Abstract
Protein contributions to the substrate-triggered cleavage of the cobalt-carbon (Co-C) bond and formation of the cob(II)alamin-5′-deoxyadenosyl radical pair in the adenosylcobalamin (AdoCbl)-dependent ethanolamine ammonia-lyase (EAL) from Salmonella typhimurium have been studied by using pulsed-laser photolysis of AdoCbl in the EAL-AdoCbl-substrate ternary complex, and time-resolved probing of the photoproduct dynamics by using ultraviolet-visible absorption spectroscopy on the 10−7 − 10−1 s time scale. Experiments were performed in a fluid dimethylsulfoxide/water cryosolvent system at 240 K, under conditions of kinetic competence for thermal cleavage of the Co-C bond in the ternary complex. The static ultraviolet-visible absorption spectra of holo-EAL and ternary complex are comparable, indicating that the binding of substrate does not labilize the cofactor cobalt-carbon (Co-C) bond by significantly distorting the equilibrium AdoCbl structure. Photolysis of AdoCbl in EAL at 240 K leads to cob(II)alamin-5′-deoxyadenosyl radical pair quantum yields of <0.01 at 10−6 s in both holo-EAL and ternary complex. Three photoproduct states are populated following a saturating laser pulse, and labeled, Pf, Ps, and Pc. The relative amplitudes and first-order recombination rate constants of Pf (0.4-0.6; 40-50 s−1), Ps, (0.3-0.4; 4 s−1) and Pc (0.1-0.2; 0) are comparable in holo-EAL and in the ternary complex. Time-resolved, full-spectrum electron paramagnetic resonance (EPR) spectroscopy shows that visible irradiation alters neither the kinetics of thermal cob(II)alamin-substrate radical pair formation, nor the equilibrium between ternary complex and cob(II)alamin-substrate radical pair, at 246 K. The results indicate that substrate binding to holo-EAL does not “switch” the protein to a new structural state, which promptly stabilizes the cob(II)alamin-5′-deoxyadenosyl radical pair photoproduct, either through an increased barrier to recombination, a decreased barrier to further radical pair separation, or lowering of the radical pair state free energy, or a combination of these effects. Therefore, we conclude that such a change in protein structure, which is independent of changes in the AdoCbl structure, and specifically the Co-C bond length, is not a basis of Co-C bond cleavage catalysis. The results suggest that, following the substrate trigger, the protein interacts with the cofactor to contiguously guide the cleavage of the Co-C bond, at every step along the cleavage coordinate, starting from the equilibrium configuration of the ternary complex. The cleavage is thus represented by a diagonal trajectory across a free energy surface, that is defined by chemical (Co-C separation) and protein configuration coordinates.
Introduction
The first step in the native catalytic cycle of all adenosylcobalamin-dependent enzymes is the thermally activated homolytic cleavage of the cobalt-carbon (Co-C) bond in adenosylcobalamin (AdoCbl; coenzyme B12; Figure 1A), which results in the formation of the cob(II)alamin-5′-deoxyadenosyl radical pair.1-3 Following Co-C bond cleavage, the C5′ radical center of the 5′-deoxyadenosyl moiety migrates through the protein to abstract a hydrogen atom from the substrate, which activates the substrate for rearrangement. High-resolution electron paramagnetic resonance (EPR) spectroscopic studies of the cob(II)alamin-substrate radical pair formed from the substrates, (S)-2-aminopropanol4-7 or aminoethanol,8-10 in the ethanolamine ammonia-lyase (EAL) [EC 4.3.1.7; cobalamin (vitamin B12)-dependent enzyme superfamily; native reaction: conversion of aminoethanol and 2-aminopropanol to the corresponding aldehydes and ammonia11]12,13 from Salmonella typhimurium,1,14,15 have shown that the migration of the C5′ radical center occurs over 5-7 Å. The canonical states and steps in the radical pair separation sequence are depicted in Figure 1B. A long-standing issue in AdoCbl-dependent enzyme catalysis is the molecular mechanism of the >1011-fold16-18 rate acceleration of Co-C bond cleavage,19,20 relative to solution. In particular, the mechanism of the substrate binding-induced transformation (the “substrate trigger”), from the quiescent Co-C bond in the EAL holoenzyme to a state in the EAL/AdoCbl/substrate ternary complex, in which the Co-C bond lifetime is <10−2 s at 298 K,21 has not been characterized experimentally,22 although a mechanism based on X-ray crystallographic structures of EAL has been proposed.15 Here, we apply the photolysis technique to address the nature of the substrate trigger and the contributions of chemical and protein coordinates to cobalt-carbon bond cleavage and radical pair separation in the EAL ternary complex. The ternary complex is prepared in a dimethylsulfoxide (DMSO)/water cryosolvent, and studied at 240 K.21
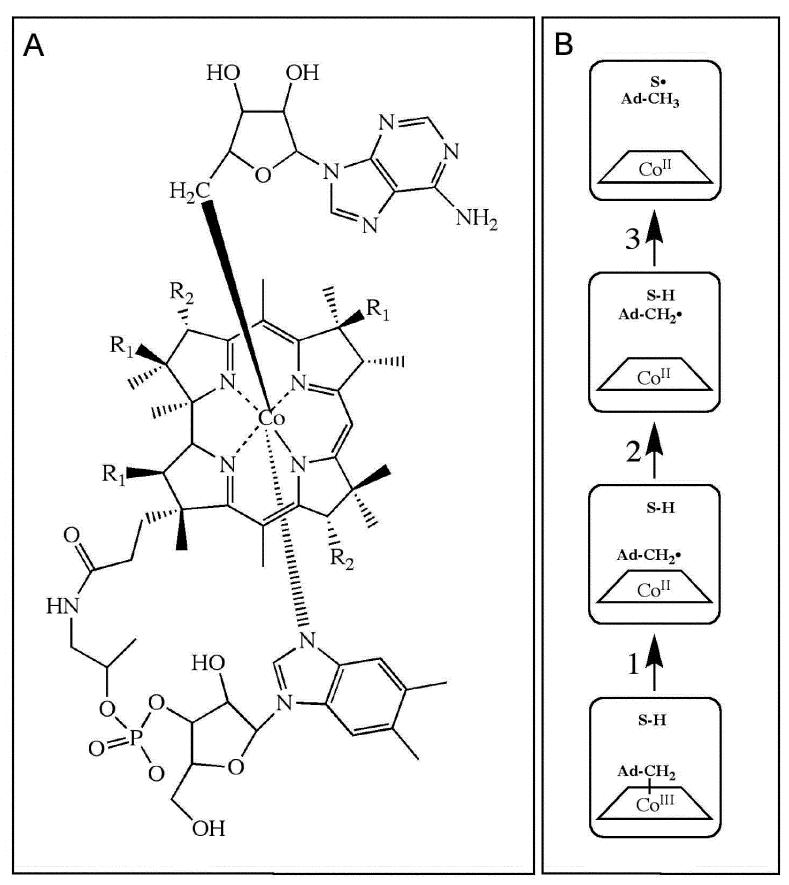
Figure 1.
Depiction of the structure of adenosylcobalamin and canonical states and steps in the radical pair creation and separation reaction sequence in EAL. (A) Structure of adenosylcobalamin. The β-axial ligand is 5′-deoxyadenosyl. The dimethylbenzimidazole α-axial ligand of the coenzyme remains coordinated when the coenzyme is bound to EAL.63,64 R1 and R2 represent acetamide and propionamide side chains, respectively. (B) Canonical states and steps in the native radical pair creation and separation reaction sequence in EAL.1,14 The steps are as follows: (1) Cobalt-carbon bond cleavage, (2) radical pair migration, and (3) hydrogen atom transfer. Substrate-derived species are designated S-H (bound substrate) and S• (substrate radical). The 5′-deoxyadenosyl β-axial ligand is represented as Ad-CH2- in the intact coenzyme, and as Ad-CH2• (5′-deoxyadenosyl radical) or Ad-CH3 (5′-deoxyadenosine) following cobalt-carbon bond cleavage. The cobalt ion and its formal oxidation states are depicted, but the corrin ring and the dimethylbenzimidazole α-axial ligand of the coenzyme are not shown for clarity.
