Abstract
Multiple genome-wide association studies (GWASs) and two large scale meta-analyses have been performed for Crohn's disease and have identified 71 susceptibility loci. These findings have contributed greatly to our current understanding of the disease pathogenesis. Yet, these loci only explain approximately 23% of the disease heritability. One of the future challenges in this post-GWAS era is to identify potential sources of the remaining heritability. Such sources may include common variants with limited effect size, rare variants with higher effect sizes, structural variations, or even more complicated mechanisms such as epistatic, gene-environment and epigenetic interactions. Here, we outline potential sources of this hidden heritability, focusing on Crohn's disease and the currently available data. We also discuss future strategies to determine more about the heritability; these strategies include expanding current GWAS, fine-mapping, whole genome sequencing or exome sequencing, and using family-based approaches. Despite the current limitations, such strategies may help to transfer research achievements into clinical practice and guide the improvement of preventive and therapeutic measures.
Background
Crohn's disease (CD) is one of the two main forms of inflammatory bowel disease (IBD), the other being ulcerative colitis (UC). It is a chronic disease characterized by recurring inflammation of the gut, and is thought to arise in response to the commensal microflora in a genetically susceptible host [1]. It can affect the entire gastrointestinal tract, although the most common locations are the terminal ileum and the colon. Symptoms can be diffuse, and include (bloody) diarrhea, abdominal discomfort, weight loss and anemia, and there may also be extra-intestinal symptoms such as arthritis, and eye and skin disorders. Complications such as strictures often occur in CD, and since the inflammation is transmural, fistulas and abscesses can develop, and these eventually require surgical treatment [2]. Most of the medications have significant side effects, and they are expensive, and often ineffective. CD is a major burden on healthcare services, with a prevalence of 100 to 150 cases per 100,000 persons per year in the western world and with a peak age of onset between 10 and 30 years of age [3]. CD is partly heritable; this is reflected in the higher concordance rate in monozygotic twins compared with dizygotic twins. The concordance for CD in dizygotic twins is 4%, and for monozygotic twins it is as high as 56% [4].
Prior to the introduction of genome-wide association studies (GWASs), only a few genetic factors (for example, NOD2, which encodes nucleotide binding oligomerization domain 2) had unequivocally been associated with CD. However, multiple GWASs have now been performed for CD, and a recent meta-analysis carried out by Franke et al. [5] has unveiled 71 genetic variants as associated with CD; Table 1 highlights some noteworthy genes from that study. Many of the genes cluster in several different molecular pathways and gene networks. In particular, results from GWASs have indicated the importance of the immune system in disease pathogenesis by identifying genes involved in innate and adaptive immunity. Hence, the association of IRGM, encoding immunity-related GTPase family M, and ATG16L1, encoding autophagy-related 16-like 1, with CD has implicated the process of autophagy [6]. The association of NOD2, CARD9, which encodes caspase recruitment domain family member 9, and TLR4, which encodes Toll-like receptor 4, indicates the involvement of pattern recognition mechanisms of the innate immune system [7]. Other genes are involved in pro-inflammatory pathways (T helper 1 cells and T helper 17 cells) and in anti-inflammatory pathways (regulatory T cells and IL-10), indicating that adaptive immunity also plays a role in CD pathogenesis (Figure 1) [8]. Another interesting association mapped to the FUT2 gene, which encodes secretor type fucosyltransferase and regulates secretion of A and B blood group antigens in intestinal mucosa [9]. Recent functional studies have suggested that fucosylation of mucin proteins is involved in interception and exclusion of bacteria; thus, association of FUT2 with CD might imply a role for the functional state of mucin in CD pathogenesis [10]. Although 5 years of GWASs have identified a substantial number of CD susceptibility loci, as much as 77% of the estimated heritability for CD is still considered to be unexplained [5].
Table 1.
Notable genes within regions associated with Crohn's disease
| Gene | Odds ratio (95% CI) | Function |
|---|---|---|
| Innate immunity | ||
| NOD2 (nucleotide binding oligomerization domain 2) | 2.2-4.0 [58] | Involved in pattern recognition |
| ATG16L1 (ATG16 autophagy related 16-like 1) | 1.34 (1.29-1.40) [5] | Involved in autophagy |
| IRGM (immunity-related GTPase family, M) | 1.37 (1.28-1.47) [5] | Involved in autophagy |
| TLR4 (Toll-like receptor 4) | 1.29 (1.08-1.54) [59] | Involved in pattern recognition |
| CARD9 (caspase recruitment domain family, member 9) | 1.18 (1.13-1.22) [5] | Involved in pattern recognition |
| VAMP3 (vesicle-associated membrane protein 3) | 1.05 (1.01-1.10) [5] | Involved in autophagy and TNF-α metabolism |
| REL (reticuloendotheliosis viral oncogene homolog) | 1.14 (1.09-1.19) [5] | Transcriptional activator of NF-кB |
| ERAP2 (endoplasmic reticulum aminopeptidase 2) | 1.05 (1.02-1.09) [5] | Involved in peptide trimming upon NF-кB stimulation; required for the generation of HLA binding peptides |
