Abstract
Biogenesis of eukaryotic tRNAs requires transcription by RNA polymerase III and subsequent processing. 5′ processing of precursor tRNA occurs by a single mechanism, cleavage by RNase P, and usually occurs before 3′ processing although some conditions allow observation of the 3′-first pathway. 3′ processing is relatively complex and is the focus of this review. Precursor RNA 3′ end formation begins with pol III termination generating a variable length 3′ oligo(U) tract that represents an underappreciated and previously unreviewed determinant of processing. Evidence that the pol III-intrinsic 3′ exonuclease activity mediated by Rpc11p affects 3′ oligo(U) length is reviewed. In addition to multiple 3′ nucleases, pre-tRNA processing involves La and Lsm, distinct oligo(U)-binding proteins with proposed chaperone activities. 3′ processing is performed by the endonuclease RNase Z or the exonuclease Rex1p (possibly others) along alternate pathways conditional on La. We review a S. pombe tRNA reporter system that has been used to distinguish two chaperone activities of La protein to its two conserved RNA binding motifs. Pre-tRNAs with structural impairments are degraded by a nuclear surveillance system that mediates polyadenylation by the TRAMP complex followed by 3′ digestion by the nuclear exosome which appears to compete with 3′ processing. We also try to reconcile limited data on pre-tRNA processing and Lsm proteins which largely affects precursors but not mature tRNAs. A pathway is proposed in which 3′ oligo(U) length is a primary determinant of La binding with subsequent steps distinguished by 3′ endo- vs. exo- nucleases, chaperone activities and nuclear surveillance.
Keywords: RNase P, tRNase Z, La protein, Lsm8, Lhp1, Sla1, Rex1, Rrp6, Rpc11, Trf4, TRAMP complex
Introduction
Nuclear tRNA genes are transcribed by RNA polymerase III (pol III) as nascent precursor tRNAs (pre-tRNAs) that undergo multiple processing and modification steps. Collective data indicate significant variability in the temporal order of cleavages and modifications among eukaryotes, for different tRNAs, and under different conditions. Since there is inherent variability in the sequence and structure of different tRNAs it should be expected that some will differ from others in their activity as substrate for a given enzyme, binding protein or process. Complexity in the collective ‘pathway’ presumably reflects that some events differ in the extent to which they are abundance-driven, kinetics-limited, spatially organized, and to which they can be substituted by alternate mechanisms that lead toward the end product.
Early steps in tRNA biosynthesis are common to most tRNAs while steps individualized for specific tRNAs more often occur later (1). The universal steps in tRNA biogenesis are transcription, 5′ and 3′ end processing, the common nucleotide modifications, addition of CCA to the processed 3′ end, and nuclear export. Individualized steps include splicing of those pre-tRNAs that contain introns, modifications that occur on specific subsets of tRNAs, and aminoacylation. Some tRNA biogenesis steps are temporally ordered and/or segregated to different subcellular locales. These aspects of the tRNA maturation process including tRNA gene localization, subcellular compartmentalization of tRNA processing activities, as well as tRNA nuclear export and retrograde transport (from cytoplasm to nucleus), have benefited from a recent review (1) as well as a more comprehensive review (2). A recent review of the role of modifications in tRNA stability and function is also available (3).
This review will focus on end processing, the removal of 5′ leaders and 3′ trailers from nascent pre-tRNAs, which occur in the nucleus relatively early during tRNA maturation. It is well accepted that all pre-tRNAs undergo 5′ leader removal by the highly conserved endonuclease, RNase P, and that for most nuclear pre-tRNA species, this appears to occur before 3′ end processing (2). Detailed reviews of 5′ processing by RNase P are available (4-6). Our focus on 5′ processing will be limited to conditions that appear to alter the temporal order of 5′ and 3′ end processing. At the other end, 3′ trailers are removed by the endonuclease RNase Z. However, in contrast to a single mechanism of 5′ leader removal, 3′ trailers can also be removed by one or more 3′ exoribonucleases and the determinants of the endo vs. exo pathways appear to be the availability and access of certain ancillary factors and other conditions.