Photolysis induces homolytic cleavage of the Co-C bond of AdoCbl in solution at ambient temperatures, which produces the cob(II)alamin-5′-deoxyadenosyl radical pair, in yields that vary from near unity on the picosecond time scale to 0.2-0.3 on the microsecond time scale.23-25 Figure 2 shows a simplified kinetic scheme of the canonical states and steps involved in the AdoCbl photolysis experiment.26 Following photo-excitation, the excited state photoproducts relax to the ground state in <10−9 s,23,27 forming the geminate cob(II)alamin-5′-deoxyadenosyl radical pair with near unit quantum yield. The excited state formation, decay and sequence of early photoproduct intermediates for alkylcobalamins in solution, have been described in detail by Sension and coworkers.23-25,27-37 In solution, the geminate radical pair exists within a cage of surrounding solvent molecules, and can recombine promptly (geminate recombination; first-order rate constant, kgr), or diffuse apart (cage escape, kce) and recombine on a slower time scale (cage escape recombination, kcer).38 The radical pair formation and decay are detected optically, by monitoring the UV-visible absorption maxima of cobalamin in the Co(III) state (visible wavelength maximum, λmax=528 nm in water) and Co(II) state (λmax=470 nm in water).39 Following AdoCbl photolysis in water, values of kgr and kce are measured to be 1.4×109 s−1 and 5.7×108 s−1, respectively.24,27 A fraction of radical pairs (0.29) escape the solvent cage.24,27
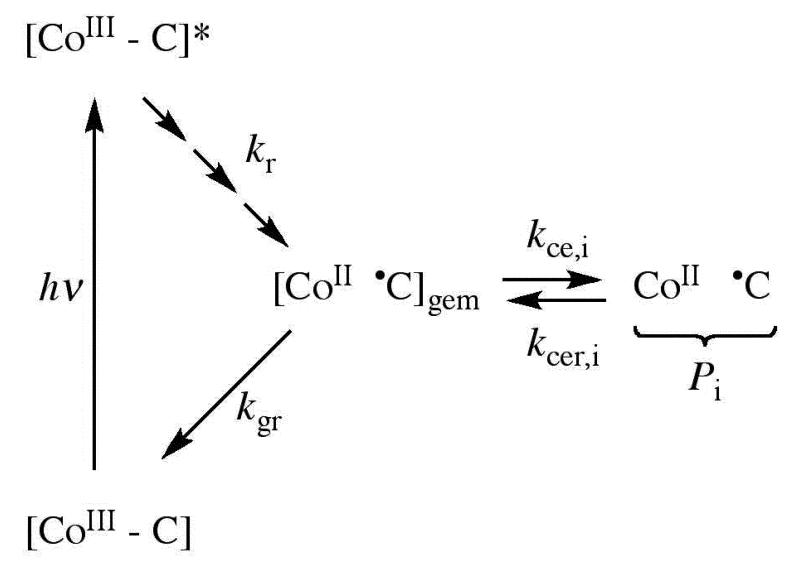
Figure 2.
Simplified schematic diagram of the states and pathways of formation following photolysis of AdoCbl in solution and in holo-EAL. The cobalt in cobalamin and the C5′-methylene center in the 5′-deoxyadenosyl moiety are represented, as follows: [CoIII-C], intact coenzyme; [CoIII-C]*, excited singlet state; [CoII •C]gem, geminate radical pair; CoII •C, cage escaped radical pair. Intermediate excited and relaxed states,27,32 which are not shown, are represented by the sequence of arrows leading from [CoIII-C]*. Rate constants are defined as follows: kr, excited state to ground state relaxation; kgr, geminate recombination; kce, cage escape; kcer, reformation of geminate radical pair from cage escaped radical pair. The subscript, i, refers to three different cage escaped photoproduct radical pair species, Pi, that were identified in holo-EAL.26
Photolysis of protein-bound AdoCbl provides a method for overcoming the large Co-C bond dissociation energy of ~30 kcal/mol 40 on a sub-nanosecond time scale. Calculations for methylcobalamin photolysis suggest that the separation of Co(II) and C5′, rCoC, in the ground state, following photocleavage and excited state relaxation, is 2.7-3.0 Å.41 These rCoC values correspond to calculated Co-C bond cleavage extents of >80%.42-44 Thus, photolysis creates a mimick of the thermal cob(II)alamin-5′-deoxyadenosyl radical pair on the nanosecond time scale, whose yield and fate can be used to probe the mechanism of the thermal radical pair generation, stabilization, and separation processes in the protein. If the “substrate trigger” involves a substrate-induced change in protein structure to a state, in which the barrier to recombination is increased, the barrier to further radical pair separation is decreased, or the cob(II)alamin-5′-deoxyadenosyl radical pair is stabilized, or a combination of the these effects, then we hypothesize that the yield of the photoproduct will increase in the ternary complex, relative to holo-EAL. In this case, photo-induced cleavage of the Co-C bond would be sufficient to significantly populate the putative stable cob(II)alamin-5′-deoxyadenosyl radical pair state. The photolysis measurements are performed on the time scale of the thermal Co-C bond cleavage and cob(II)alamin-substrate radical pair formation reaction, which is characterized by τobs=8.3×102 s at 240 K.21 In the DMSO/water cryosolvent system over 234-248 K, a quasi-equilibrium between the ternary complex-intact coenzyme state and the cob(II)alamin-substrate radical pair state is established, because the rearrangement reaction of the substrate radical to form the product radical is >102-fold slower, relative to the formation of the cob(II)alamin-substrate radical pair.21 At 240 K, the equilibrium fraction of the ternary complex-intact AdoCbl state is 0.57.21 Therefore, we assume that any protein structural changes associated with Co-C bond cleavage catalysis will be manifested under the conditions of the photolysis and continuous-wave absorption spectroscopy experiments.
We previously used pulsed-laser photolysis of AdoCbl in holo-EAL and in an inhibitor-holo-EAL complex at room temperature, to prepare the cob(II)alamin-5′-deoxyadenosyl radical pair population, followed by a visible absorption probe of its time evolution on the 10−7 to 10−2 s time scale.26 We observed that AdoCbl binding to EAL lowered the quantum yield of the room temperature photolysis, from 0.23 in solution, to a value of 0.08, at 10−6 s. Similar results were obtained for the holo-enzyme of AdoCbl-dependent glutamate mutase (GluM), where a quantum yield of 0.05 was observed at ≤9×10−9 ns.45,46 In GluM, this was caused primarily by a decrease in kce to 5-6 ×107 s−1,45,46 relative to the value for pure water, because the protein reduced kgr by only 30%, relative to pure water.29 Three different cage escape photoproduct states were identified, following room temperature photolysis of holo-EAL (Pi, in Figure 2). Contrary to expectation, a substrate analog inhibitor, (S)-1-amino-2-propanol, which binds at the substrate binding site, further decreased the quantum yield, to 0.04.
Here, we report the results of visible light, pulsed-laser photolysis of the EAL ternary complex, formed with the substrate, (S)-2-amino-1-propanol. The ternary complex is formed by mixing holo-EAL and substrate in an optically transparent, fluid dimethylsulfoxide (DMSO)/water cryosolvent.21,47 We observe that substrate binding to holo-EAL under conditions of kinetic competence for thermally-activated turnover at 240 K21 does not significantly perturb the absorption spectrum of AdoCbl, and that the optically-detected photoproduct amplitudes and recombination rates in holo-EAL and the ternary complex do not differ significantly. Continuous visible irradiation also has no effect on the kinetics of thermally-activated cob(II)alamin-substrate radical pair formation, or the subsequent equilibrium level of this state, at 246 K, as detected by EPR spectroscopy. Overall, the results do not support the simple hypothesis of a substrate binding-induced change in the protein structure to a new state, which is capable of prompt stabilization (<10 ns) of the radical pair photoproduct. Therefore, we conclude that such a change in protein structure, which is independent of changes in the AdoCbl structure, and specifically the Co-C bond length, is not a basis of Co-C bond cleavage catalysis. The results suggest that, following the substrate trigger, the protein interacts with the cofactor to contiguously guide the cleavage of the Co-C bond, at every step along the cleavage coordinate, starting from the equilibrium configuration of the ternary complex. The cleavage is thus represented by a diagonal trajectory across a free energy surface, that is defined by chemical (Co-C separation) and protein configuration coordinates.
Materials and Methods
Materials
All chemicals were obtained from commercial sources, and were used without further purification. Adenosylcobalamin, (S)-1-amino-2-propanol and (S)-2-amino-1-propanol were purchased from Sigma-Aldrich Chemical Co. EAL was purified from the Escherichia coli over expression strain incorporating the cloned S. typhimurium EAL coding sequence48 essentially as described,49 with the exception that the enzyme was dialyzed against buffer containing 100 mM HEPES (pH 7.5), 10 mM potassium chloride, 5 mM dithiothreitol, 10 mM urea, and 10% glycerol,50 and neither Triton X-100 nor urea were used during the purification. Enzyme activity was determined as described51 by using the coupled assay with alcohol dehydrogenase/NADH. The specific activity of the purified enzyme with aminoethanol as substrate was 20-30 μmol/min/mg.
Enzyme Sample Preparation
Adenosylcobalamin, (S)-1-amino-2-propanol and (S)-2-amino-1-propanol were purchased from Sigma-Aldrich Chemical Co. The preparation of EAL in the cryosolvent system has been described in detail.21 Briefly, samples with a 2.0-fold excess of EAL enzyme active sites (60 μM active site concentration) with cofactor (30 μM) were prepared in water buffered with 10 mM potassium cacodylate (pH 7.1), to form holoenzyme at 295 K. A cryosolvent pH value of 7.1 at this stage leads to a pH value of 7.5 ±0.4 at 230-240 K in the low temperature cryosolvent.21 Potassium cacodylate buffer was used, owing to the relatively small temperature dependence of the pKa47 The solution was sonicated at 277 K to minimize light scattering. Small volumes of 70% (volume/volume, v/v) dimethysulfoxide (DMSO)/water, which represented less than 15% of the volume of the holoenzyme-containing solution, were then added with continuous slow stirring, in four steps, at decreasing temperatures from 273 to 240 K. The solution was then transferred, with the protection of cold isopentane, to a quartz cuvette containing a solution of 54% (v/v) DMSO/water at 240 K, to achieve a final 50% (v/v) DMSO/water solution of EAL holoenzyme. The quartz cuvette was positioned in the low temperature optical cryostat, which was mounted inside either the transient, or the static, absorption spectrometer. The substrate, (S)-2-amino-1-propanol, or substrate analog, (S)-1-amino-2-propanol, at a concentration of 5 mM final concentration in 50% (v/v) DMSO/water cryosolvent, was introduced after further lowering of the temperature to 230 K. The system was then incubated at 230 K for at least 5 min to allow substrate equilibration with the active site. All procedures were performed under a dim red safe light.