| UBE2L3 (ubiquitin-conjugating enzyme E2L 3) | 0.70 [15] | Ubiquitinates, among others, the NF-кB precursor |
| Adaptive immunity | ||
| IL23R (IL-23 receptor) | 2.66 (2.36-3.00) [5] | Activates Th17 cells |
| IL12B (IL-12β) | 1.18 (1.13-1.24) [5] | Stimulates Th0 differentiation to Th1 cells |
| CCR6 (chemokine (C-C motif) receptor 6) | 1.17 (1.12-1.22) [5] | Chemoattractant receptor of immune cells |
| HLA-DQA2 (major histocompatibility complex, class II, DQα2) | 1.19 (1.13-1.25) [5] | Antigen presenting to Th0 |
| TNFSF11 (tumor necrosis factor super family 11) | 1.10 (1.05-1.15) [5] | Augments the ability of dendritic cells to stimulate naive T-cell proliferation |
| TNFSF15 (tumor necrosis factor super family 15) | 1.21 (1.15-1.27) [5] | Mediates activation of NF-кB |
| ICOSLG (inducible T-cell co-stimulator ligand) | 1.18 (1.13-1.23) [5] | Acts as a co-stimulatory signal for T-cell proliferation and cytokine secretion |
| IL2RA (IL receptor α) | 1.11 (1.05-1.16) [5] | Th0 activation |
| TAGAP (T-cell activation GTPase-activating protein) | 1.10 (1.05-1.14) [5] | May function as a GTPase activating protein and may play important roles during T-cell activation |
| IL10 (IL-10) | 1.12 (1.07-1.17) [5] | Inhibits synthesis of pro-inflammatory cytokines |
| IL18RAP (IL-18 receptor accessory protein) | 1.19 (1.14-1.26) [5] | Protein required for NF-кB activation |
| TYK2 (tyrosine kinase 2) | 1.12 (1.06-1.19) [5] | Probably involved in intracellular signal transduction by initiation of IFN signaling |
| JAK2 (Janus kinase 2) | 1.18 (1.13-1.23) [5] | Involved in JAK/STAT pathway; mediates signal transduction of many cytokines |
| STAT3 (signal transducer and activator of transcription 3) | 1.15 (1.10-1.21) [5] | Involved in JAK/STAT pathway; mediates signal transduction of many cytokines |
| SMAD3 (SMAD family member 3) | 1.12 (1.07-1.16) [5] | Involved in Treg activation through TGF-β signal transduction |
| ICAM1,3 (intercellular adhesion molecule) | 1.12 (1.06-1.19) [5] | Homing of leukocytes to inflammation |
| Other genes of interest | ||
| MUC1,19 (mucin) | 1.74 (1.55-1.95) [5] | Involved in mucus production, to protect the epithelial barrier |
| FUT2 (fucosyltransferase 2) | 1.07 (1.04-1.11) [5] | Involved in the A and B antigen synthesis pathway |
| PUS10 (pseudouridylate synthase 10) | 1.16 [19] | Post-transcriptional nucleotide modification of structural RNAs, including tRNA, rRNA and sRNAs |
Genes that we consider to be noteworthy in the Crohn's disease associated loci. Further investigation is necessary to identify the causal variants. CI, confidence interval; HLA, human leukocyte antigen; IFN, interferon; IL, interleukin; JAK, Janus kinase; NF, nuclear factor; rRNA, ribosomal RNA; sRNA, splicing RNA; STAT, signal transducer and activator of transcription; TGF, transforming growth factor; Th, T helper cell; TNF, tumor necrosis factor; Treg, regulatory T cell; tRNA transferRNA.
Figure 1.
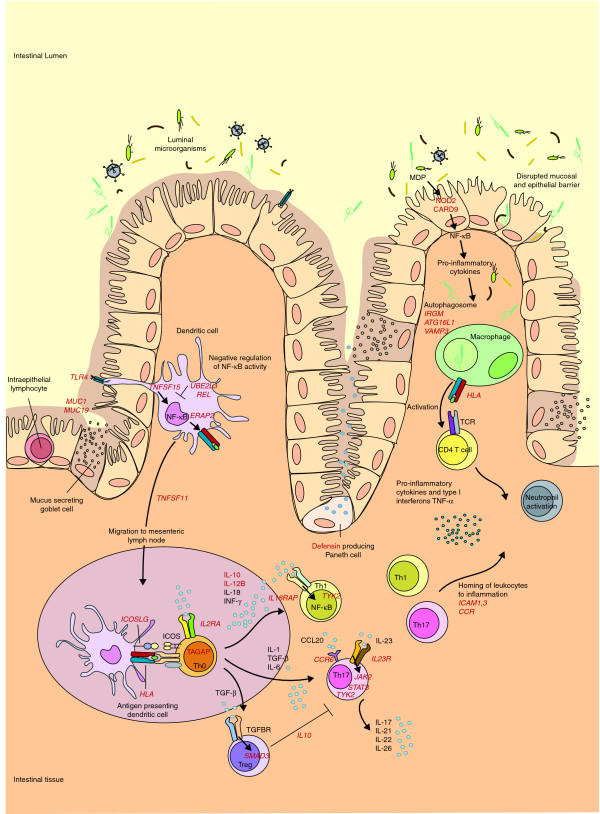
Schematic representation of the genes and pathways associated with Crohn's disease pathogenesis. The ongoing inflammatory response in the gastrointestinal tract in patients with Crohn's disease (CD) is thought to be caused by an aberrant immune response to commensal microflora in the gut. In patients with CD, defects in first defense mechanisms (that is, disrupted epithelial and mucosal barrier) contribute to increased bacterial penetration (MUC1 and MUC19). Genes involved in pattern recognition (NOD2, TLR4 and CARD9) suggest an increased response of antigen-presenting cells to commensal microbes. Consequently, the NF-кB cascade is activated (TNFSF15), leading to production of pro-inflammatory cytokines. Association of REL and UBE2L3 suggest an impaired NF-кB negative feedback. Antigen-presenting cells migrate to Peyer's patches (intestinal mesenteric lymph nodes) (TNFSF11) to present antigens and stimulate T-cell proliferation (IL2RA and TAGAP) and differentiation. T cells of patients with CD, in turn, respond more intensely. Th0 cells are stimulated to differentiate into T-cell subtypes regulated by a variety of the produced cytokines and their receptors. Th17 cells are involved in many immune-related diseases, and they are activated through IL-23R, which, in turn, activates the JAK-STAT-TYK (Janus kinase- signal transducer and activator of transcription-tyrosine kinase) pathway that enhances pro-inflammatory cytokine production (JAK2, STAT3 and TYK2). Th1 and Th17 cells are pro-inflammatory, whereas Treg cells downregulate the immune response. Another major contribution to CD pathogenesis comes from autophagy. In autophagosomes, intracellular components, including phagocytosed microbes, are degraded, after which their antigens are presented to CD4+ cells. Autophagy is at least partly regulated by the CD risk genes ATG16L1, IRGM and VAMP3. The activation of CD4+ cells leads to the production of pro-inflammatory cytokines and the maintenance of the inflammation. All the displayed processes could finally lead to homing of leukocytes to inflammation sites (ICAM1,3, CCR cluster), and neutrophil recruitment. Consequently, chronic inflammation, ulceration and deeper microbial penetrance occur. The known associated genes are shown in red. Table 1 summarizes the associated loci shown here. CCL20, chemokine (C-C motif) ligand 20; ICOS, inducible T-cell co-stimulator; MDP, muramyl dipeptide; NF, nuclear factor; TCR, T-cell receptor; TGF, transforming growth factor; TGFBR, TGF β receptor; Th, T helper cell; TNF, tumor necrosis factor; Treg, regulatory T cell.