The pre-tRNA processing factors La and RNase P have been found associated with tRNA genes in human cells (7, 8), suggesting continuity between transcription and end processing. However, in contrast to mRNA biogenesis in which 5′ capping and splicing occur in a polar manner during synthesis by pol II, we are unaware of evidence that any 5′ processing event begins prior to release of the nascent transcript by pol III. As will be detailed below, pol III produces nascent RNAs with oligo(U) tracts on their 3′ ends which is a sequence-specific binding site for the conserved La protein (9) and for the Lsm protein complex, both of which have been shown to be involved in pre-tRNA metabolism in yeast. Considerably more work has been on the effects of La protein on pre-tRNA processing than the Lsm proteins, the latter of which is limited to one solid publication. We will focus only on the early steps in eukaryotic pre-tRNA 3′ processing, removal of the 3′ trailer.
Does the 5′-before-3′ processing pathway overshadow a simultaneous 3′-before-5′ pathway?
The 5′-before-3′ cleavage order appears to reflect a major pathway for tRNA processing (10-12, reviewed in 13). In addition to the endonuclease RNase Z, the exonuclease Rex1p (and perhaps others) can also process 3′ ends (14, 15). The 5′-before-3′ order is reversed in yeast mutants that lack the La protein (16). However, this is not a simple reversal since in this case the 3′ ends are matured by exonuclease(s) rather than RNase Z (16). La protein-RNA crystal structures reveal that La sequesters the RNA 3′ end in its binding pocket (17), apparently imposing a steric block to the 3′ exonucleases. The 5′-before-3′ end processing appears to apply most clearly when RNase Z is used for 3′ processing.
A change in 5′ and 3′ processing order may be explained by the relative robustness of these processes in the presence or absence of La. It would appear that La binding stabilizes nascent pre-tRNAs and delays their processing, since in its absence the 3′ exonuclease(s) pathway is so robust that nascent transcripts with intact leaders and trailers do not accumulate as distinct species (16). A consideration raised in this review is that La-dependent and La-independent pathways of pre-tRNA processing occur simultaneously for different subsets of pre-tRNAs in normal cells. Some of the individual transcripts from a single tRNA gene may use the La-dependent pathway while others from the same gene use the La-independent pathway. Observing pre-tRNAs of the La-independent pathway is challenging because the intermediates differ by only a few nucleotides at their 3′ ends (13, 16, 18, 19). While distinct species of the La-dependent pathway are concentrated as a band on Northern blots, those of the La-independent pathway are dispersed into a smear, attributed to variable 3′ exonucleolytic nibbling (16), some of which may be masked by the overlying distinct band. The 5′-before-3′ order is most clearly reversed in mutants lacking La (16). However, we suspect that the La-independent pathway is simultaneously active for some pre-tRNAs in La replete cells but these intermediates are more difficult to discern in the presence of an intact La-dependent pathway.
The 5′-before-3′ order is reversed for S. cerevisiae pre-tRNATrp (13), and possibly other pre-tRNAs, even when La is present. Since La binds pre-tRNAs in a 3′ oligo(U) length-dependent manner, we suggest that the 5′-before-3′ order would be reversed for pre-tRNAs that do not bind La due to a limitation of La and/or an insufficient number of 3′ terminal U residues (20) because these would be subjected to robust 3′ exonucleolytic processing to indistinct intermediates difficult to discern. As will be reviewed in detail below, current data argue that La is indeed limiting for the maturation of some pre-tRNAs and that some pre-tRNAs are synthesized with too few Us for La binding when La is limited (20).
In a later section we will return to the issue of 5′-before-3′ processing, considering other kinetic aspects of the interactions between La, RNase P, RNase Z and the pre-tRNA, and then discuss the limited published data on Lsm proteins and pre-tRNA metabolism and offer suggestions for future studies. Before that however, since a significant portion of this review will focus on the relationship between pol III termination, La function and early pre-tRNA processing, we will first introduce some features of the S. pombe system that has been used to examine this.
S. pombe suppressor tRNA alleles for the study of pol III termination and early processing
An opal suppressor tRNA gene whose functional readout is suppression of a UGA stop codon in ade6-704 mRNA and suppression of red pigment can be used to follow tRNA biogenesis (21-23). Various alleles of the suppressor tRNA have been examined (24). An allele was developed such that only accurate pol III termination leads to functional tRNA (25). Some allelic variants of this require only the 3′ end protection activity of the La protein (either S. pombe La protein, Sla1p, or human La), while other tRNA alleles also require La's secondary chaperone-like activity, for functional suppressor tRNA (24). Thus a tRNA pathway that operates in S. pombe independently of La is nonfunctional for these pre-tRNAs which are processed exclusively along the La-dependent pathway(s).