Adenosylcobalamin Sample Preparation
Anaerobic solutions of AdoCbl in cryosolvent were prepared by nitrogen gas bubbling for 15-30 min at 295 K, in a 3 ml quartz anerobic cuvette through a septum. A positive pressure of nitrogen (3 psi) was maintained inside the cuvette during subsequent manipulations. The temperature of the sample was then lowered to either 230 (static spectrum acquisition experiments) or 240 K (photolysis experiments).
Low Temperature Optical Cryostat
A low temperature (77 K to 300 K) cryostat system was designed and constructed for optical monitoring of the cryosolvent system. Dry nitrogen gas was flowed through a stainless-steel heat exchanging coil which was immersed in liquid nitrogen. A home-made temperature controller, based on a 1/16 din MICROMEGA® autotune PID temperature/process controller (Omega CN77000), heats the cold nitrogen gas to the desired temperature by using an electric heating element along the nitrogen gas pathway. The temperature is monitored by using a T-band thermocouple (Omega 5STRC-TT-T-30-36). The nitrogen gas flows through a brass cuvette holder where the sample is held in a quartz cuvette (Hellma QS 282 1.000 ml). An outer housing is used to eliminate the moisture condensation along the optical path. The pre-cooled nitrogen gas is used to purge the inside of the outer housing after passing through the sample holder. Two on-axis heated quartz windows provide optical access to the cooled sample. The system has >90% transmittance from 250 nm to 2500 nm at 230 K. The top of the housing is covered by a glass hatch for easy sample access. The temperature stability at 230 K is ± 1 K over a 2 h time interval. This system is used with both the transient and static absorption spectrometers.
Low Temperature Static Absorption Spectra
Low temperature static absorption spectra from 300 nm to 650 nm were collected by using a Shimadzu UV-1601 absorption spectrometer (0.5 nm wavelength accuracy), which was fitted with the low temperature optical cryostat. The average of three spectra were taken for all low temperature spectra. Three enzyme sample spectra without cofactor were taken and averaged for a background scattering baseline, and this baseline was later subtracted from the holoenzyme and ternary complex spectra. All measurements were performed at 230 ±1 K.
Low Temperature Transient Absorption Spectroscopy
Transient absorption measurements were made by using the low temperature optical cryostat, and a transient spectrophotometer of home design and construction, which has been previously described.26 The transient absorption spectrometer has a sensitivity to change in absorbance (A) of 2×10−3 over 300-650 nm, and a deadtime of ≤20 ns. The second harmonic output (532 nm) of a Nd:YAG laser (SpectraPhysics GCR-10; 10 ns pulse width), with pulse energy adjusted by a glan prism polarizer/half-wave plate, was used as actinic source. The spectrometer's pump and probe pulse timing sequences and data collection were controlled by a Macintosh computer with LabVIEW software (National Instruments) via a GPIB/IEEE-488.2 interface. Data fitting and analysis was performed by using Matlab (Natick, MA) or OriginPro (OriginLab Corporation, Northampton, MA), routines that were run on PC computers. Transient absorption of the cob(II)alamin state was monitored at 470 nm (probe) following laser pulse photolysis at 532 nm. Measurements were made with a 100 μs dwell time between acquired data points and a corresponding time constant for the detector of 10 μs. The average of 12 photolysis events was taken for each sample. All measurements were performed at 240 ±1 K.
EPR Spectroscopy of Cob(II)alamin-Substrate Radical Pair Formation
X-band continuous-wave EPR spectra were obtained by using a Bruker E500 ElexSys EPR spectrometer, equipped with a Bruker ER4123 SHQE X-band cavity. Temperature was controlled with a Bruker ER4131VT liquid nitrogen/gas flow cryostat system, with ER4121VT-1011 evaporator/transfer line, ER4121VT-1013 heater/thermocouple, and 26 liter liquid nitrogen reservoir. EAL samples in 41% v/v DMSO/water cryosolvent were prepared for EPR spectroscopy in 2 mm outer diameter EPR tubes (Wilmad/Lab Glass, 712-SQ-250M), as described briefly above, and previously.21 Holo-EAL was mixed with the substrate, (S)-2-amino-1-propanol, at 230 K. As described in Results, photolysis of the ternary complex at 230 K was carried out by using the focused 532 nm output of the pulsed Nd-YAG laser (50 mJ/pulse, 10 Hz, 90 seconds). A subsequent experiment was performed in which the reaction of the ternary complex to form the cob(II)alamin-substrate radical state was initiated by a temperature step from 230 K to 246 K. During the rise of the cob(II)alamin-substrate radical EPR signal, the sample was subjected to a protocol of intermittent irradiation by using the filtered output (250 nm – 380 nm) of a 200 W mercury lamp (Oriel Optics Corp., C-60-30). All samples where frozen in liquid nitrogen following photolysis and EPR measurements were performed at 120 K, under dim light.
Quantum Yield Measurements
Quantum yield measurements were obtained by using laser pulse energies of ≤3 mJ, in order to eliminate multiple photon absorption by AdoCbl during the laser pulse. The 470 nm probe beam was used to detect the formation of cob(II)alamin. Quantum yield measurements were made by taking the average of 24 individual photolysis events, with a 1.0 μs dwell time and a time constant of 0.1 μs. The quantum yield (ϕ) is defined as the concentration of cob(II)alamin photoproduct formed by the laser pulse, divided by the absorbed photon (hν) concentration, [hν]abs. Details of the calculation of ϕ values have been described.26
Temperature-Dependence of the First-Order Rate Constant
The temperature dependence of the first order rate constant, k, is given by the Arrhenius expression:52
| (1) |
where Ea is the activation energy, R is the gas constant, and A is a prefactor that represents the value of k as Ea→0. The value of A is typically approximated as , where kB and h are the Boltzmann and Planck constants, respectively, and T is absolute temperature.
RESULTS
UV/visible Absorption Spectra of Holoenzyme and Ternary Complex at 230 K
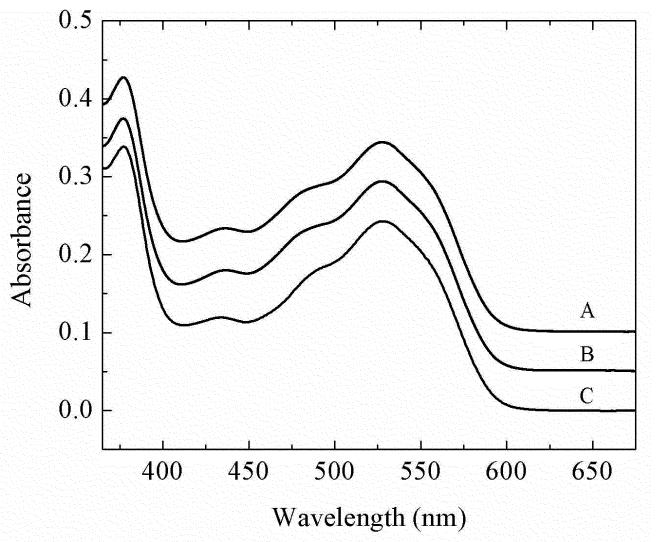
The absorption spectra of AdoCbl at 230 K in the 50% v/v DMSO-water cryosolvent, and for AdoCbl in holo-EAL and in the ternary complex with (S)-2-amino-1-propanol bound, are presented in Figure 3. The AdoCbl spectra show the characteristic visible wavelength absorption maximum (α/β band) and the UV maximum (γ-band).19 In the cryosolvent system at 240 K, the λmax value of the α/β band was found to be 528.3 ± 0.3 nm for free AdoCbl, 527.7 ± 0.2 nm for holo-EAL, and 527.9 ± 0.3 nm for the EAL ternary complex. The λmax value of the γ band was found to be 377.0 ± 0.1 nm for free AdoCbl, 376.9 ± 0.1 nm for holo-EAL, and 376.9 ± 0.1 nm for the EAL ternary complex. There is small hypsochromic shift of 0.6 ±0.4 nm of the α/β band following AdoCbl binding to EAL, which is caused by the different cofactor environments in aqueous solution and in EAL, as found for other AdoCbl-dependent enzymes.19,20,53 No significant shift in the positions or absorbance values of the visible or UV absorption maxima of AdoCbl are observed following binding of the substrate to holo-EAL.
Figure 3.
UV/visible absorption spectra of adenosylcobalamin in free solution, and in EAL in the holo-enzyme and ternary complex. The spectra were collected at 230 K for samples in the 50% dimethylsulfoxide/water cryosolvent system. (A) AdoCbl in EAL with bound (S)-2-amino-propanol substrate (ternary complex). (B) AdoCbl in EAL (holoenzyme). (C) AdoCbl in solution. Concentrations were as follows: 60 μM EAL, 30 μM AdoCbl, 5 mM (S)-2-amino-propanol.