Thus, one of the current challenges in the study of CD, like other complex diseases, is to identify potential sources of this hidden heritability. These might be additional common variants with very limited effect size, or rare variants with a higher effect size. Part of the hidden heritability may lie in structural variations such as copy number variations (CNVs; a type of structural DNA sequence alteration, including deletions, duplications, insertions and inversions, that results in varying numbers of copies of a particular gene or DNA sequence from one person to the next) or even more complicated mechanisms, such as epistatic, gene-environment and epigenetic interactions.
In this review, we discuss the known genetic risk factors for CD, the potential sources of the hidden heritability, and strategies to investigate these.
Further exploration of GWAS results
Thus far, the GWASs performed for CD have implicated many genes, and have thereby provided valuable insights into the etiology of CD. However, there are several ways to explore GWAS results in more depth that might lead to solving a part of the hidden heritability puzzle. The design of GWASs holds several limitations, with the first being the extensive correction needed for multiple testing. Hence, many true-positive findings are discarded because of the stringent significance thresholds, and large amounts of data are therefore ignored. Several methods have been applied successfully to overcome this statistical power issue. A major step to overcoming this problem has been taken by the International IBD Genetics Consortium (IIBDGC) [11], which performed a novel meta-analysis of six index GWASs and a follow-up study in independent cohorts. This study increased the number of confirmed CD loci to 71, although the explained heritability only increased from 20% to 23% [5].
Another way to overcome the lack of power inherent in GWASs is to follow-up specific SNPs (variation in a single base in the DNA sequence; the most common type of variation in the human genome) identified by them. Following up the top 1,000 less-strongly associated loci, for example, could yield new true associations. Meta-analysis of these results with the results from the index GWASs leads to a gain of power, as shown by a study of celiac disease [12]. Another approach is to prioritize genes from the top associated loci based on interaction or functional analyses. This has proven to be a successful strategy in rheumatoid arthritis, where genes were prioritized based on network analysis or interaction analysis [13]. For CD, Wang et al. [14] used a different prioritizing criterion based on pathway analysis and they uncovered a significant association between susceptibility to CD and the IL-12/IL-23 pathway, harboring 20 genes. Prioritizing SNPs based on their effect on gene expression (for example, expression quantitative trait locus, a locus at which genetic allelic variation(s) correlates with variation in gene expression) led to identification of potentially novel associations of CD with UBE2L3, encoding ubiquitin-conjugating enzyme E2L 3 (involved in ubiquitinating the NF-кB precursor), and BCL3, encoding B-cell lymphoma 3-encoded protein (involved in downregulation of the NF-кB pathway) [15].
Results of GWASs and their meta-analyses have revealed that multiple autoimmune diseases have a common genetic architecture [16]. Several studies have been successful in identifying new CD risk variants by testing previously established loci for other immune-related diseases [17,18]. Festen et al. [19] developed a new method to identify shared risk loci of two immune-mediated diseases with a partially shared genetic background, namely celiac disease and CD. To increase the statistical power, they performed a combined analysis of GWAS results from celiac disease and CD, and identified TAGAP, which encodes T-cell activation GTPase-activating protein, and PUS-10, which encodes tRNA pseudouridylate synthase, as new shared loci [19].
The second limitation of the GWAS design is that it does not lead to the identification of causal variants, since the tested SNPs are merely tagging SNPs in linkage disequilibrium (LD; a non-random association of alleles at two or more loci as a result of a recent mutation, genetic drift, selection, or non-random mating) with the causal variants. Therefore, the effect sizes of known CD loci may be an underestimation of their actual relative risk. To further investigate the known risk loci and identify new SNPs, either as causal or close-to-causal variants, extensive fine-mapping is currently being performed by the IIBDGC using a custom-made GWA chip. In addition, cross-ethnicity fine-mapping has proven successful in exploring conserved haplotype structures (that is, LD blocks) [20]. The most common LD blocks occur in all populations; however, their frequencies vary among different ethnicities [20]. For example, common NOD2 and IL23R variants that are well established in Caucasians could not be replicated in an Indian population, implying that additional variants in these or other candidate genes may play a role in the pathogenesis of CD in Indians [21]. This principle was also successfully applied in analyzing the IL2/IL21 LD block, which is strongly conserved in Caucasians as opposed to Han Chinese, in which the IL2 and IL21 genes reside on two distinct LD blocks. Both IL2 and IL21 could be identified as separate UC risk loci in Han Chinese [22].
Park et al. [23] proposed a method to evaluate statistical power and risk prediction of future GWASs. They estimated that there are, in total, 142 CD susceptibility loci with effect sizes similar to the loci reported in the current GWASs, and that a sample size of approximately 50,000 would be needed to uncover them. However, even if a GWAS with hundreds of thousands of cases were to provide new CD susceptibility loci and explain more of the genetic variance, it seems unlikely that it would capture even half of the estimated heritability since 142 loci only explain 20% of the sibling relative risk for CD. We can speculate that identification of the true causal variants could amplify the effect size for some of the known loci and could consequently increase the discriminatory power of risk models.
