Abstract
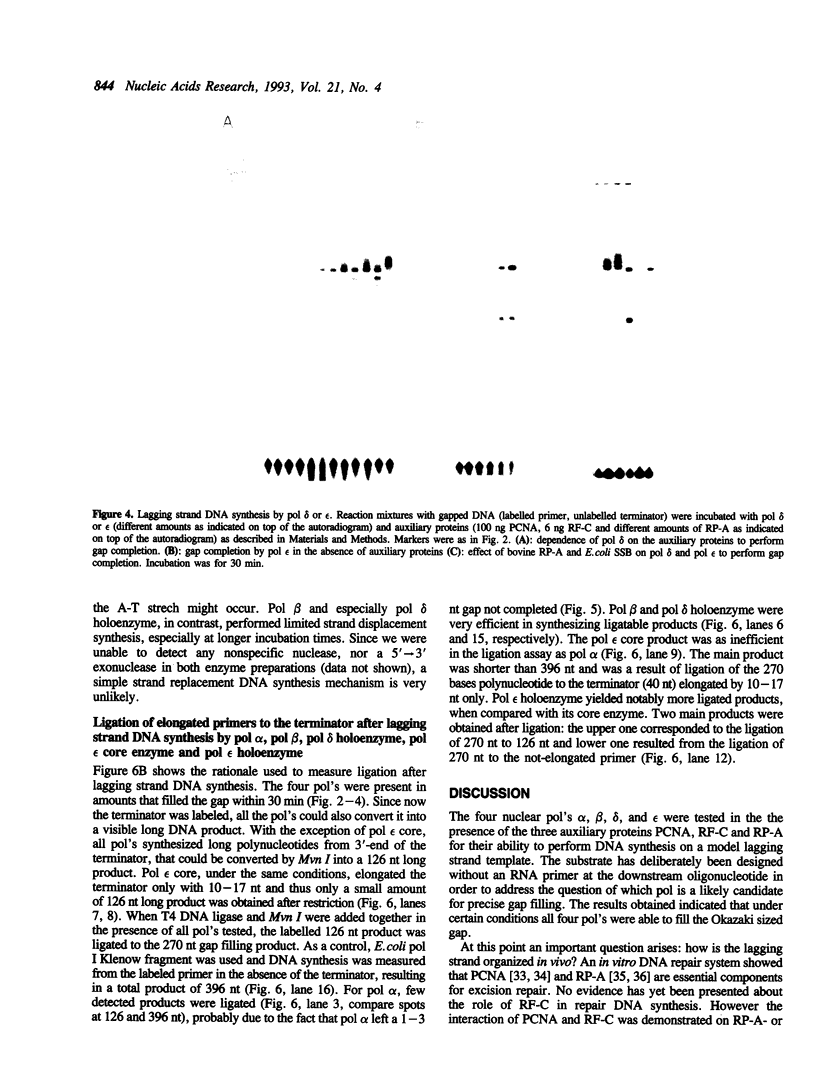
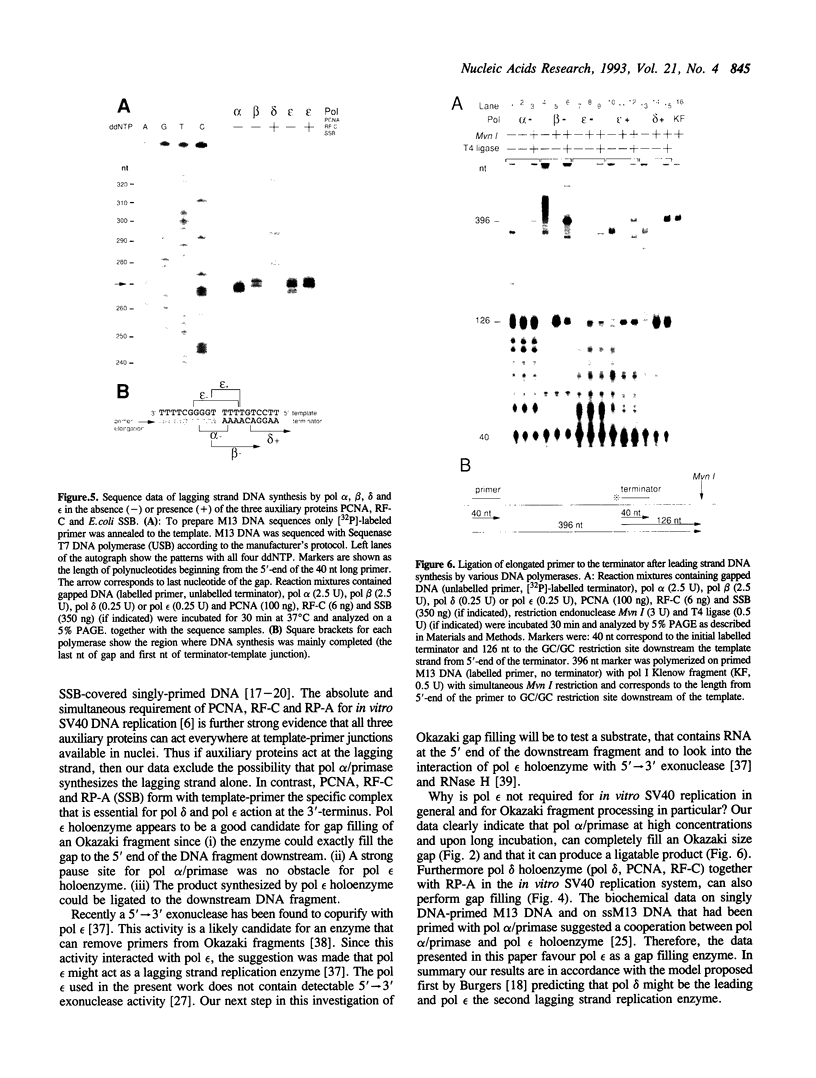
By using a defined gapped DNA substrate that mimics a lagging strand of 230 nucleotides and that contains a defined pause site, we have analyzed calf thymus DNA polymerases (pol) alpha, beta, delta, and epsilon in the presence of the three auxiliary proteins proliferating cell nuclear antigen (PCNA), replication factor C (RF-C) and replication protein A (RP-A) for their ability to complete an Okazaki fragment. Pol alpha alone could fill the gap to near completion, but was strongly stopped by the pause site. Addition of low amounts of RP-A resulted in an increased synthesis by pol alpha past the pause site. In contrast, high amounts of RP-A strongly inhibited gap filling by pol alpha. Further inhibition was evident when the two other auxiliary proteins, PCNA and RF-C, were added in addition to RP-A. Pol beta could completely fill the gap without specific pausing and also was strongly inhibited by RP-A. PCNA and RF-C had no detectable effect on pol beta. Pol delta, relied as expected, on all three auxiliary proteins for complete gap filling synthesis and could, upon longer incubation, perform a limited amount of strand displacement synthesis. Pol epsilon core enzyme was able to fill the gap completely, but like pol alpha, essentially stopped at the pause site. This pausing could only be overcome upon addition of PCNA, RF-C and E. coli single-stranded DNA binding protein. Thus pol epsilon holoenzyme preferentially synthesized to the end of the gap without pausing. Ligation of the DNA products indicated that pol beta core enzyme, pol delta and pol epsilon holoenzymes (but not pol alpha and pol epsilon core enzyme) synthesized products that were easily ligatable. Our results indicate that pol epsilon holoenzyme fills a defined lagging strand gapped template to exact completion and is able to pass a pause site. The data favour the hypothesis of Burgers (Burgers, P.M.J. (1991) J. Biol. Chem. 266, 22698-22706) that pol epsilon might be a candidate for the second replication enzyme at the lagging strand of the replication fork.