Abstract
Plausible genome-wide associations for episodic neurological diseases (such as migraine, epilepsy and ataxias) have been slow to emerge. The first such association was reported in a recent genome-wide association study of migraine, with quantitative expression analysis linking the variant to a nearby regulatory gene, MTDH/AEG-1. This putative mechanism, regulating the expression of the primary glutamate transporter in the brain, EAAT2/GLT-1, has interesting implications bridging the gap between Mendelian and common forms in this key group of disorders.
Research highlight
The recent boom in genome-wide association (GWA) studies has resulted in a vast expansion of our knowledge of common variants influencing human diseases. So far, 1,888 single nucleotide polymorphism (SNP) associations have been reported, 1,435 of which are unique to the SNP [1]. Neurological diseases are underrepresented among successful GWA study findings. They account for only 36 publications, reporting 63 unique SNPs, out of a total of 812 publications and 3,942 unique SNPs [1]. If we exclude the APOE locus and the HLA region of chromosome 6p21 (which are associated with multiple diseases), the number of unique significant neurological loci found drops to 42.
A probable explanation for the underrepresentation is the cost and difficulty of obtaining large sample sets; for anthropomorphic traits such as height and readily quantifiable measures such as blood lipids, sample sizes of up to 100,000 have been analyzed in widely successful GWA studies. A further complication in neurological diseases is the complexity of phenotyping brain phenotypes (description-based only, as in migraine), and the myriad diagnostic divisions (as in epilepsy), which together make collecting sufficiently large samples of any given subtype a hard task. Estimates from several diseases suggest that sample sizes exceeding 10,000 are required to reach sufficient statistical power to detect new loci [2].
Gene loci predisposing to migraine
The recent publication of the large-scale GWA study by the International Headache Genetics Consortium [3] in Nature Genetics was the first GWA study reported for a headache disorder. As the primary finding, it reported the first genome-wide significant association in the group of paroxysmal neurological diseases, which comprises epilepsy, migraine, episodic ataxia and various cerebrovascular and sleep disorders. In this study [3], a total of 5,933 cases and 50,809 controls were used to establish a single significant locus. The discovery sample consisted of 2,731 migraine cases collected from four European headache centers located in three countries. The initial finding was replicated in 3,202 cases and 40,062 independent controls from four European countries. Using RNA expression data from lymphoblastoid cell lines, the identified variant rs1835740 was found to be an expression quantitative locus (eQTL) affecting glutamate metabolism. As glutamate is a major excitatory neurotransmitter in the central nervous system (CNS), the eQTL finding provides a promising link between the genetic association and function. The eQTL data suggest that the predisposing allele of rs1835740 stimulates the expression of metadherin (MTDH, also called astrocyte elevated gene-1, AEG-1), which downregulates excitatory amino-acid transporter 2 (EAAT2, also called glutamate transporter 1, GLT-1), the main glutamate transporter in the brain. This finding stimulates the hypothesis that accumulation of glutamate, a potent neurotransmitter, in the synaptic cleft might contribute to the occurrence of the migraine attack.
A problem in uncovering the genetic component of migraine has been the dearth of unambiguous results. After the early encouraging findings of the mutations causing the rare Mendelian forms of migraine (familial hemiplegic migraine, FHM) and the loci identified in the first linkage scan for common migraine [4], progress has been slow. The genes identified in FHM, similar to the corresponding findings for epilepsy, pinpointed ion channel defects, grouping these diseases into channelopathies. However, neither targeted ion channel association studies [5] nor the first GWA studies have provided evidence of the involvement of common variants in ion channel genes in common forms of migraine or epilepsy [3,6]. Candidate gene studies covering obvious targets (the ion channels and the estrogen system) and up to 1,000 migraine cases and controls turned up conflicting results at best [4]. Although a number of loci using family-based linkage studies were identified, no genes for the common forms were found. Thus, the genetic and functional link between FHM and common forms of migraine remained unclear.
Interestingly, the GWA association and the eQTL data point to the same synaptic transmission mechanism (the imbalance in glutamate release and its clearance from the synaptic cleft) that has been shown to be a key component of FHM pathogenesis in migraine with aura (Figure 1), although through a more peripheral part of the pathway in the latter [3]. Although this provides a potential link between FHM and common migraine pathophysiology, it should be noted that because of the ascertainment of the GWA study cases from tertiary headache clinics, it is not possible to estimate the effect of the identified variant on a population level from the current data. What we can conclude is that among patients whose migraine is severe enough to need tertiary care, this variant is significantly overrepresented. Furthermore, mutations in the gene encoding the EAAT1 transporter also result in an episodic disease phenotype [7]. The diversity of pathways resulting in similar synaptic events might reflect one of the challenges we might encounter in dissecting the genetic predisposition of paroxysmal CNS disorders.