Another potential source of hidden heritability could lie in sample mix-ups that occur accidentally during sample collection, genotyping or data management. Some genetic variants influence gene expression phenotypes (expression quantitative trait loci); this allows checking for concordance between phenotypic measurements and genetic variants that affect these phenotypes. Westra et al. (personal communication) found that 3% of sample mix-ups decrease the number of loci normally discovered by 23% for a trait with a heritability of 50% and 500 loci explaining the total heritability. Thus, sample mix-ups may explain part of the hidden heritability and it will be possible to detect them as long as databases encompass sufficient numbers of phenotypes that are strongly determined by known genetic variants.
GWASs are most likely to remain an important approach for investigating the hidden heritability, since the potential of their results can be enhanced by: performing meta-analyses (for example, between multiple GWASs or between similar disease phenotypes); following-up prioritized SNPs based on pathway, functional or interaction analyses; studying SNPs that have been associated with other immune-related diseases; and expanding the design of GWASs to include samples from non-Caucasians.
Low frequency and rare variants
Common variants identified by GWASs represent only a small fraction of the phenotypic variation. Thus, much speculation about the hidden heritability has focused on the contribution of variants with low allele frequencies, defined as 0.5% <minor allele frequency (MAF; proportion of the less common of two alleles in a population) <5%, or from rare variants with MAF <0.5%, that are not sufficiently frequent to be captured by current GWA arrays, nor sufficiently penetrant to be captured by traditional, family-based linkage studies [24]. Detecting such variants will be facilitated by advances in high-throughput sequencing technologies and by the wide-ranging catalog of variants with MAF >1% generated by the 1000 Genomes Project [25]. Current efforts to identify rare variants by sequencing are likely to focus on the regions of most significant GWAS SNPs and around genes already implicated in CD pathogenesis or treatment. Resequencing of selected susceptibility loci has led recently to the discovery of three IL23R (the gene encoding IL-23 receptor) coding variants that offer protection against CD [26]. The results of this particular study confirmed an increase in effect size with decreasing variant frequency, although rare variants explained less of the heritability than common variants.
In addition to resequencing efforts, whole-genome/exome sequencing will be needed to detect rare high-risk variants beyond the LD reach of tag SNPs. Although the costs of next-generation sequencing remain high, they are dropping fairly rapidly as the technologies improve and the process time per sample is becoming shorter; so this method is becoming more and more feasible and accessible for researchers. Evaluating such signals and determining the real causal variant will, however, be a difficult task. Feng and Zhu [27] developed an alternative method for searching for rare variants in previously published GWAS datasets. Their method relies on haplotype analysis across the genome and the hypothesis that multiple rare variants can be captured by many haplotypes. Using this method, they confirmed nine previously established loci and also discovered four new CD susceptibility loci [27].
Another approach that may prove to be important is performing resequencing studies of individuals with extreme phenotypes in lipid levels; these studies have shown that such individuals seem more likely to be the carriers of rare, yet non-synonymous, variants [28]. A large number of rare variants may have distinct effects on the phenotype. Therefore, pooling variants of similar effect and locus-specific matching of cases with specific CD subphenotypes and controls throughout the genome may help to reveal some of the hidden heritability [29].
Structural variation
It has been estimated that chromosomal rearrangements (that is, duplications, deletions, insertions and inversions), collectively named CNVs, comprise 12% of the human genome [30]. Currently, more than 15,000 CNV loci are catalogued in the Database of Genomic Variants [31]. Some CNVs have been linked to complex disorders, such as autism, neuroblastoma and systematic lupus erythematosus [32-34]. A recent study suggested that CNVs are enriched in genomic regions containing genes that influence immunity [35]. In particular, low and high copy numbers of the β-defensin gene (HBD2), which acts as an antimicrobial peptide and as a cytokine, have been found to predispose to colonic CD [36,37]. Yet, in a recent study, Aldhous et al. [38] failed to replicate both of the previously published associations. Moreover, they argued that these two associations could be due to measurement error because of a general deficiency of real-time PCR to distinguish multiple CNV clusters. In addition to the β-defensins, a fine-mapping study of the IRGM susceptibility locus revealed a 20-kb deletion polymorphism immediately upstream of IRGM that was associated with CD risk and IRGM expression [39]. Furthermore, a recent GWAS of CNVs from the Wellcome Trust Case Control Consortium has confirmed these CNVs for CD, and also discovered new CNVs in the IRGM and human leukocyte antigen (5.1 kb) regions [40]. The Wellcome Trust Case Control Consortium study also showed that the most common CNVs are well tagged by SNPs in current GWAS chips, and that they are unlikely to make much contribution to the hidden heritability in common diseases. More work is needed to elucidate the functional consequences and impact of high copy-number repeats (for example, long interspersed nuclear elements), and of rare CNVs on clinical phenotypes, such as CD.
Family-based approaches
Since the possibility of chip-based GWASs became available, linkage analysis and family-based approaches have been largely discarded. However, now that the opportunities for gene detection by conventional GWASs have been almost exhausted, researchers are shifting back towards family-based approaches. These approaches can be helpful when GWASs fail to detect signals from rare variants and are biased by population stratification, which is defined as a presence of subpopulations in a supposedly homogeneous population. Subpopulations arise from differences in allele frequencies between individuals as a consequence of distinct ancestral and/or demographic origin. Family-based studies may also be advantageous since the low frequency risk alleles (SNPs with MAF <5%) are likely to be more prevalent in large families with several affected members and should therefore be easier to detect. By assessing GWAS data in such families, large regions of identity-by-descent may be identified and found to include genes associated with CD; this approach has already proved to be a powerful tool in classical linkage analysis. However, the shared environment of family members is an alternative explanation for familial clustering that should be taken into account. Glocker et al. [41] identified loss-of-function mutations in two loci by considering early onset colitis as a monogenic trait in two consanguineous families. They performed a genetic linkage analysis followed by candidate gene sequencing and identified the IL10RA (the gene encoding IL-10 receptor α) and IL10RB (the gene encoding IL-10 receptor β) loci as being associated with early-onset enterocolitis. However, it is most likely that in this particular case a private variant, not present in the general population, is responsible for the disease.