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Araki H., Ropp P. A., Johnson A. L., Johnston L. H., Morrison A., Sugino A. DNA polymerase II, the probable homolog of mammalian DNA polymerase epsilon, replicates chromosomal DNA in the yeast Saccharomyces cerevisiae. EMBO J. 1992 Feb;11(2):733–740. doi: 10.1002/j.1460-2075.1992.tb05106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bambara R. A., Jessee C. B. Properties of DNA polymerases delta and epsilon, and their roles in eukaryotic DNA replication. Biochim Biophys Acta. 1991 Jan 17;1088(1):11–24. doi: 10.1016/0167-4781(91)90147-e. [DOI] [PubMed] [Google Scholar]
- Boulet A., Simon M., Faye G., Bauer G. A., Burgers P. M. Structure and function of the Saccharomyces cerevisiae CDC2 gene encoding the large subunit of DNA polymerase III. EMBO J. 1989 Jun;8(6):1849–1854. doi: 10.1002/j.1460-2075.1989.tb03580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers P. M., Bambara R. A., Campbell J. L., Chang L. M., Downey K. M., Hübscher U., Lee M. Y., Linn S. M., So A. G., Spadari S. Revised nomenclature for eukaryotic DNA polymerases. Eur J Biochem. 1990 Aug 17;191(3):617–618. doi: 10.1111/j.1432-1033.1990.tb19165.x. [DOI] [PubMed] [Google Scholar]
- Burgers P. M. Saccharomyces cerevisiae replication factor C. II. Formation and activity of complexes with the proliferating cell nuclear antigen and with DNA polymerases delta and epsilon. J Biol Chem. 1991 Nov 25;266(33):22698–22706. [PubMed] [Google Scholar]
- Coverley D., Kenny M. K., Lane D. P., Wood R. D. A role for the human single-stranded DNA binding protein HSSB/RPA in an early stage of nucleotide excision repair. Nucleic Acids Res. 1992 Aug 11;20(15):3873–3880. doi: 10.1093/nar/20.15.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coverley D., Kenny M. K., Munn M., Rupp W. D., Lane D. P., Wood R. D. Requirement for the replication protein SSB in human DNA excision repair. Nature. 1991 Feb 7;349(6309):538–541. doi: 10.1038/349538a0. [DOI] [PubMed] [Google Scholar]
- DePamphilis M. L., Wassarman P. M. Replication of eukaryotic chromosomes: a close-up of the replication fork. Annu Rev Biochem. 1980;49:627–666. doi: 10.1146/annurev.bi.49.070180.003211. [DOI] [PubMed] [Google Scholar]
- Fairman M. P., Stillman B. Cellular factors required for multiple stages of SV40 DNA replication in vitro. EMBO J. 1988 Apr;7(4):1211–1218. doi: 10.1002/j.1460-2075.1988.tb02933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgaki A., Strack B., Podust V., Hübscher U. DNA unwinding activity of replication protein A. FEBS Lett. 1992 Aug 24;308(3):240–244. doi: 10.1016/0014-5793(92)81283-r. [DOI] [PubMed] [Google Scholar]