Evidence for binding of the cofactor and substrate to the active site in the low temperature cryosolvent system is as follows. It has been established that AdoCbl binds relatively tightly to EAL at ambient temperatures (KD≈1 μM),49,54 and remains bound during multiple turnovers of the enzyme.14,55 In the cryosolvent system preparation procedure, holo-EAL is formed at 295 K. A stable holo-EAL complex in the low-temperature cryosolvent system is shown by the difference in photoproduct yield and transient decay kinetics for AdoCbl in DMSO/water solution and in EAL (Figure 4, A and D; described below), and by the reproduction of the characteristic room temperature UV/visible spectra for the free solution and EAL-bound forms of aquocob(III)alamin (Figure S1, Supporting Information). Our previous results21 indicate that the substrate is bound to the protein in the ternary complex, under the conditions of the static and transient spectroscopy experiments. We demonstrated that the kobs value for the monoexponential thermal cob(II)alamin-substrate radical pair formation reaction, and the final, equilibrium concentration of the cob(II)alamin-substrate radical pair, are independent of the substrate concentration over a range of substrate/active site ratios of 1 to 100.21 These results were shown to be consistent with (S)-2-aminopropanol binding at the substrate site, and the condition, KD << [active sites], in the low-temperature DMSO/water cryosolvent system.21 Photolysis measurements were performed under conditions of kinetic competence for thermal substrate-induced Co-C bond cleavage, either for times short compared to the thermal reaction (t< τobs=8.3×102 s), or at equilibrium (t>3τobs; fraction of intact AdoCbl-ternary complex of 0.57, relative to total concentration of ternary complex).
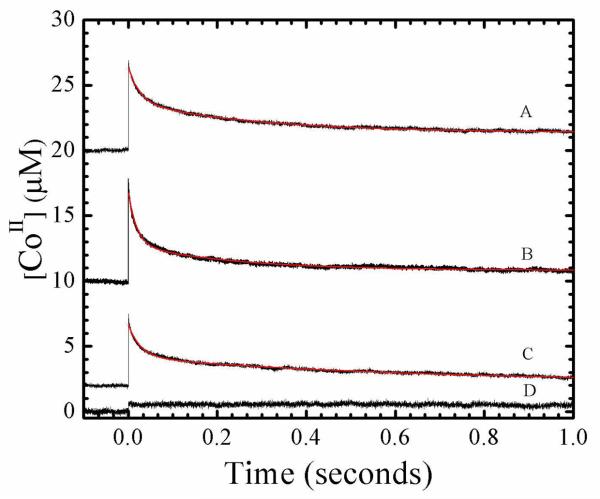
Figure 4.
Time dependence of cob(II)alamin concentration following pulsed laser photolysis of adenosylcobalamin at 240 K, and overlaid best-fit biexponential plus constant decay functions (red, smooth curves). Data was collected at times, t < τobs. Fitting parameters are collected in Table 1. (A) Holo-EAL in aerobic solution. (B) Holo-EAL in aerobic solution, with bound (S)-2-amino-1-propanol bound (ternary complex). (C) Holo-EAL in aerobic solution, with bound (S)-1-amino-2-propanol bound (inhibitor complex). (D) Anaerobic solution. Concentrations were as follows: 60 μM EAL, 30 μM AdoCbl, 5 mM (S)-2-amino-propanol, 5 mM (S)-1-amino-2-propanol.
Quantum Yield of Cob(II)alamin Formation following Low Temperature Photolysis of AdoCbl in Solution, Holoenzyme and Ternary Complex
The quantum yield of AdoCbl photolysis at 240 K was measured on the 10−6 s time scale by using low, sub-saturating pulse energy (≤3 mJ) excitation from the 532 nm output of the pulsed-Nd-YAG laser, and a continuous-wave probe for cob(II)alamin formation at 470 nm. The low energies were selected to prevent multiple photon absorption by AdoCbl. The ratio of the concentration of AdoCbl to EAL active sites was ≤0.5, to avoid possible interference from photolysis of free cofactor. The measured quantum yield at 10−6 s for AdoCbl photolysis in cryosolvent is <10−2, which is the detection limit of the instrument. Measurements were performed for times, t< τobs=8.3×102 s, for thermal Co-C bond cleavage at 240 K.21 Measurements performed for t>3τobs, which corresponds to equilibrium between the AdoCbl-intact state of the ternary complex and the cob(II)alamin-substrate radical pair, gave the same result. The small quantum yield of AdoCbl photolysis in cryosolvent is consistent with the viscosity dependence of kgr and kce.56 The quantum yield was also <10−2 for AdoCbl in holo-EAL and in the ternary complex. Therefore, binding of the substrate does not enhance the quantum yield above the level of 10−2.
Time Dependence of Photoproduct Cob(II)alamin Recombination Following Photolysis Under Saturating Pulsed-Laser Irradiation
Figure 4 shows the transient decay kinetics of cob(II)alamin at 240 K in the cryosolvent solution on the milliseconds time scale, following photolysis of AdoCbl in solution, in holo-EAL, in holo-EAL with the inhibitor (S)-1-amino-2-propanol bound, and in the ternary complex with (S)-2-amino-1-propanol bound. A relatively high laser pulse energy of 10 mJ/pulse was used to enhance the population of cob(II)alamin, by photolyzing AdoCbl that had undergone geminate recombination on the timescale of the 10 ns laser pulse width. Measurements of the decay on shorter time scales of 1.0×10−7 s do not show additional kinetic transients. The kinetics in Figure 4 therefore represent the events that occur on the time scale of ≥1.0×10−7 s for each condition.
The transient decays for holo-EAL, ternary complex, and holo-EAL with inhibitor bound (Figure 4 A-C) were well fit by using a biexponential plus constant function. The biexponential and monoexponential plus constant functions did not provide satisfactory fits to the transient decays (Figure S2 and Figure S3, Supporting Information). The results for holo-EAL in Table 1 show that the normalized relative amplitudes of the fast decay population (Pf), the slow decay population (Ps), and the constant amplitude population (Pc) are 0.57, 0.33, and 0.11, respectively. Table 1 also shows that binding of the substrate or inhibitor to holo-EAL does not significantly alter the relative populations, or significantly influence the first-order rate constants for the fast (kf) and slow (ks) decay populations, relative to holo-EAL.
Table 1.
Relative cob(II)alamin photoproduct amplitudes, limits on quantum yield and cage escape rate constant values, and observed recombination decay rate constants for cage escape populations at 240 K, following photolysis of AdoCbl with a saturating laser flash, in holo-EAL, holo-EAL with bound substrate, (S)-2-amino-propanol (ternary complext), and holo-EAL with bound substrate analog, (S)-1-amino-2-propanol. Standard deviations for determinations on at least three separate samples are shown in parentheses.
| Condition |
Pi |
Arela |
ϕ
ib |
kce,ic (s−1) |
kdecay,i (s−1) |
|---|---|---|---|---|---|
| EAL•AdoCbl | P f | 5.7 (1.1) ×10−1 | <6×10−3 | <5.8×105 | 5.0 (1.4) ×101 |
| P s | 3.3 (0.6) ×10−1 | <3×10−3 | <2.9×105 | 4.3 (1.3) ×100 | |
| P c | 1.1 (0.7) ×10−1 | <1×10−3 | <9.6×104 | - | |
| EAL•AdoCbl | P f | 6.1 (0.5) ×10−1 | <6×10−3 | <5.8×105 | 6.0 (1.4) ×101 |
| •Substrate | P s | 3.0 (0.3) ×10−1 | <3×10−3 | <2.9×105 | 4.9 (2.0) ×100 |
| P c | 0.9 (0.5) ×10−1 | <1×10−3 | <9.6×104 | - | |
| EAL•AdoCbl | P f | 5.7 (0.4)×10−1 | <6×10−3 | <5.8×105 | 6.0 (2.1) ×101 |
| •Inhibitor | P s | 3.4 (0.7) ×10−1 | <3×10−3 | <2.9×105 | 4.0 (1.9) ×100 |
| P c | 0.9 (0.5) ×10−1 | <1×10−2 | <9.6×104 | - |
Normalized relative amplitude at 10−6 s, obtained by using saturating laser flash (10 mJ).
Upper limit on quantum yield, obained from relative amplitude and upper limit on absolute quantum yield at 10−6 s of <1×10−2.
Upper limit on cage escape rate constant, obtained by using kgr=9.5×107 s−1.
Comparable decay kinetics and relative amplitudes of the Pi were obtained for measurements performed at times of t<τobs and t>3τobs. Thus, the presence of the bound substrate or inhibitor does not significantly effect either the Pi or the kdecay,i values, relative to holo-EAL.