It is important to note that despite the popular impression that La is an abundant protein, the normal amount of endogenous Sla1p is limiting for maturation of suppressor pre-tRNA. This is evident since overexpression of ectopic Sla1p increases mature suppressor tRNA levels with concomitant increase in ade6-704 suppression activity (20, 22). This limitation presumably reflects the ability of the many other endogenous cellular pre-tRNAs and other pol III products to out compete the suppressor pre-tRNA for Sla1p.
Pre-tRNA 3′ end formation begins with pol III transcription termination
The earliest event in the processing pathway is the point at which the transcription phase ends and the nascent pre-tRNA is released from the pol III enzyme complex. Although the way pol III creates RNA 3′ ends prior to transcript release is very similar in vertebrates, S. pombe and S. cerevisiae, relevant differences will be reviewed. The relevant issues discussed in this and the following three sections are species-specific length differences in the oligo(dT) termination signals recognized by pol III, the minimal oligo(U) length required for recognition by La protein, which also appears to be species-specific, and 3′ oligo(U) length heterogeneity as a conserved intrinsic property of pol III and a possible mechanism for its generation.
A distinguishing feature of pol III is its termination mechanism, which occurs at a simple oligo(dT) tract at the 3′ ends of its transcribed genes (26). It is imperative to note relevant differences among several model organisms regarding nascent RNA 3′ end formation that result from pol III termination. While an oligo(dT) tract of four Ts leads to highly efficient termination by vertebrate pol III, four Ts are insufficient for S. pombe and S. cerevisiae pols III which require progressively longer dT tracts (25). This is reflected in the distribution of the oligo(dT) lengths of tRNA gene terminators by genome-wide analysis. Most tRNA genes end with four or five dTs in H. sapiens, five or six dTs in S. pombe, and six or more dTs in S. cerevisiae (27), nicely matching the oligo(dT) length dependent termination activities of the these pols III in vitro (25).
Pol III termination-associated RNA 3′ oligo(U) length heterogeneity
For most intents here, RNA 3′ oligo(U) length heterogeneity refers to the fact that the pol III transcripts produced from a single gene make up a population of RNAs, usually consisting of U(1-3)U-3′OH. The distribution pattern of oligo(U) length can vary in a gene-specific manner, limited by the length of the oligo(dT) tract. High resolution gels or two-dimensional systems revealed 3′ length heterogeneity after in vitro transcription (28-34). Although some of this could be due to contaminating exonuclease activities in the extract used, 3′ heterogeneity was also observed using purified pol III (in some cases noted not to have contaminating nuclease activity) (20, 35, 36). Nascent RNAs with 3′ oligo(U) length heterogeneity of 1-3 nt were synthesized and released by S. cerevisiae pol III observed using fast kinetics analysis (37, 38). Moreover, mutations in the second largest subunit of yeast pol III cause greater variations in the 3′ oligo(U) length distribution relative to wildtype in the RNAs released from pol III (see figure 7 in 38), providing additional support for the idea that RNA 3′ oligo(U) heterogeneity is a conserved intrinsic property of pol III itself.
Nascent RNA 3′ oligo(U) metabolism by pol III subunit Rpc11p
RNA 3′ oligo(U) length heterogeneity is generated during the slowing down or pausing phase of pol III termination. A well-characterized, pol III-intrinsic processive 3′ exoribonuclease activity cleaves RNA in the active center of pol III during pausing (38-41). This 3′-5′ nt removal activity is distinct (and ‘opposite’) from the principal activity of pol III, which is 5′-3′ nt polymerization, and is not simply a chemical reversal of the forward reaction. This 3′ exoribonuclease activity is mediated by a 12.5 kDa subunit of pol III, Rpc11p, that is homologous to TFIIS a factor that helps pol II overcome pausing and other blocks to elongation (42). Mutants defective for pol III termination show alterations in this activity (38). In a report that examined tRNA maturation monitored by functional suppression, Huang et al. proposed that 3′ terminal oligo(U) length distribution results in part from cleavage by Rpc11p (38). This was supported by the observation that rpc11-mutants differing in their deficiency of pol III-associated 3′-5′ RNA cleavage activity showed progressive lengthening of the 3′ oligo(U) tracts in the nascent transcripts (20). This lengthening presumably reflects alterations of Rpc11p-mediated 3′-5′ RNA cleavage activity during the pausing phase of transcription termination that precedes transcript release.