Akolkar et al. [42] found that CD is subject to a parent-of-origin effect, indicating that loci affected by genomic imprinting play a role in CD pathogenesis. In genomic imprinting, the expression of an inherited variant is determined by the parent from whom that variant is inherited. If the maternal allele, for instance, is inactivated by genomic imprinting, then expression of the locus is determined by the paternal allele only. If this effect is not taken into account, a significant loss in the statistical power of the study might develop [43].
Family-based approaches may be useful in the search for the hidden heritability since low-frequency variants accumulate in families with multiple affected individuals; moreover, low-frequency variants are not affected by population stratification and they also include parent-of-origin effects. However, the causal variants identified in such families may prove to be private variants or the shared environment may play a major role.
GWAS aftermath: epistatic, gene-environment and epigenetic interactions
Given that a large proportion of the heritability of CD and its complex architecture is as yet unexplained, one might speculate other aspects of inheritance, such as epistasis, gene-environment interactions or epigenetic effects, might be involved. GWASs may be missing higher-order genetic effects that arise from the interaction of two or more SNPs [44]. The underlying idea for such epistatic effects is that a significant proportion of the hidden heritability is not due to single common variants, nor to single rare variants, but rather to rare combinations of common variants. Since typical GWASs examine the association of single SNPs with a phenotype, SNPs that contribute epistatically will not be revealed by such an analysis. A recent pair-wise analysis of variants related to the IL17-IL23 pathway showed an increasing odds ratio for CD when the 'risk' haplotypes for these genes were combined [45]. Analysis of epistatic interactions in better-powered datasets, and the use of more efficient computational approaches that can account for the complex nature of biomolecular networks, may yield new genetic risk factors for CD [46,47].
An even more complex source for the hidden heritability might lie in gene-environment interactions, which are defined as the joint effect of one or more genes with one or more environmental factors that cannot be readily explained by their separate marginal effects [48]. The strongest and best replicated environmental risk factor for CD is smoking, which increases both the risk and severity of CD. However, a recent, moderately sized study found remarkable differences in associated loci between smoking and non-smoking CD patients, thereby implying that a complex gene-environment interaction must be at work [49]. Another example of the complex interaction between genetic and environmental factors was shown in a study by Cadwell et al. [50] where Atg16L1-deficient mice infected with a specific strain of norovirus developed CD-like phenotypes in a model of intestinal injury induced by dextran sodium sulfate. In particular, structural Paneth cell abnormalities and decreased production of antimicrobial granules in the mice resembled those found in CD patients who are homozygous carriers of the ATG16L1 risk alleles. Remarkably, the severity of intestinal injury induced by dextran sodium sulfate was not only dependent on aberrant Atg16L1 function and norovirus infection, but also on the timing of infection, secretion of the pro-inflammatory cytokines TNF-α and IFN-γ, and the presence of commensal bacteria in the mouse intestine.
Other environmental factors, such as appendectomy, diet and domestic hygiene habits, may also play a role in CD, but the evidence for each of these factors is much weaker. To study gene-environment interactions will require careful consideration of the epidemiologic study design, exposure assessment, and methods of analysis, paying particular attention to ways of harmonizing these features across consortia.
An additional source of the hidden heritability might not lie in the genome sequence itself, but in subtle mechanisms interfering with genome functions, such as gene expression. These mechanisms include histone modification, methylation and gene inactivation, and are covered by the study of epigenetics. However, there is much controversy on this topic. Its role in CD is unknown, but there are some hints that methylation plays a role in other complex diseases: type 2 diabetes, rheumatoid arthritis and neurodegenerative diseases [51-53]. Epigenetics is also correlated with age, gender and nutrition, and it is likely that there are other environmental factors to be discovered [54,55]. It has been shown that changes in DNA methylation in mice can be provoked by dietary alterations and subsequently transmitted across generations [56]. Thus, sequence-independent epigenetic effects (beyond imprinting) that might be environmentally induced and transmitted across several generations [57] could represent a revolutionary glimpse into the enigmatic world of the heritability of complex diseases.
Conclusions
CD is a complex genetic disorder with an estimated heritability of 50% and it is characterized by a recurring inflammation of the gastrointestinal tract. Two decades of research have led to the discovery of 71 risk loci, which have improved our understanding of the disease pathogenesis. At the moment, approximately 23% of the heritability can be explained. To fully understand the disease pathogenesis and link current insights to clinically relevant knowledge, it is important to continue our quest to identify more genetic risk factors in CD. In this review, we have presented various potential sources for the hidden heritability of complex diseases given the current knowledge on CD.
It is unlikely that conventional GWASs alone can solve the puzzle of the hidden heritability. They are not powerful enough to detect signals from common variants with low impact, nor extensive enough to capture rarer variants with high impact. The resources of GWASs are expected to be exhausted fairly soon, although new loci have recently been identified by replicating prioritized SNPs and meta-analysis of GWAS results.
Identification of causal variants may elucidate a substantial part of the hidden heritability; however, current GWASs are insufficient for the purpose of identifying causal variants since the identified SNPs are merely the surrogates for causal variants. However, fine-mapping can uncover SNPs closer to the causal variants, since SNPs can then be tested beyond the scope of GWASs. The true causal variants might be identified by whole genome sequencing or exome sequencing. More sources than the linear DNA sequence have to be investigated to unravel the total heritability. Epigenetics and gene-environment studies have been shown to be worthwhile, but the study of epistatic effects in CD is still needed, and results from other complex genetic diseases seem to be promising.
To fully unravel the hidden heritability of CD, collaborations between genome research centers are crucial, since the solutions to identify the hidden heritability are either costly or require a huge number of cases and controls. The IIBDGC is a good example of what can be achieved by performing large meta-analyses, and it is currently performing dense fine-mapping and replication studies to identify causal variants and additional risk loci in CD.