Visible Light Irradiation of the EAL Ternary Complex Does Not Generate the Cob(II)alamin-Substrate Radical Pair
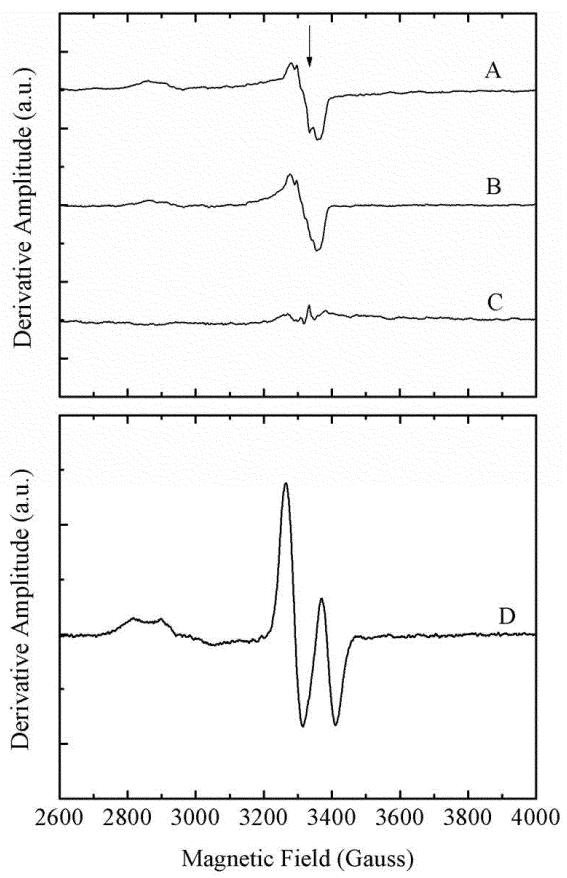
The ability of long-term irradiation to generate the cob(II)alamin-substrate radical pair was addressed by using EPR spectroscopy. Samples of holo-EAL and the ternary complex were irradiated for 1.5 min at 230 K by using the output of the pulsed-Nd-YAG laser at 532 nm. The relatively low temperature was chosen to suppress thermal radical pair formation, during the irradiation. The individual photoproduct spectra and the difference spectrum for ternary complex minus holo-EAL are shown in Figure 5A-C. The cob(II)alamin-substrate radical pair spectrum, generated by the native, thermally activated pathway, by using (S)-2-aminopropanol substrate, is presented in Figure 5D. Comparison of the spectra in Figure 5 shows that irradiation does not generate a significant level of cob(II)alamin-substrate radical pair state. Therefore, light does not drive the productive formation of the ultimate product of the radical pair separation reaction.
Figure 5.
EPR spectra for EAL ternary complex, holo-EAL, and difference spectrum for ternary complex minus holo-EAL, following long-term photolysis, and control EPR spectrum of the (S)-2-amino-2-propanol-generated cob(II)alamin-substrate radical pair generated by the native, thermal reaction. Note that the ordinate scale in (A)-(C) is expanded by a factor of 80, relative to the scale for (D). (A) Holo-EAL. (B) EAL ternary complex. (C) Difference EPR spectrum; ternary complex (B) minus holo-EAL (A). (D) Cob(II)alamin-substrate radical pair EPR spectrum, following 15 min incubation at 275 K (EAL active site concentration, 150 μM; AdoCbl, 300 μM; (S)-2-amino-1-propanol, 10 mM). Photolysis conditions: EAL active sites, 120 μM; AdoCbl, 60 μM. Iirradiation for 1.5 min with the 532 nm output of the pulsed Nd-YAG laser (10 Hz, 50 mJ/pulse) at 230 K. EPR conditions: Temperarure, 120 K; microwave frequency, 9.436 GHz; microwave power, 20 mW; magnetic field modulation, 10 Gauss peak-peak; modulation frequency, 100 kHz; field sweep rate, 1.5 Gauss s−1; time constant, 200 ms; average of 2 sweeps, minus average of 2 baseline spectra.
Effect of Photolysis on Thermally-Activated Cob(II)alamin-Substrate Radical Pair Formation
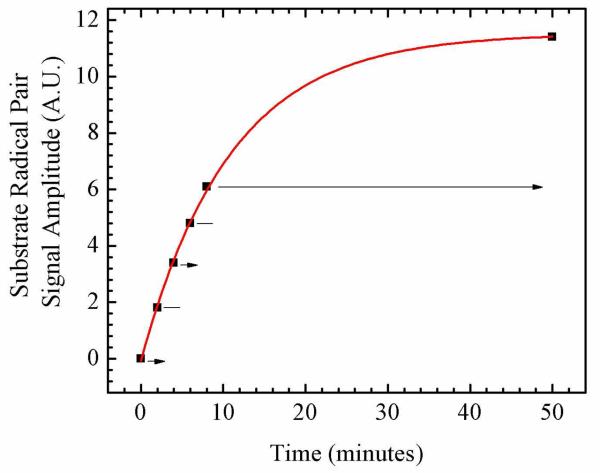
The EAL ternary complex was irradiated during the thermal cob(II)alamin-substrate radical pair formation reaction, at 246 K, in order to address effects of photolysis on the first-order rate constant for radical pair formation, and on the long-time equilibrium between the ternary complex and the radical pair states. The formation of the cob(II)alamin-substrate radical pair was detected by CW-EPR spectroscopy. The experiment was performed by using alternating light and dark time periods, that were applied during the rise of the radical pair, and following the establishment of equilibrium. This avoided uncertainties caused by comparison of different samples. The kinetics of cob(II)alamin-substrate radical formation obtained by using this protocol are shown in Figure 6. No distinguishable effect of continuous-wave visible irradiation was observed on the rate of cob(II)alamin-substrate radical pair formation. Irradiation also did not alter the long-time equilibrium populations of the ternary complex and the radical pair.
Figure 6.
Time-dependence of the EPR amplitude of the cob(II)alamin-substrate radical pair state in EAL in the cryosolvent system at T=245 K, following temperature-step initiation of reaction. The amplitude is given by the difference between the first peak and second trough amplitudes, as shown in Figure 5B. The intermittent irradiation protocol is denoted, as follows: Arrows represent incubation periods under illumination, and lines represent dark incubation periods. The experimental data points are overlaid with the best-fit exponential growth function (solid curve; kobs= 1.53×10−3 s−1). EPR conditions are as described in the legend to Figure 5.
DISCUSSION
Substrate Binding to Holo-EAL Does Not Activate the Co-C Bond for Cleavage by Significantly Distorting AdoCbl Structure
Brunold and coworkers have performed experimentally calibrated, time-domain density functional theory (TD-DFT) calculations, which predict significant spectral shifts of the α/β band maximum of AdoCbl, in response to relatively small changes in compression (approximately +15 nm/0.1 Å) or extension (approximately 115 nm/0.1 Å) of the Co-C bond length, and opposing shifts of comparable magnitude for changes in the Co-Naxial bond length.19 The spectra in Figure 3 for holo-EAL and the ternary complex show that the presence of bound substrate does not induce significant shifts in the α/β band λmax, or significant changes in the wavelength maxima and absorbance values, of other principal UV/visible absorption bands of the bound AdoCbl cofactor. Our results for the ternary complex are obtained under conditions in which the mechanisms that promote the thermal cleavage of the Co-C bond cleavage are poised.21 Therefore, the EAL protein does not activate the Co-C bond for cleavage by significantly distorting the structure of the AdoCbl cofactor. This result is consistent with studies of other AdoCbl-dependent enzymes. Ultra-violet/visible absorption, magnetic circular dichroism (MCD) and resonance Raman studies showed that the protein in AdoCbl-dependent methylmalonyl-CoA mutase (MMCM) does not significantly distort AdoCbl in the holo-enzyme, or in holo-enzyme with bound substrate analogs.19 An absence of Co-C bond length changes by the protein in MMCM was also concluded from infrared studies.57 Picosecond optical studies suggested an absence of structural perturbations of the Co-C bond in glutamate mutase (GM).45,46 Therefore, the substrate trigger in AdoCbl-dependent enzymes does not appear to be mediated by protein-induced structural distortion of the cofactor.
Substrate Binding to Holo-EAL Does Not Elicit Prompt Stabilization of the Cob(II)alamin-Radical Pair Photoproduct
Table 1 shows the relative amplitudes of the cob(II)alamin-radical pair states on the 10−6 s time scale, following saturating laser pulse photolysis of the following enzyme states: holo-EAL, inhibitor-bound holo-EAL, and the EAL ternary complex. The relative amplitudes of each of the photoproduct populations, Pf, Ps, and Pc, are comparable for each enzyme state. The upper limits on the absolute quantum yields, ϕi, for each population, Pi, which are presented in Table 1, are also uniform and small for each enzyme state. In combination, these results suggest that the binding of substrate to holo-EAL does not induce, or “switch,” the protein to a new state, in which a cob(II)alamin-5′-deoxyadenosyl radical pair, in a configuration characteristic of the geminate radical pair photoproduct, is promptly stabilized. Therefore, a substrate binding-induced, protein-imposed barrier to recombination, a protein-mediated lowering of the free energy of the cob(II)alamin-radical pair state, or both, are not observed. Thus, the substrate does not induce protein-mediated mechanisms which promptly stabilize the photoproduct over Co(II)-C5′ separation distances, rCoC, in the range from >2.0 Å (greater than the Co-C bond length) to approximately 4 Å (the distance at which Co-C bond scission is >90% complete 42-44).