- Goulian M., Richards S. H., Heard C. J., Bigsby B. M. Discontinuous DNA synthesis by purified mammalian proteins. J Biol Chem. 1990 Oct 25;265(30):18461–18471. [PubMed] [Google Scholar]
- Hagemeier A., Grosse F. A distinct form of ribonuclease H from calf thymus stimulates its homologous DNA-polymerase-alpha-primase complex. Eur J Biochem. 1989 Nov 20;185(3):621–628. doi: 10.1111/j.1432-1033.1989.tb15158.x. [DOI] [PubMed] [Google Scholar]
- Hübscher U., Thömmes P. DNA polymerase epsilon: in search of a function. Trends Biochem Sci. 1992 Feb;17(2):55–58. doi: 10.1016/0968-0004(92)90499-y. [DOI] [PubMed] [Google Scholar]
- Johnson L. M., Snyder M., Chang L. M., Davis R. W., Campbell J. L. Isolation of the gene encoding yeast DNA polymerase I. Cell. 1985 Nov;43(1):369–377. doi: 10.1016/0092-8674(85)90042-x. [DOI] [PubMed] [Google Scholar]
- Lee S. H., Pan Z. Q., Kwong A. D., Burgers P. M., Hurwitz J. Synthesis of DNA by DNA polymerase epsilon in vitro. J Biol Chem. 1991 Nov 25;266(33):22707–22717. [PubMed] [Google Scholar]
- Linn S. How many pols does it take to replicate nuclear DNA? Cell. 1991 Jul 26;66(2):185–187. doi: 10.1016/0092-8674(91)90608-2. [DOI] [PubMed] [Google Scholar]
- Morrison A., Araki H., Clark A. B., Hamatake R. K., Sugino A. A third essential DNA polymerase in S. cerevisiae. Cell. 1990 Sep 21;62(6):1143–1151. doi: 10.1016/0092-8674(90)90391-q. [DOI] [PubMed] [Google Scholar]
- Nethanel T., Kaufmann G. Two DNA polymerases may be required for synthesis of the lagging DNA strand of simian virus 40. J Virol. 1990 Dec;64(12):5912–5918. doi: 10.1128/jvi.64.12.5912-5918.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podust V. N., Georgaki A., Strack B., Hübscher U. Calf thymus RF-C as an essential component for DNA polymerase delta and epsilon holoenzymes function. Nucleic Acids Res. 1992 Aug 25;20(16):4159–4165. doi: 10.1093/nar/20.16.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podust V., Mikhailov V., Georgaki A., Hübscher U. DNA polymerase delta and epsilon holoenzymes from calf thymus. Chromosoma. 1992;102(1 Suppl):S133–S141. doi: 10.1007/BF02451797. [DOI] [PubMed] [Google Scholar]
