: 10.1158/1078-0432.CCR-03-0093. [DOI] [PubMed] [Google Scholar]
- Singh SK, Moretta D, Almaguel F, Wall NR, De Leon M, De Leon D. Differential effect of proIGF-II and IGF-II on resveratrol induced cell death by regulating survivin cellular localization and mitochondrial depolarization in breast cancer cells. Growth Factors. 2007;25(6):363–372. doi: 10.1080/08977190801886905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Moretta D, Almaguel F, De Leon M, De Leon D. Precursor IGF-II and mature IGF-II induce Bcl-2 and Bcl-XL expression through different signaling pathways in breast cancer cells. Growth Factors. 2008;26:92–103. doi: 10.1080/08977190802057258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song RX, Fan P, Yue W, Chen Y, Santen RJ. Role of receptor complexes in the extranuclear actions of estrogen receptor alpha in breast cancer. Endocr Relat Cancer. 2006;13 Suppl 1:S3–S13. doi: 10.1677/erc.1.01322. [DOI] [PubMed] [Google Scholar]
- Sukmi Kang-Park, Yoon Ik Lee, Young Ik Lee PTEN modulates insulin-like growth factor II (IGF-II)-mediated signaling; the protein phosphatasectivity of PTEN downregulates IGF-II expression in hepatoma cells. 2003 June 19;Volume 545(Issues 2–3):203–208. doi: 10.1016/s0014-5793(03)00535-0. [DOI] [PubMed] [Google Scholar]
- Szepeshazi K, Schally AV, Halmos G, Groot K, Radulovic S. Growth inhibition of estrogen-dependent and estrogen-independent MXT mammary cancers in mice by the bombesin and gastrin-releasing peptide antagonist RC-3095. J Natl Cancer Inst. 1992;84(24):1915–1922. doi: 10.1093/jnci/84.24.1915. [DOI] [PubMed] [Google Scholar]
- Toretsky JA, Helman LJ. Involvement of IGF-II in human cancer. J Endocrinol. 1996;149:367–372. doi: 10.1677/joe.0.1490367. [DOI] [PubMed] [Google Scholar]
- Valentinis B, Baserga R. IGF-1 receptor signalling in transformation and differentiation. Mol Path. 2001;54(3):133–137. doi: 10.1136/mp.54.3.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas S, Asmerom Y, De Leon DD. Resveratrol regulates insulin-like growth factor-II in breast cancer cells. Endocrinology. 2005;146(10):4224–4233. doi: 10.1210/en.2004-1344. [DOI] [PubMed] [Google Scholar]
- Wang J, Green PS, Simpkins JW. Estradiol protects against ATP depletion, mitochondrial membrane potential decline and the generation of reactive oxygen species induced by 3-nitroproprionic acid in SK-N-SH human neuroblastoma cells. J Neurochem. 2001;77(3):804–811. doi: 10.1046/j.1471-4159.2001.00271.x. [DOI] [PubMed] [Google Scholar]
- Webb P, Nguyen P, Valentine C, Lopez GN, Kwok GR, McInerney E, et al. The estrogen receptor enhances AP-1 activity by two distinct mechanisms with different requirements for receptor transactivation functions. Mol Endocrinol. 1999;13(10):1672–1685. doi: 10.1210/mend.13.10.0357. [DOI] [PubMed] [Google Scholar]
- Xu Y, Traystman RJ, Hurn PD, Wang MM. Membrane restraint of estrogen receptor alpha enhances estrogen-dependent nuclear localization and genomic function. Mol Endocrinol. 2004;18(1):86–96. doi: 10.1210/me.2003-0262. [DOI] [PubMed] [Google Scholar]
- Yang SH, Liu R, Perez EJ, Wen Y, Stevens SM, Jr, Valencia T, et al. Mitochondrial localization of estrogen receptor beta. Proc Natl Acad Sci U S A. 2004;101(12):4130–4135. doi: 10.1073/pnas.0306948101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yballe CM, Vu TH, Hoffman AR. Imprinting and expression of insulin-like growth factor-II and H19 in normal breast tissue and breast tumor. J Clin Endocrinol Metab. 1996;81(4):1607–1612. doi: 10.1210/jcem.81.4.8636375. [DOI] [PubMed] [Google Scholar]
- Zhang L, Kashanchi F, Zhan Q, Brady JN, Fornace AJ, Seth P, Helman LJ. Regulation of insulin-like growth factor II P3 promoter by p53: A potential mechanism for tumorigenesis. Cancer Res. 1996;56:1367–1373. [PubMed] [Google Scholar]
- Zhang LJ, Zhan QM, Zhan SL, Kashanchi F, Fornace AJ, Seth P, Helman LJ. p53 regulates human insulin-like growth factor II gene expression through active P4 promoter in rhabdomyosarcoma cells. DNA and Cell Biology. 1998;17:1. doi: 10.1089/dna.1998.17.125. [DOI] [PubMed] [Google Scholar]