Some pol III transcripts contain 3′ oligo(U) tracts too short for competitive La binding
La protein is a molecular chaperone that protects newly synthesized nascent pol III transcripts from 3′ exonucleases and also assists in pre-tRNA folding (43-45). As noted above, La binds RNA in an 3′ oligo(U) length-dependent manner (9). Human La binds stably to RNAs that contain 3 or more Us at their 3′ end (9). This was confirmed for human La by Huang et al. who also showed by comparison that the S. pombe La protein, Sla1p, requires 4 or more Us (20).
Using purified pol III Cambell & Setzer found that a majority of Xenopus nascent 5S rRNAs contained only two Us at the 3′ end, confirming earlier data from Bogenhagen & Brown and Cozzarelli et al, who reported a major pol III transcription stop after 2 Us and a minor stop after 3 Us from a highly efficient 4 T terminator (26, 36). Studies on pol III transcription of adenovirus VA1 RNA revealed more transcripts with 1 or 2 Us at the 3′ end than with 3 Us (46). It is noteworthy that the vertebrate VA1 and 5S RNA genes used for these studies contain highly efficient oligo(dT) terminators that are only 4 Ts in length.
The pol III primary transcript of a mouse B1 RNA gene terminates at its natural 5 T terminator, AATTTTTAA, and is stabilized by La protein. Replacement of this terminator with the Xenopus 5S rRNA efficient terminator of 4 Ts, GCTTTTGC, produces a primary transcript not stabilized by La, that undergoes quick 3′ processing in vitro and in vivo (47, 48). These data are consistent with the earlier results showing that most transcripts from the 5S gene contain only 2 Us (26, 36) and strongly support the idea that some pol III terminators produce transcripts with too few Us for efficient La binding.
Mathews & Francouer confirmed that VA1 RNA transcripts made by pol III in vitro that ended in 2 Us were more abundant than those with 3 Us, and further, that only the 3U RNA species was immunoprecipitable with La while the 2U RNA species was left intact in the supernatant (29). This binding preference agrees with results of Stefano who used a mixture of tRNAs with varying numbers of terminal Us and showed that the tRNAs with 3 or more terminal Us are most readily immunoprecipitable with La protein (9).
In S. pombe, nascent pol III transcripts with 4 or more terminal Us competed significantly better for Sla1p than did the RNAs with 3 or fewer Us (20). In a side-by-side analysis Sla1p required a longer oligo(U) tract for competitive binding than did human La (20). The same difference between human La and Sla1p was observed using chemically synthesized RNAs that differed in 3′ oligo(U) length (20). Thus Sla1p prefers nascent transcripts with 4 or more terminal Us while leaving disproportionately more of the RNAs with the shorter U tracts in the supernatant. The requirement of Sla1p for an oligo(U) tract that is slightly longer than is required by human La protein matches the slightly longer oligo(dT) tract lengths required for termination by the S. pombe pol III relative to human pol III noted earlier (25). Examination of S. cerevisiae Lhp1p minimal oligo(U) length has not been reported.