The Pi states may originate from shallow local minima at rCoC values of 2.613.0 Å, which capture the cob(II)alamin-5′-deoxyadenosyl radical pair photoproduct. States of this type may be similar to those found for the 4′-5′-dehydroadenosylcobalamin cofactor in AdoCbl-dependent diol dehydratase, which traps (by internal allyl radical stabilization) the 4′-5′-dehydroadenosyl radical thermal cleavage product.58 In the EAL ternary complex with AdoCbl, the Pi states could represent a small angular excursion about the ribose-adenine bond, or small degree of ribose ring flexure, which lead to weak,59 favorable interactions with the protein. The low quantum yield values and rapid cage escape-recombination rates suggest that the Pi states are not on the native path of Co-C bond cleavage (they correspond to non-native, “Path 1” trajectories, as described, below).
Substrate Binding to Holo-EAL Does Not Significantly I nfluence Cage Escape of the Radical Pair
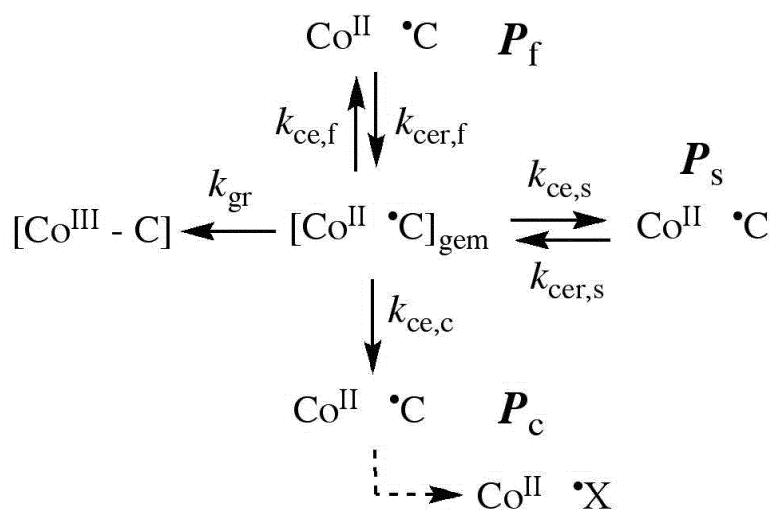
In solution, the existence of alkycobalamin radical pair photoproducts on the ≥10−6 s time scale requires escape from the geminate, caged radical pair state.27,28 The cage escape yield is determined by competition between geminate recombination and cage escape processes, as described by the classical general description.27,28,38 Our specific model for the EAL reaction, which is based on photolysis studies of holo-EAL and inhibitor-holo-EAL at 295 K,26 is shown in Figure 7. The absence of a measurable quantum yield for photolysis at 240 K allows only limits to be placed on the values of the first-order rate constant for cage escape, kce,i, for the different Pi. An upper limit on the kce,i value for each each Pi in each EAL state is estimated by using the corresponding limiting ϕi values, and the following system of equations:26
| (2) |
Figure 7.
Proposed kinetic scheme for reactions of the cob(II)alamin-5′-deoxyadenosyl radical pair states following photolysis of adenosylcobalamin in EAL. First-order rate constants and different photoproduct populations are described in the text. X• indicates a secondary radical reaction product.
In Eq. 2, the index, i, corresponds to f, s, or c, and the quantum yield values for Pf, Ps, and Pc, are ϕf, ϕs, and ϕc, respectively. The geminate recombination rate constant in the EAL ternary complex at 240 K is estimated to be kgr=9.5×107 s−1, based on extrapolation by using the Arrhenius relation [Eq. 1] and an activation energy of 5.2 kcal/mol, which corresponds to the rate of kgr=1.0×109 s−1 determined at 298 K.29 The limiting values of the kce,i are presented in Table 1. The kce,i values for holo-EAL and inhibitor-bound holo-EAL do not differ significantly from the values for the ternary complex. Therefore, substrate binding to holo-EAL does not significantly influence cage escape of the photoproduct radical pair.
The values of the activation energy for cage escape, Ea,ce, at 240 K are calculated to be 8 kcal/mol for each Pf and Ps, and 7-8 kcal/mol for each Pc. These Ea,ce values for 240 K match the values reported for holo-EAL and inhibitor-bound holo-EAL at 298 K. This suggests that the mechanisms that govern cage escape of the photoproduct geminate radical pair at 240 and 298 K are the same.
Substrate Binding to Holo-EAL Does Not Significantly Alter Stabilization of the Cage-Escaped Cob(II)alamin-Radical Pair Photoproduct
Comparison of the observed cob(II)alamin photoproduct decay rate constants, kdecay,i, in Table 1 for the ternary complex, relative to holo-EAL and inhibitor-bound holo-EAL, provides an assessment of the microscopic effects of substrate binding on the protein that influence the stability of the cage-escaped radical pair states. The kdecay,i values approximate well the values of the corresponding cage escape recombination rate constants, kcer,i, which are defined in Figure 2 and Figure 7, because kdecay,i << kgr. Table 1 shows that the kdecay,i≈kcer,i values for the three populations, Pf, Ps, and Pc, for each EAL state are comparable. Therefore, the binding of substrate to holo-EAL does not cause significant changes in the active site structure, which influence the stability of cage-escaped cob(II)alamin-5′-deoxyadenosyl radical pairs. This includes any contribution to “trapping” of the radical pair state by reaction of the cob(II)alamin-5′-deoxyadensosyl radical pair with substrate to form the cob(II)alamin-substrate radical pair, because no detectable photo-induced cob(II)alamin-substrate radical pair formation is observed by EPR spectroscopy. Over rCoC values of approximately 4-6 Å, which correspond to the full extent of the radical pair separation coordinate,4-7,15 the binding of substrate to holo-EAL does not induce factors that stabilize the photo-generated radical pair state.
Models for the Substrate-Initiated Co-C Bond Activation and Cleavage
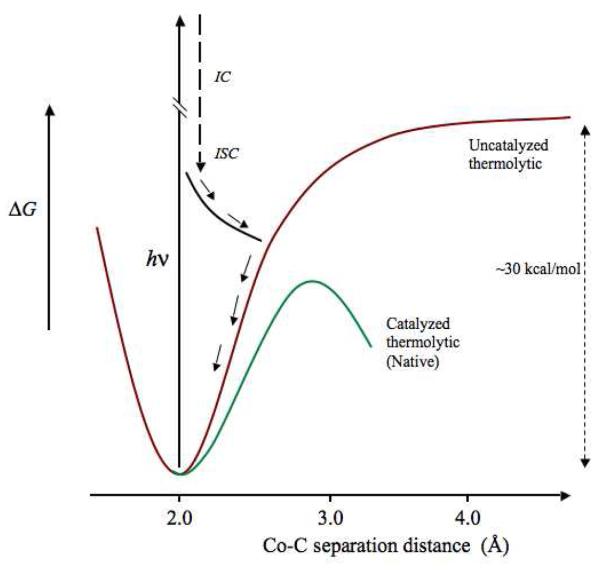
Hypothetical free energy curves and pathways for the photolytic and thermal Co-C bond cleavage and radical pair separation are depicted on a 1-dimensional reaction coordinate in Figure 8. The curve for holo-EAL, which corresponds to the absence of substrate activation, and therefore, the uncatalyzed bond dissociation process, is modeled after calculated potential energy curves for complete Co-C bond homolysis in condensed phases.42-44 The thermal free energy curve for the EAL ternary complex depicts a progressive lowering of the free energy, over the duration of the Co-C bond cleavage event. As shown in Figure 8, the contributions of the protein to Co-C bond cleavage catalysis must occur over relatively short rCoC values, in the >2.0 - 4 Å range of early radical pair separation, because calculations show that the energy changes associated with Co-C separation are approximately 70% developed by 2.7-3.0 Å 41 and essentially complete at 4.0 Å.42-44 As depicted in Figure 8, the photolysis results are consistent with a predominant decay channel of the photo-produced AdoCbl excited state that involves relaxation, through internal conversion and intersystem crossing, and eventual intersection with the uncatalyzed zero-order surface, leading to cob(II)alamin-radical pair recombination. The barrier region for the catalyzed zero-order curve is vertically aligned with Co-C separation distances achieved by photolysis in Figure 8. Therefore, if the “substrate trigger” induced a change in protein structure capable of prompt stabilization of the cob(II)alamin-5′-deoxyadenosyl radical pair, then a significant quantum yield would be expected. This was not observed. The results suggest that the catalyzed, zero-order surface is not accessed by the photogenerated radical pair in the EAL ternary complex.
Figure 8.