Abbreviations
CD: Crohn's disease; CNV: copy number variation; GWAS: genome-wide association study; IBD: inflammatory bowel disease; IFN: interferon; IIBDGC: International IBD Genetics Consortium; IL: interleukin; LD: linkage disequilibrium; MAF: minor allele frequency; NF: nuclear factor; SNP: single nucleotide polymorphism; TNF: tumor necrosis factor; UC: ulcerative colitis.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
KF and MM contributed equally to this study. KF and MM conceived the idea for the review and wrote the paper. CCD and RKW critically revised and supervised the paper. CCD prepared Figure 1. All authors read and approved the final manuscript.
Contributor Information
Karin Fransen, Email: k.fransen@medgen.umcg.nl.
Mitja Mitrovic, Email: m.mitrovic@medgen.umcg.nl.
Cleo C van Diemen, Email: c.c.van.diemen@medgen.umcg.nl.
Rinse K Weersma, Email: r.k.weersma@mdl.umcg.nl.
Acknowledgements
We thank Jackie Senior for editing the manuscript. We also thank Gosia Trynka for help with Figure 1. RW is supported by a clinical fellowship (90700281) from the Netherlands Organization for Scientific Research (NWO). KF is supported by an MD/PhD student grant from the Graduate School for Drug Exploration (GUIDE) Institute, University of Groningen. MM is supported by a research grant from the Slovenian Research Agency.
References
- Nell S, Suerbaum S, Josenhans C. The impact of the microbiota on the pathogenesis of IBD: lessons from mouse infection models. Nat Rev Microbiol. 2010;8:564–577. doi: 10.1038/nrmicro2403. [DOI] [PubMed] [Google Scholar]
- Baumgart DC, Sandborn WJ. Inflammatory bowel disease: clinical aspects and established and evolving therapies. Lancet. 2007;369:1641–1657. doi: 10.1016/S0140-6736(07)60751-X. [DOI] [PubMed] [Google Scholar]
- Logan I, Bowlus CL. The geoepidemiology of autoimmune intestinal diseases. Autoimmun Rev. 2010;9:A372–A378. doi: 10.1016/j.autrev.2009.11.008. [DOI] [PubMed] [Google Scholar]
- Brant SR. Update on the heritability of inflammatory bowel disease: the importance of twin studies. Inflamm Bowel Dis. 2011;17:1–5. doi: 10.1002/ibd.21385. [DOI] [PubMed] [Google Scholar]
- Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R, Anderson CA, Bis JC, Bumpstead S, Ellinghaus D, Festen EM, Georges M, Green T, Haritunians T, Jostins L, Latiano A, Mathew CG, Montgomery GW, Prescott NJ, Raychaudhuri S, Rotter JI, Schumm P, Sharma Y, Simms LA, Taylor KD, Whiteman D. et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stappenbeck TS, Rioux JD, Mizoguchi A, Saitoh T, Huett A, Darfeuille-Michaud A, Wileman T, Mizushima N, Carding S, Akira S, Parkes M, Xavier RJ. Crohn's disease: A current perspective on genetics, autophagy and immunity. Autophagy. 2010;7:1–20. doi: 10.4161/auto.7.4.13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham C, Cho J. Inflammatory bowel disease. N Engl J Med. 2009;361:2066–2078. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM, Bitton A, Dassopoulos T, Datta LW, Green T, Griffiths AM, Kistner EO, Murtha MT, Regueiro MD, Rotter JI, Schumm LP, Steinhart AH, Targan SR, Xavier RJ. NIDDK IBD Genetics Consortium. Libioulle C, Sandor C, Lathrop M, Belaiche J, Dewit O, Gut I. et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–962. doi: 10.1038/ng.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM, Bitton A, Dassopoulos T, Datta LW, Green T, Griffiths AM, Kistner EO, Murtha MT, Regueiro MD, Rotter JI, Schumm LP, Steinhart AH, Targan SR, Xavier RJ. NIDDK IBD Genetics Consortium. Libioulle C, Sandor C, Lathrop M, Belaiche J, Dewit O, Gut I. et al. Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn's disease. Hum Mol Genet. 2010;19:3468–3476. doi: 10.1093/hmg/ddq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden SK, Sutton P, Karlsson NG, Korolik V, McGuckin MA. Mucins in the mucosal barrier to infection. Mucosal Immunol. 2008;1:183–197. doi: 10.1038/mi.2008.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Inflammatory Bowel Disease Genetics Consortium (IIBDGC). http://www.ibdgenetics.org
- Hunt KA, Zhernakova A, Turner G, Heap GA, Franke L, Bruinenberg M, Romanos J, Dinesen LC, Ryan AW, Panesar D, Gwilliam R, Takeuchi F, McLaren WM, Holmes GK, Howdle PD, Walters JR, Sanders DS, Playford RJ, Trynka G, Mulder CJ, Mearin ML, Verbeek WH, Trimble V, Stevens FM, O'Morain C, Kennedy NP, Kelleher D, Pennington DJ, Strachan DP, McArdle WL. et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet. 2008;40:395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, Thomson BP, Remmers EF, Eyre S, Hinks A, Guiducci C, Catanese JJ, Xie G, Stahl EA, Chen R, Alfredsson L, Amos CI, Ardlie KG. BIRAC Consortium. Barton A, Bowes J, Burtt NP, Chang M, Coblyn J, Costenbader KH, Criswell LA, Crusius JB, Cui J, De Jager PL, Ding B, Emery P, Flynn E, Harrison P, Hocking LJ, Huizinga TW. et al. Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat Genet. 2009;41:1313–1318. doi: 10.1038/ng.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Zhang H, Kugathasan S, Annese V, Bradfield JP, Russell RK, Sleiman PM, Imielinski M, Glessner J, Hou C, Wilson DC, Walters T, Kim C, Frackelton EC, Lionetti P, Barabino A, Van Limbergen J, Guthery S, Denson L, Piccoli D, Li M, Dubinsky M, Silverberg M, Griffiths A, Grant SF, Satsangi J, Baldassano R, Hakonarson H. Diverse genome-wide association studies associate the IL12/IL23 pathway with Crohn Disease. Am J Hum Genet. 2009;84:399–405. doi: 10.1016/j.ajhg.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransen K, Visschedijk MC, van Sommeren S, Fu JY, Franke L, Festen EA, Stokkers PC, van Bodegraven AA, Crusius JB, Hommes DW, Zanen P, de Jong DJ, Wijmenga C, van Diemen CC, Weersma RK. Analysis of SNPs with an effect on gene expression identifies UBE2L3 and BCL3 as potential new risk genes for Crohn's disease. Hum Mol Genet. 2010;19:3482–3488. doi: 10.1093/hmg/ddq264. [DOI] [PubMed] [Google Scholar]