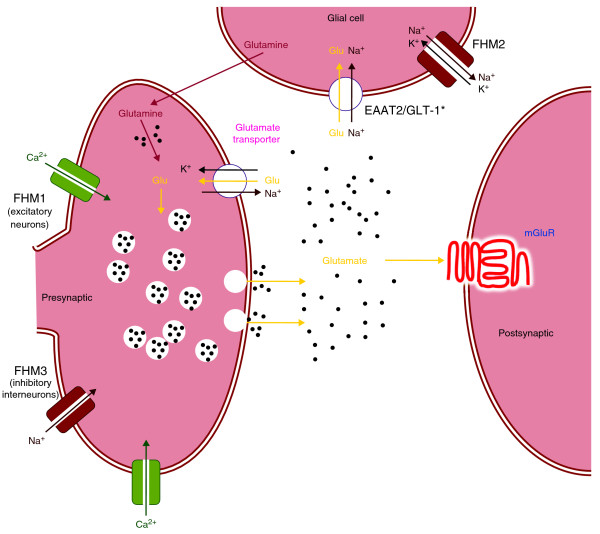
Figure 1.
Localization and effect of a variant identified in the recent genome-wide association study of migraine in synaptic transmission, together with the previously known mutations in familial hemiplegic migraine (FHM: genes are FHM1, CACNA1A; FHM2, ATP1A2; FHM3, SCN1A). The asterisk indicates the excitatory amino acid transporter 2 (EAAT2/GLT-1) recently linked to migraine. Glu, glutamate; FHM1-3, products of genes reported for familial hemiplegic migraine; mGluR, metabolic glutamate receptor. Black dots indicate the accumulation of the neurotransmitter glutamate in the synaptic cleft.
Does the clinical classification facilitate gene identification?
The obvious next steps for GWA studies are to address different migraine subtypes (such as migraine without aura) and from different recruitment settings (such as population-based, instead of clinic based migraine cases). As mentioned above, many neurological diseases lack such easily quantifiable phenotypes as serum lipid or blood sugar levels that are available elsewhere. The lack of quantitative phenotypes is also reflected in dissecting the umbrella diagnosis into subphenotypes, as they may similarly be based on non-objective measures. Thus, there is a chance that clinical subtypes might be of limited help and traditional diagnostic boundaries might not correlate with identified susceptibility genes. Although data are limited, there is emerging evidence that susceptibility loci can cross diagnostic boundaries and be associated with multiple traits, such as schizophrenia and bipolar disorder [8]. There are similar overlaps with many autoimmune diseases and immunologically mediated traits [9]. There is evidence that the same locus or even same variant can contribute to the susceptibility of different disease outcomes.
A case in point is that in another major paroxysmal CNS disorder, epilepsy, there is a long tradition of rigorous clinical subgrouping. If secondary factors contribute to the precise form of the disease, such strong divisions might distort from the ultimate goal of identification of the biological basis of seizure susceptibility, as was discussed in a recent editorial [10]. In the case of migraine, the diagnostic classification is perhaps more dynamic than for epilepsies. The classification recognizes that most patients suffer from various different forms of attacks, such as a combination of migraine attacks with and without aura. A migraine patient may, over the course of decades, pass through different types of migraine. Diagnostic flexibility accounts better for the varying severity spectrum both between patients and along a timeline. Although this flexibility does not abolish the phenotyping challenges, it is likely to be helpful in large GWA meta-analyses. When subsequent migraine association studies have identified more loci and we start to tackle their interplay in disease susceptibility, this might provide us with new tools for disease classification.
It should perhaps not be surprising that a highly adaptable system such as the brain, with its myriad, cross-compensating pathways, can react with a very limited set of overt phenotypes, such as headache, to a variety of biological challenges. Therefore, summing over all possible symptom backgrounds in a large-scale GWA study is likely to result in only small odds ratios and thus sample sizes exceeding 10,000 may well be necessary. These large samples should also allow secondary analyses addressing questions related to disease subclassification and co-morbidity.