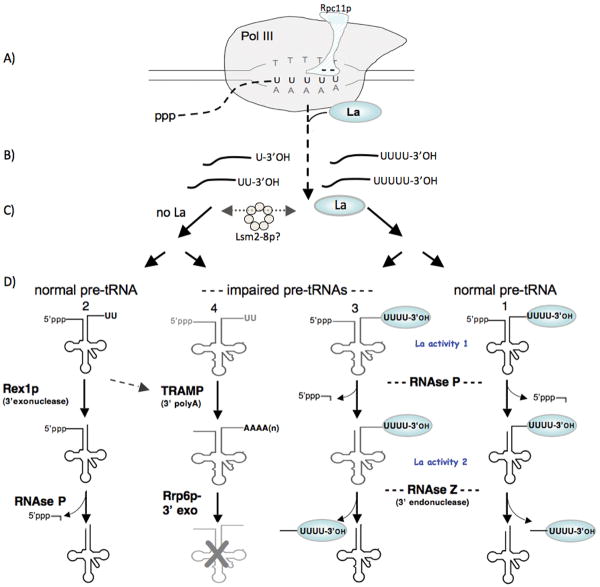
Huang et al. mapped the oligo(U) length of in vivo synthesized nascent suppressor pre-tRNAs in wild type S. pombe and in rpc11 mutants (20). In wild type S. pombe only ∼25% of the nascent suppressor pre-tRNAs had 4 or more 3′ Us, while this was increased to 46% and 57% in the two rpc11 mutants examined (see above). Moreover, for the La-dependent suppressor tRNA allele used the increases in 3′ oligo(U) length were accompanied by corresponding increases in the efficiency of conversion of pre-tRNA to mature tRNA, mature suppressor tRNA levels, and suppression activity (20). A straightforward model that emerged is that a relatively small fraction (∼25%) of suppressor pre-tRNAs is bound by La in wild type cells while a larger fraction is bound in the rpc11 mutants, with congruent effects on tRNA maturation and functional suppression (20). The unbound fraction presumably has too few 3′ U residues for La binding which apparently leads them to be degraded (20). Thus, by affecting 3′ oligo(U) length, the presence or absence of the RNA 3′ cleavage activity of Rpc11p can exert a most early effect on nascent RNA 3′ end formation and subsequent pre-tRNA metabolism (Fig. 1). This model predicts that tRNA genes with longer oligo(dT) tracts may produce nascent transcripts that compete better for La protein than other tRNA genes with relatively short oligo(dT) tracts. Another hypothesis would be that genes with the shortest oligo(dT) tracts may have adapted to be less dependent on La function than genes with longer oligo(dT) tracts.
Figure 1.
Alternative pathways for pre-tRNA processing begin during transcription termination by RNA polymerase III. This figure incorporates protein nomenclature from multiple organisms (see text). A) Pol III pauses at the tRNA gene terminator. According to structure modeling the integral pol III subunit, Rpc11p, contains a N-terminal domain that resides on the surface of pol III and an acidic hairpin that enters through a pore to the catalytic center. The acidic tip of the Rpc11p hairpin (designated “- -”) mediates RNA 3′ cleavage of nascent pre-tRNA transcripts during pausing by pol III. B) Nascent pre-tRNA transcripts with different length 3′ oligo(U) termini are released from pol III and sorted into those that bind La and those that do not, depicted on the right and left sides of the vertical dashed arrow respectively. C) Lsm proteins (presumably Lsm2p-8p) can affect the degree to which nascent pre-tRNAs are associated with La and may function at this point (see text). Pre-tRNA transcripts with sufficient 3′ oligo(U) length maintain stable association with La protein. D) Pre-tRNAs move along alternative pathways, numbered 1-4, depicted for normal pre-tRNAs (pathways 1 & 2) and structurally-impaired pre-tRNAs (pathways 3 & 4), distinguished by 3′ endonucleolytic (pathways 1 & 3), or 3′ exonucleolytic (pathways 2 & 4), as well as nuclear surveillance involving TRAMP and Rrp6p (pathway 4). La protein gains early dominance over the pre-tRNAs with sufficient 3′ oligo(U) length (see text). The exact point in this scheme at which the RNA chaperone-like activity of La (La activity 2) appears to function is unclear from current data but occurs sometime before RNase Z separates the trailer from the tRNA body.
La, RNase Z and RNase P interactions during pre-tRNA processing
La protein and RNase P have been found localized with the pol III machinery and at tRNA genes (7, 8, 49, 50), consistent with their function in the early phases of processing (28, 51-55). Early binding by La fits nicely with the facts that La exhibits RNA chaperone-like activity possibly to assist pre-tRNA folding in vivo and that RNase P requires some tRNA-like structure in its substrates (23, 56-59). Mutations that affect pre-tRNA folding or structure can decrease the substrate activity of pre-tRNA for RNase P (4-6), even when the mutation is in the intron of the pre-tRNA (60).