- Zhernakova A, van Diemen CC, Wijmenga C. Detecting shared pathogenesis from the shared genetics of immune-related diseases. Nat Rev Genet. 2009;10:43–55. doi: 10.1038/nrg2489. [DOI] [PubMed] [Google Scholar]
- Wang K, Baldassano R, Zhang H, Qu HQ, Imielinski M, Kugathasan S, Annese V, Dubinsky M, Rotter JI, Russell RK, Bradfield JP, Sleiman PM, Glessner JT, Walters T, Hou C, Kim C, Frackelton EC, Garris M, Doran J, Romano C, Catassi C, Van Limbergen J, Guthery SL, Denson L, Piccoli D, Silverberg MS, Stanley CA, Monos D, Wilson DC, Griffiths A. et al. Comparative genetic analysis of inflammatory bowel disease and type 1 diabetes implicates multiple loci with opposite effect. Hum Mol Gen. 2010;19:2059–2067. doi: 10.1093/hmg/ddq078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danoy P, Pryce K, Hadler J, Bradbury LA, Farrar C, Pointon J. Australo-Anglo-American Spondyloarthritis Consortium; Ward M, Weisman M, Reveille JD, Wordsworth BP, Stone MA. Spondyloarthritis Research Consortium of Canada. Maksymowych WP, Rahman P, Gladman D, Inman RD, Brown MA. Association of variants at 1q32 and STAT3 with Ankylosing Spondilitis suggests genetic overlap with Crohn's disease. PLoS Genet. 2010;6:e1001195. doi: 10.1371/journal.pgen.1001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festen EAM, Goyette P, Green T, Beauchamp C, Boucher G, Trynka G. A meta-analysis of genome wide association scans identifies TAGAP and PUS10 as shared risk loci for Crohn's disease and celiac disease. PLoS Genet. 2011;7:e1001283. doi: 10.1371/journal.pgen.1001283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Mahurkar S, Banerjee R, Rani SV, Thakur N, Guduru VR, Duvvuru NR, Chandak GR. Common variants in NOD2 and IL23R are not associated with inflammatory bowel disease in Indian patients. J Gastroenterol Hepatol. 2010. in press . [DOI] [PubMed]
- Shi J, Lu Z, Zhernakova A, Qian J, Zhu F, Sun G, Zhu L, Ma X, Dijkstra G, Wijmenga C, Faber KN, Lu X, Weersma RK. Haplotype-based analysis of ulcerative colitis risk loci identifies both IL2 and IL21 as susceptibility genes in Han Chinese. Inflamm Bowel Dis. 2010. in press . [DOI] [PubMed]
- Park JH, Wacholder S, Gail MH, Peters U, Jacobs KB, Chanock SJ, Chatterjee N. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat Genet. 2010;42:570–575. doi: 10.1038/ng.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex disease. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes - A Deep Catalog of Human Genetic Variation. http://www.1000genomes.org
- Momozawa Y, Mni M, Nakamura K, Coppieters W, Almer S, Amininejad L. Resequencing of positional candidates identifies low frequency IL23R coding variants protecting against inflammatory bowel disease. Nat Genet. 2011;43:43–47. doi: 10.1038/ng.733. [DOI] [PubMed] [Google Scholar]
- Feng T, Zhu X. Genome-wide searching of rare genetic variants in WTCCC data. Hum Genet. 2010;128:269–280. doi: 10.1007/s00439-010-0849-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo S, Pennacchio LA, Fu Y, Boerwinkle E, Tybjaerg-Hansen A, Hobbs HH, Cohen JC. Population-based resequencing of ANGPTL4 uncovers variations that reduce triglycerides and increase HDL. Nat Genet. 2007;39:513–516. doi: 10.1038/ng1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, González JR, Gratacòs M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F. et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Database of Genomic Variants - a curated catalogue of structural variation in the human genome. http://projects.tcag.ca/variation/ [DOI] [PMC free article] [PubMed]
- Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, Zhang H, Estes A, Brune CW, Bradfield JP, Imielinski M, Frackelton EC, Reichert J, Crawford EL, Munson J, Sleiman PM, Chiavacci R, Annaiah K, Thomas K, Hou C, Glaberson W, Flory J, Otieno F, Garris M, Soorya L, Klei L, Piven J, Meyer KJ, Anagnostou E, Sakurai T. et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diskin SJ, Hou C, Glessner JT, Attiyeh EF, Laudenslager M, Bosse K, Cole K, Mossé YP, Wood A, Lynch JE, Pecor K, Diamond M, Winter C, Wang K, Kim C, Geiger EA, McGrady PW, Blakemore AIF, London WB, Shaikh TH, Bradfield J, Grant SFA, Li H, Devoto M, Rappaport ER, Hakonarson H, Maris JM. Copy number variation at 1q21.1 associated with neuroblastoma. Nature. 2009;459:987–991. doi: 10.1038/nature08035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcocks LC, Lyons PA, Clatworthy MR, Robinson JI, Yang W, Newland SA, Plagnol V, McGovern NN, Condliffe AM, Chilvers ER, Adu D, Jolly EC, Watts R, Lau YL, Morgan AW, Nash G, Smith KG. Copy number of FCGR3B, which is associated with systematic lupus erythematosus, correlates with protein expression and immune complex uptake. J Exp Med. 2008;205:1573–1582. doi: 10.1084/jem.20072413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaschl H, Aitman TJ, Vyse TJ. Copy number variation in the human genome and its implication in autoimmunity. Clin Exp Immunol. 2009;156:12–16. doi: 10.1111/j.1365-2249.2008.03865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellermann K, Stange DE, Schaeffeler E, Schmalzl H, Wehkamp J, Bevins CL, Reinisch W, Teml A, Schwab M, Lichter P, Radlwimmer B, Stange EF. A chromosome 8 gene-cluster polymorphism with low human β-defensin 2 gene copy number predisposes to Crohn's disease of the colon. Am J Hum Genet. 2006;79:439–448. doi: 10.1086/505915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley R, Pearson J, Gearry R, Barclay M, McKinney C, Merriman T, Roberts R. Association of higher DEFB4 genomic copy number with Crohn's disease. Am J Gastroenterol. 2010;105:354–359. doi: 10.1038/ajg.2009.582. [DOI] [PubMed] [Google Scholar]