Free energy curves for Co-C bond cleavage that depict the uncatalyzed thermolytic and native catalyzed thermolytic conditions, and representative trajectory of the cob(II)alamin-5′-deoxyadenosyl radical pair photoproduct, in EAL. The uncatalyzed thermolytic curve is based on calculated energy curves for Co-C bond cleavage for AdoCbl in solution and in proteins,42-44 and the native catalyzed thermolytic curve is based on the limit on the maximum of the free energy barrier of −15 kcal/mol.21 The representative photoproduct trajectory is based on calculations.41
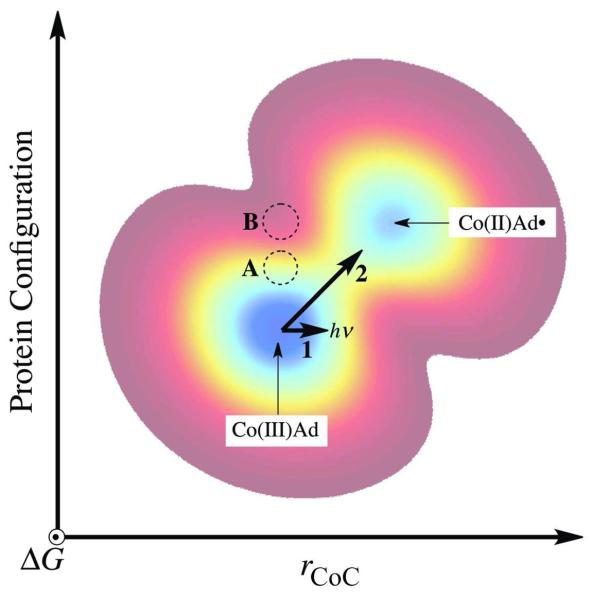
The interpretation of the EAL photolysis results, in relation to the native reaction, requires the explicit introduction of protein structural changes, that develop during Co-C bond cleavage and radical pair separation. The protein structural changes can be represented by using a protein configuration coordinate that is orthogonal to the chemical Co-C bond cleavage coordinate. A simple two-dimensional free energy surface is shown in Figure 9. Path 1 on the free energy surface in Figure 9 depicts the light-induced, photolytic transition of the bound cofactor from AdoCbl to the cob(II)alamin-5′-deoxyadenosyl radical pair. As concluded in the preceding paragraph, the protein on Path 1 is not in a configuration that can stabilize the nascent radical pair, and this leads to rapid recombination. Figure 9 also illustrates the effect of a simple substrate-induced switch of the protein configuration from holo-EAL (represented by position “A”) to a state capable of prompt stabilization of the photoproduct (position “B”). Our results do not support the existence of a substrate-induced switch of the protein to a new state, such as “B.” Instead, we propose that substrate binding to holo-EAL (position “A”), induces a change in protein configuration to the position in Figure 9, that is represented by the ternary complex state. Path 2 represents a thermally-activated path, which involves the adjustment of the protein in concert with the formation of the cob(II)alamin-5′-deoxyadenosyl radical pair. The concommitant lowering of the free energy, as Co-C bond cleavage occurs, facilitates transition to the cob(II)alamin-radical pair state.
Figure 9.
Two-dimensional contour representation of the ground state free energy surface for the ternary complex and radical pair formation and separation in EAL as a function of chemical (Co-C bond cleavage) and protein configuration coordinates. The two minima represent the ternary complex [Co(III)Ad] and the cob(II)alamin-5′-deoxyadenosyl radical pair [Co(II)Ad•] states. The one-dimensional trajectory for photolysis (Path 1) is represented by a horizontal bold arrow. The trajectory for the native thermal Co-C bond cleavage is represented by the diagonal bold arrow (Path 2). A representative position of the free energy minimum for the holo-EAL state is given by the region marked “A.” The region, “B,” marks a representative position for the free energy minimum of a protein state, that might be obtained by a simple substrate binding-induced switch of the holo-EAL protein configuration to one that favors prompt stabilization of the photoproduct radical pair.
In principle, the nascent cob(II)alamin-5′-deoxyadenosyl radical pair formed by photolysis on Path 1 in Figure 9 could execute a Brownian or biased walk over the free energy surface to the region of Path 2, and thus react to form a stable cob(II)alamin-5′-deoxyadenosyl radical pair. A trajectory of the photolyzed cob(II)alamin-5′-deoxyadenosyl radical pair towards the thermal reaction path requires a structural change along the protein coordinate, that occurs on a time-scale comparable to, or faster than, the time scale of geminate recombination, or ≤10−8 s. The undetectable photolysis quantum yield values indicate that the thermal fluctuations in the protein, that would drive a trajectory from Path 1 to the native thermal Path 2, occur on time scales at least 10-fold longer than τgr, or >10−7 s. This is consistent with the seconds time scale of radical pair formation at 240 K.21
Conclusions
Photolysis of AdoCbl in the EAL ternary complex overcomes the demanding microscopic event of Co-C bond homolysis (bond dissociation energy in solution, ~30 kcal/mol40) in the process of radical pair separation, and presents the protein with a cob(II)alamin-5′-deoxyadenosyl radical pair that mimicks an early thermal cleavage state, in terms of the Co-C separation distance of 2.7-3.0 Å.41 The results indicate that substrate binding to EAL does not “switch” the protein to a new structural state, which promptly stabilizes the cob(II)alamin-5′-deoxyadenosyl radical pair photoproduct, either through increased barriers to recombination, decreased barriers to forward radical pair separation, or through lowering of the free energy of the radical pair state, or a combination of these effects. The absence of significant distortion of the AdoCbl in the ternary complex, and the absence of a protein structure state capable of prompt stabilization of the cob(II)alamin-5′-deoxyadenosyl radical pair photoproduct, indicate that the substrate trigger induces a change in protein structure and/or dynamics, that is not detected by our spectroscopic probes of the equilibrium protein state in the ternary complex. The detailed microscopic mechanism of the “substrate trigger” thus remains unclear. The results further suggest that, following the substrate trigger, the protein interacts with the cofactor to contiguously guide the cleavage of the Co-C bond, at every step along the cleavage coordinate, starting from the equilibrium configuration of the ternary complex. This situation is represented in Figure 9 by the diagonal trajectory across the free energy surface, which requires progress along coupled chemical and protein coordinates. In support of this model, protein involvement in rapid stabilization of the nascent radical pair has been proposed, based on magnetic field effects on the radical pair yield,60,61 and mechanisms that involve the development of favorable cofactor-protein interactions as Co-C bond cleavage proceeds, have been proposed for EAL 15 and other AdoCbl-dependent enzymes.62 Future application of the AdoCbl photolysis approach to the elucidation of the structural and dynamical bases of the native radical pair creation and separation in EAL and other AdoCbl-dependent enzymes must involve a concommitant perturbation of the protein coordinate.
Supplementary Material
ACKNOWLEDGMENT
The project described was supported by Grant Number R01DK054514 from the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health (NIH). The purchase of the Bruker E500 EPR spectrometer was funded by NIH NCRR grant RR17767 and by Emory University.
Footnotes
The project described was supported by Grant Number R01DK054514 from the National Institute of Diabetes and Digestive and Kidney Diseases.
SUPPORTING INFORMATION AVAILABLE
Static UV/visible spectra of aquocobalamin in solution and in EAL, and transient decay data for holo-EAL, ternary complex, and holo-EAL with inhibitor bound, with overlaid fits obtained by using monoexponential and biexponential functions. This information is available free of charge via the internet at http://pubs.acs.org/.