Conclusions
The first genetic link to common forms of migraine was recently reported, pointing the way towards alterations in glutamate homeostasis. This finding is the first step to uncovering the genetic background. Upcoming larger migraine GWA studies, across different ascertainment schemes, are likely to identify more genes and cellular pathways contributing to migraine susceptibility and will hopefully also shed light on the potentially different genetic background of different migraine subtypes. The emerging wealth of GWA data from other neurological and neuropsychiatric traits will also provide an opportunity to study whether some loci or pathways contribute to the susceptibility of co-morbid traits and whether genetic data will cross traditional diagnostic boundaries. However, it is evident that not all of genetic predisposition is explained by common variants, and future exome and whole-genome sequencing studies in large datasets will probably identify contributing highly penetrant low-frequency variants and expand our knowledge of the migraine susceptibility landscape.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
All authors participated in writing the manuscript.
Contributor Information
Verneri Anttila, Email: verneri.anttila@sanger.ac.uk.
Maija Wessman, Email: maija.wessman@helsinki.fi.
Mikko Kallela, Email: mikko.kallela@pp.fimnet.fi.
Aarno Palotie, Email: aarno.palotie@sanger.ac.uk.
References
- A Catalog of Published Genome-Wide Association Studies. http://www.genome.gov/gwastudies
- Park JH, Wacholder S, Gail MH, Peters U, Jacobs KB, Chanock SJ, Chatterjee N. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat Genet. 2010;42:570–575. doi: 10.1038/ng.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anttila V, Stefansson H, Kallela M, Todt U, Terwindt GM, Calafato MS, Nyholt DR, Dimas AS, Freilinger T, Muller-Myhsok B, Artto V, Inouye M, Alakurtti K, Kaunisto MA, Hämäläinen E, de Vries B, Stam AH, Weller CM, Heinze A, Heinze-Kuhn K, Goebel I, Borck G, Göbel H, Steinberg S, Wolf C, Björnsson A, Gudmundsson G, Kirchmann M, Hauge A, Werge T. et al. Genome-wide association study of migraine implicates a common susceptibility variant on 8q22.1. Nat Genet. 2010;42:869–873. doi: 10.1038/ng.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessman M, Terwindt GM, Kaunisto MA, Palotie A, Ophoff RA. Migraine: a complex genetic disorder. Lancet Neurol. 2007;6:521–532. doi: 10.1016/S1474-4422(07)70126-6. [DOI] [PubMed] [Google Scholar]
- Nyholt DR, LaForge KS, Kallela M, Alakurtti K, Anttila V, Farkkila M, Hamalainen E, Kaprio J, Kaunisto MA, Heath AC, Montgomery GW, Göbel H, Todt U, Ferrari MD, Launer LJ, Frants RR, Terwindt GM, de Vries B, Verschuren WM, Brand J, Freilinger T, Pfaffenrath V, Straube A, Ballinger DG, Zhan Y, Daly MJ, Cox DR, Dichgans M, van den Maagdenberg AM, Kubisch C. et al. A high-density association screen of 155 ion transport genes for involvement with common migraine. Hum Mol Genet. 2008;17:3318–3331. doi: 10.1093/hmg/ddn227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasperaviciūte D, Catarino CB, Heinzen EL, Depondt C, Cavalleri GL, Caboclo LO, Tate SK, Jamnadas-Khoda J, Chinthapalli K, Clayton LM, Shianna KV, Radtke RA, Mikati MA, Gallentine WB, Husain AM, Alhusaini S, Leppert D, Middleton LT, Gibson RA, Johnson MR, Matthews PM, Hosford D, Heuser K, Amos L, Ortega M, Zumsteg D, Wieser HG, Steinhoff BJ, Krämer G, Hansen J. et al. Common genetic variation and susceptibility to partial epilepsies: a genome-wide association study. Brain. 2010;133:2136–2147. doi: 10.1093/brain/awq130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jen JC, Wan J, Palos TP, Howard BD, Baloh RW. Mutation in the glutamate transporter EAAT1 causes episodic ataxia, hemiplegia, and seizures. Neurology. 2005;65:529–534. doi: 10.1212/01.WNL.0000172638.58172.5a. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhernakova A, van Diemen CC, Wijmenga C. Detecting shared pathogenesis from the shared genetics of immune-related diseases. Nat Rev Genet. 2009;10:43–55. doi: 10.1038/nrg2489. [DOI] [PubMed] [Google Scholar]
- Johnson MR. Comment on "Blinders, phenotype, and fashionable genetic analysis: A critical examination of the current state of epilepsy genetic studies. Epilepsia. 2011;52:190–191. doi: 10.1111/j.1528-1167.2010.02791.x. [DOI] [PubMed] [Google Scholar]