- Aldhous MC, Abu Bakar S, Prescott NJ, Palla R, Soo K, Mansfield JC, Mathew CG, Satsangi J, Armour JA. Measurement methods and accuracy in copy number variation: failure to replicate associations of beta-defensin copy number with Crohn's disease. Hum Mol Gen. 2010;19:4930–4938. doi: 10.1093/hmg/ddq411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, Goyette P, Zody MC, Hall JL, Brant SR, Cho JH, Duerr RH, Silverberg MS, Taylor KD, Rioux JD, Altshuler D, Daly MJ, Xavier RJ. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. Nat Genet. 2008;40:1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Wellcome Trust Consortium. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010;464:713–720. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glocker EO, Kotlarz D, Boztug K, Gertz EM, Schäffer AA, Noyan F, Perro M, Diestelhorst J, Allroth A, Murugan D, Hätscher N, Pfeifer D, Sykora KW, Sauer M, Kreipe H, Lacher M, Nustede R, Woellner C, Baumann U, Salzer U, Koletzko S, Shah N, Segal AW, Sauerbrey A, Buderus S, Snapper SB, Grimbacher B, Klein C. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361:2033–2045. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akolkar PN, Gulwani-Akolkar B, Heresbach D, Lin XY, Fisher S, Katz S, Silver J. Differences in risk of Crohn's disease in offspring of mothers and fathers with inflammatory bowel disease. Am J Gastroenterol. 1997;92:2241–2244. [PubMed] [Google Scholar]
- Hanson RL, Kobes S, Lindsay RS, Knowler WC. Assessment of parent-of-origin effects in linkage analysis of quantitative traits. Am J Hum Genet. 2001;68:951–962. doi: 10.1086/319508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JH, Williams SM. Epistasis and its implications for personal genetics. Am J Hum Genet. 2009;85:309–320. doi: 10.1016/j.ajhg.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern DP, Rotter JI, Mei L, Haritunians T, Landers C, Derkowski C, Dutridge D, Dubinsky M, Ippoliti A, Vasiliauskas E, Mengesha E, King L, Pressman S, Targan SR, Taylor KD. Genetic epistasis of IL23/IL17 related genes in Crohn's disease. Inflamm Bowel Dis. 2009;15:883–889. doi: 10.1002/ibd.20855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Donnelly P, Cardon LR. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat Genet. 2005;37:413–417. doi: 10.1038/ng1537. [DOI] [PubMed] [Google Scholar]
- Cordell HJ. Detecting gene-gene interactions that underlie human diseases. Nat Rev Genet. 2009;10:392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D. Gene-environment-wide association studies: emerging approaches. Nat Rev Genet. 2010;11:259–272. doi: 10.1038/nrg2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Heide F, Nolte IM, Kleibeuker JH, Wijmenga C, Dijkstra G, Weersma RK. Differences in genetic background between active smokers, passive smokers, and non-smokers with Crohn's disease. Am J Gastroenterol. 2010;105:1165–1172. doi: 10.1038/ajg.2009.659. [DOI] [PubMed] [Google Scholar]
- Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, Head RD, Xavier R, Stappenbeck TS, Virgin HW. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier S, Olek A. Diabetes: a candidate disease for efficient DNA methylation profiling. J Nutr. 2002;132:2440S–2443S. doi: 10.1093/jn/132.8.2440S. [DOI] [PubMed] [Google Scholar]
- Kim YI, Logan JW, Mason JB, Roubenoff R. DNA hypomethylation in inflammatory arthritis: reversal with methotrexate. J Lab Clin Med. 1996;128:165–172. doi: 10.1016/S0022-2143(96)90008-6. [DOI] [PubMed] [Google Scholar]
- Cara Terribas CJ, Gonzalez Guijarro L. Hypomethylation and multiple sclerosis, the susceptibility factor? Neurologia. 2002;17:132–135. [PubMed] [Google Scholar]
- Issa JP. Epigenetic variation and human disease. J Nutr. 2002;132:2388S–2392S. doi: 10.1093/jn/132.8.2388S. [DOI] [PubMed] [Google Scholar]
- Ahuja N, Issa JP. Aging, methylation and cancer. Histol Histopathol. 2000;15:835–842. doi: 10.14670/HH-15.835. [DOI] [PubMed] [Google Scholar]
- Nadeau JH. Transgenerational genetic effects on phenotypic variation and disease risk. Hum Mol Genet. 2009;18:202–210. doi: 10.1093/hmg/ddp366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- Economou M, Trikalinos TA, Loizou KT, Tsianos EV, Ioannidis JP. Differential effects of NOD2 variants on Crohn's disease risk and phenotype in diverse populations: a metaanalysis. Am J Gastroenterol. 2004;99:2393–2404. doi: 10.1111/j.1572-0241.2004.40304.x. [DOI] [PubMed] [Google Scholar]
- Shen X, Shi R, Zhang H, Li K, Zhao Y, Zhang R. The Toll-like receptor 4 D299G and T399I polymorphisms are associated with Crohn's disease and ulcerative colitis: a meta-analysis. Digestion. 2010;81:69–77. doi: 10.1159/000260417. [DOI] [PubMed] [Google Scholar]