REFERENCES
- 1.Toraya T. Chemical Reviews. 2003;103:2095–2127. doi: 10.1021/cr020428b. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee R, Ragsdale SW. Annual Review of Biochemistry. 2003;72:209–247. doi: 10.1146/annurev.biochem.72.121801.161828. [DOI] [PubMed] [Google Scholar]
- 3.Brown KL. Chem. Rev. 2005;105:2075–2149. doi: 10.1021/cr030720z. [DOI] [PubMed] [Google Scholar]
- 4.Warncke K, Utada AS. Journal of the American Chemical Society. 2001;123:8564–8572. doi: 10.1021/ja003658l. [DOI] [PubMed] [Google Scholar]
- 5.Canfield JM, Warncke K. Journal of Physical Chemistry B. 2002;106:8831–8841. [Google Scholar]
- 6.Bandarian V, Reed GH. Biochemistry-Us. 2002;41:8580–8588. doi: 10.1021/bi0201217. [DOI] [PubMed] [Google Scholar]
- 7.LoBrutto R, Bandarian V, Magnusson OT, Chen X, Schramm VL, Reed GH. Biochemistry-Us. 2001;40:9–14. doi: 10.1021/bi001865s. [DOI] [PubMed] [Google Scholar]
- 8.Canfield JM, Warncke K. Journal of Physical Chemistry B. 2005;109:3053–3064. doi: 10.1021/jp046167m. [DOI] [PubMed] [Google Scholar]
- 9.Warncke K, Schmidt JC, Ke SC. J. Am. Chem. Soc. 2008;130:6055. [Google Scholar]
- 10.Bender G, Poyner RR, Reed GH. Biochemistry. 2008;47:11360–11366. doi: 10.1021/bi801316v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bradbeer C. J Biol Chem. 1965;240:4669–4674. [PubMed] [Google Scholar]
- 12.Hubbard BK, Gulick AM, Babbitt PC, Rayment I, Gerlt JA. Faseb Journal. 1999;13:A1446–A1446. [Google Scholar]
- 13.Sun L, Warncke K. Proteins-Structure Function and Bioinformatics. 2006;64:308–319. doi: 10.1002/prot.20997. [DOI] [PubMed] [Google Scholar]
- 14.Bandarian V, Reed GH. In: Chemistry and Biochemistry of B12. Banerjee R, editor. John Wiley and Sons; New York: 1999. pp. 811–833. [Google Scholar]
- 15.Shibata N, Tamagaki H, Hieda N, Akita K, Komori H, Shomura Y, Terawaki S-I, Mori K, Yasuoka N, Higuchi Y, Toraya T. J Biol Chem. 2010 doi: 10.1074/jbc.M110.125112. JBC Papers in Press: Manuscript M110.125112 (June 1, 2010): [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hay BP, Finke RG. Inorg. Chem. 1984;23:3041–3043. [Google Scholar]
- 17.Halpern J, Kim S-H, Leung TW. J. Am. Chem. Soc. 1984;106:8317–8319. [Google Scholar]
- 18.Hay BP, Finke RG. Polyhedron. 1988;7:1469–1481. [Google Scholar]
- 19.Brooks AJ, Vlasie M, Banerjee R, Brunold TC. J. Am. Chem. Soc. 2004;126:8167–8180. doi: 10.1021/ja039114b. [DOI] [PubMed] [Google Scholar]
- 20.Brooks AJ, Vlasie M, Banerjee R, Brunold TC. J. Am. Chem. Soc. 2005;127:16522–16528. doi: 10.1021/ja0503736. [DOI] [PubMed] [Google Scholar]
- 21.Wang M, Warncke K. J. Am. Chem. Soc. 2008;130:4846–4858. doi: 10.1021/ja710069y. [DOI] [PubMed] [Google Scholar]
- 22.Brown K. Chem. Rev. 2005;105:2075–2149. doi: 10.1021/cr030720z. [DOI] [PubMed] [Google Scholar]
- 23.Endicott JF, Netzel TL. Journal of the American Chemical Society. 1979;101:4000–4002. [Google Scholar]
- 24.Chen EF, Chance MR. Abstracts of Papers of the American Chemical Society. 1990;200:196-INOR. [Google Scholar]
- 25.Cole AG, Yoder LM, Shiang JJ, Anderson NA, Walker LA, Holl MMB, Sension RJ. Journal of the American Chemical Society. 2002;124:434–441. doi: 10.1021/ja011628s. [DOI] [PubMed] [Google Scholar]
- 26.Robertson WD, Warncke K. Biochemistry-Us. 2009;48:140–147. doi: 10.1021/bi801659e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shiang JJ, Walker LA, Anderson NA, Cole AG, Sension RJ. Journal of Physical Chemistry B. 1999;103:10532–10539. [Google Scholar]
- 28.Shiang JJ, Cole AG, Sension RJ, Hang K, Weng Y, Trommel J, Marzilli L, Lian T. J. Am. Chem. Soc. 2006;128:801–808. doi: 10.1021/ja054374+. [DOI] [PubMed] [Google Scholar]
- 29.Yoder LM, Cole AG, Walker LA, Sension RJ. Journal of Physical Chemistry B. 2001;105:12180–12188. [Google Scholar]
- 30.Walker LA, Shiang JJ, Anderson NA, Pullen SH, Sension RJ. Journal of the American Chemical Society. 1998;120:7286–7292. [Google Scholar]
- 31.Walker LA, Jarrett JT, Anderson NA, Pullen SH, Matthews RG, Sension RJ. Journal of the American Chemical Society. 1998;120:3597–3603. [Google Scholar]
- 32.Shiang JJ, Cole AG, Sension RJ, Hang K, Weng YX, Trommel JS, Marzilli LG, Lian TQ. Journal of the American Chemical Society. 2006;128:801–808. doi: 10.1021/ja054374+. [DOI] [PubMed] [Google Scholar]
- 33.Sension RJ, Walker LA, Shiang JJ. Abstracts of Papers of the American Chemical Society. 1998;216:U684–U684. [Google Scholar]
- 34.Sension RJ, Harris DA, Cole AG. Journal of Physical Chemistry B. 2005;109:21954–21962. doi: 10.1021/jp053202w. [DOI] [PubMed] [Google Scholar]
- 35.Cole AG, Anderson N, Shiang JJ, Sension RJ. Abstracts of Papers of the American Chemical Society. 2000;220:U223–U223. [Google Scholar]
- 36.Brownawell AM, Chen E, Chance MR. Biophysical Journal. 1993;64:A161–A161. [Google Scholar]
- 37.Lott WB, Chagovetz AM, Grissom CB. J. Am. Chem. Soc. 1995;117:12194–12201. [Google Scholar]
- 38.Noyes RM. Prog React Kinet Mec. 1961;1:129–160. [Google Scholar]
- 39.Chen E, Chance MR. Biochemistry-Us. 1993;32:1480–1487. doi: 10.1021/bi00057a011. [DOI] [PubMed] [Google Scholar]
- 40.Halpern J. Accounts of Chemical Research. 1982;15:238–244. [Google Scholar]
- 41.Jaworska M, Lodowski P, Andruniow T, Kozlowski PM. J. Chem. Phys. B. 2007;111:2419–2422. doi: 10.1021/jp0685840. [DOI] [PubMed] [Google Scholar]
- 42.Dölker N, Maseras F, Lledos A. J. Phys. Chem. B. 2001;105:7564–7571. [Google Scholar]
- 43.Dölker N, Maseras F, Siegbahn PEM. Chem. Phys. Lett. 2004;386:174–178. [Google Scholar]
- 44.Kwiecien RA, Khavrutskii IV, Musaev DG, Morokuma K, Banerjee R, Paneth P. J. Am. Chem. Soc. 2006;128:1287–1292. doi: 10.1021/ja056333j. [DOI] [PubMed] [Google Scholar]
- 45.Sension RJ, Cole AG, Harris AD, Fox CC, Woodbury NW, Lin S, Marsh ENG. Journal of the American Chemical Society. 2004;126:1598–1599. doi: 10.1021/ja0396910. [DOI] [PubMed] [Google Scholar]
- 46.Sension RJ, Harris DA, Stickrath A, Cole AG, Fox CC, Marsh ENG. Journal of Physical Chemistry B. 2005;109:18146–18152. doi: 10.1021/jp052492d. [DOI] [PubMed] [Google Scholar]
- 47.Douzou P. Cryobiochemistry: An Introduction. Academic Press; New York: 1977. [Google Scholar]
- 48.Faust LRP, Connor JA, Roof DM, Hoch JA, Babior BM. Journal of Biological Chemistry. 1990;265:12462–12466. [PubMed] [Google Scholar]
- 49.Faust LP, Babior BM. Archives of Biochemistry and Biophysics. 1992;294:50–54. doi: 10.1016/0003-9861(92)90135-j. [DOI] [PubMed] [Google Scholar]
- 50.Harkins TT, Grissom CB. Journal of the American Chemical Society. 1995;117:566–567. [Google Scholar]
- 51.Kaplan BH, Stadtman ER. Journal of Biological Chemistry. 1968;243:1787. [PubMed] [Google Scholar]
- 52.Moore JW, Pearson RG. Kinetics and Mechanism. Wiley and Sons; New York: 1981. [Google Scholar]
- 53.Huhta MS, Chen H-P, Hemann C, Hille CR, Marsh ENG. Biochem. J. 2001;355:131–137. doi: 10.1042/0264-6021:3550131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Babior BM. J Biol Chem. 1970;245:6125–6133. [PubMed] [Google Scholar]
- 55.Babior BM. In: B12. Dolphin D, editor. Vol. 2. Wiley; New York: 1982. pp. 263–287. [Google Scholar]
- 56.Stickrath AB, Carroll EC, Dai X, Harris A, Rury A, Smith B, Tang K-C, Wert J, Sension RJ. J. Phys. Chem. A. 2009;113:8513–8522. doi: 10.1021/jp9017986. [DOI] [PubMed] [Google Scholar]
- 57.Dong SL, Padmakumar R, Banerjee R, Spiro TG. J. Am. Chem. Soc. 2004;121:7063–7070. [Google Scholar]
- 58.Mansoorabadi SO, Magnusson OT, Poyner RR, Frey PA, Reed GH. Biochmistry. ASAP; Web 110906. [Google Scholar]
- 59.Khoroshu DV, Warncke K, Ke S-C, Musaev DG, Morokuma K. J. Am. Chem. Soc. 2003;125:570–579. doi: 10.1021/ja028393k. [DOI] [PubMed] [Google Scholar]
- 60.Jones AR, Hay S, Woodward JR, Scrutton NS. J. Am. Chem. Soc. 2007;129:15718–15727. doi: 10.1021/ja077124x. [DOI] [PubMed] [Google Scholar]
- 61.Jones AR, Woodward JR, Scrutton NS. J. Am. Chem. Soc. 2009;131:17246–17253. doi: 10.1021/ja9059238. [DOI] [PubMed] [Google Scholar]
- 62.Sharma PK, Chu ZT, Olsson MHM, Warshel A. Proc. Natl. Acad. Sci. 2007;104:9661–9666. doi: 10.1073/pnas.0702238104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ke SC, Torrent M, Museav DG, Morokuma K, Warncke K. Biochemistry. 1999;38:12681–12689. doi: 10.1021/bi983067w. [DOI] [PubMed] [Google Scholar]
- 64.Abend A, Bandarian V, Nitsche R, Stupperich E, Retey J, Reed GH. Archives of Biochemistry and Biophysics. 1999;370:138–141. doi: 10.1006/abbi.1999.1382. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.