- Prelich G., Tan C. K., Kostura M., Mathews M. B., So A. G., Downey K. M., Stillman B. Functional identity of proliferating cell nuclear antigen and a DNA polymerase-delta auxiliary protein. Nature. 1987 Apr 2;326(6112):517–520. doi: 10.1038/326517a0. [DOI] [PubMed] [Google Scholar]
- Siegal G., Turchi J. J., Myers T. W., Bambara R. A. A 5' to 3' exonuclease functionally interacts with calf DNA polymerase epsilon. Proc Natl Acad Sci U S A. 1992 Oct 15;89(20):9377–9381. doi: 10.1073/pnas.89.20.9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitney K. C., Budd M. E., Campbell J. L. DNA polymerase III, a second essential DNA polymerase, is encoded by the S. cerevisiae CDC2 gene. Cell. 1989 Feb 24;56(4):599–605. doi: 10.1016/0092-8674(89)90582-5. [DOI] [PubMed] [Google Scholar]
- Tsurimoto T., Melendy T., Stillman B. Sequential initiation of lagging and leading strand synthesis by two different polymerase complexes at the SV40 DNA replication origin. Nature. 1990 Aug 9;346(6284):534–539. doi: 10.1038/346534a0. [DOI] [PubMed] [Google Scholar]
- Tsurimoto T., Stillman B. Multiple replication factors augment DNA synthesis by the two eukaryotic DNA polymerases, alpha and delta. EMBO J. 1989 Dec 1;8(12):3883–3889. doi: 10.1002/j.1460-2075.1989.tb08567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser T., Gassmann M., Thömmes P., Ferrari E., Hafkemeyer P., Hübscher U. Biochemical and functional comparison of DNA polymerases alpha, delta, and epsilon from calf thymus. J Biol Chem. 1991 Jun 5;266(16):10420–10428. [PubMed] [Google Scholar]
- Wintersberger U., Wintersberger E. Studies on deoxyribonucleic acid polymerases from yeast. 1. Parial purification and properties of two DNA polymerases from mitochondria-free cell extracts. Eur J Biochem. 1970 Mar 1;13(1):11–19. doi: 10.1111/j.1432-1033.1970.tb00893.x. [DOI] [PubMed] [Google Scholar]
- Wobbe C. R., Weissbach L., Borowiec J. A., Dean F. B., Murakami Y., Bullock P., Hurwitz J. Replication of simian virus 40 origin-containing DNA in vitro with purified proteins. Proc Natl Acad Sci U S A. 1987 Apr;84(7):1834–1838. doi: 10.1073/pnas.84.7.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold M. S., Kelly T. Purification and characterization of replication protein A, a cellular protein required for in vitro replication of simian virus 40 DNA. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2523–2527. doi: 10.1073/pnas.85.8.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]