That La protein is linked to the earliest phase of pre-tRNA biogenesis is consistent with the phenotypes of two classes of S. pombe mutants. In nonphosphorylatable La mutants in which La exhibits increased affinity for the nascent suppressor pre-tRNA, the latter accumulates to high levels relative to the phosphorylatable La control, but with inefficient 5′ processing by RNase P and failure to produce mature suppressor tRNASerUCA (22). This is consistent with earlier data that showed that nonphosphorylated human La binds to the 5′ end of pre-tRNA (as well as the UUU-3′OH end) and interferes with processing by RNAse P, an inhibitory activity that is attenuated by phosphorylation on serine-366 (61). However this would appear not to reflect an activity that is used during tRNA biogenesis in vivo since most nuclear La is in the phosphorylated form and it is this form that is found associated with pre-tRNAs in HeLa cells (62). Nonetheless, these mutants provide evidence that La gains control of the nascent pre-tRNA even before what is normally considered the first processing step, 5′ cleavage by RNase P. Also in support of this conclusion, a second class of mutants, in which subcellular trafficking of La is aberrant, La-associated pre-tRNAs are prematurely exported from the nucleus before RNAse P or Z cleaves the pre-tRNA (63, 64). Thus although La and RNAse P are localized at tRNA genes, it would appear in these cases that La gains control over the nascent pre-tRNAs.
Why RNase P cleaves before RNase Z when La is present is not entirely understood. Purified recombinant La protein can impede 3′ cleavage by RNase Z in a purified in vitro system, dependent on UUU-3′OH trailer binding (65). Initially it might seem that inhibition of RNase Z activity by La would be somewhat at odds with the conclusion that yeast La protein is required for pre-tRNA 3′ endonucleolytic cleavage (16). However, we believe that it fits nicely with the in vivo data if we consider that La binding stabilizes nascent pre-tRNAs, as evidenced by their accumulation as distinct species (16). A model to accommodate these data is that La binding to pre-tRNA would be inhibitory both to 3′ exo- and 3′ endo-nucleolytic processing, but more so to the 3′ exonuclease(s) than to RNase Z. This hierarchy seems quite plausible knowing that La sequesters the 3′-OH end of oligo(U) RNA in a largely hydrophobic binding pocket (17). In either case, inhibition of 3′ end processing might allow RNase P to act first. Moreover, it is conceivable that by sequestering the 3′ trailer La could actually enhance reactivity of the 5′ leader with RNase P (for example see Fig. 6 in 23). Other aspects of the relative affinities of RNAses P and Z for a given pre-tRNA substrate may contribute to processing order in some cases.
Isoforms of RNase Z
Both short and long forms of RNase Z are present in eukaryotes, designated tRNase ZS and tRNase ZL respectively, whereas bacteria and archaea have only the short form (reviewed in 66). tRNase ZL is ∼850 amino acids in length and contains mitochondrial or chloroplast targeting sequences while the ZS form is ∼350 amino acids in length (67). The C-terminal part of tRNase ZL has sequence homology with tRNase ZS. Understanding the roles of tRNases Z is also important because independent roles for tRNase ZL in gene silencing, including involvement of siRNA-related enzymes, have emerged (68-71) and because human tRNase ZL is encoded by ELAC2, a prostate cancer susceptibility gene (72, 73).
Humans and S. cerevisiae have one copy each of tRNase ZS and tRNase ZL genes, with different isoforms of tRNase ZL localized to nucleus and mitochondria. Curiously, S. pombe has no apparent RNase ZS but contains two tRNase ZL genes, sptrz1+ and sptrz2+ (66). These localize to nuclei and mitochondria respectively, suggesting that the RNase Z functions of nuclei and mitochondria may be more readily dissected in S. pombe than in other organisms (66). Both sptrz1+ and sptrz2+ are required for cell growth (66) and consistent with its nuclear localization, sptrz1+ was shown to promote 3′ processing of a nuclear encoded suppressor-tRNASerUCA resulting in a functional increase in the mature tRNA and suppression of red pigment (66).
Unexpected finding in S. pombe La and RNase Z double mutant
La promotes pre-tRNA 3′ endonucleolytic processing (16) by blocking 3′ exonucleases (14, 15). Because the 3′ exonuclease Rex1p and/or other exonucleases can functionally process pre-tRNAs in the absence of La (14, 15), it may have been predicted that nuclear RNAse Z, i.e., encoded by sptrz1+, would be nonessential in La-deleted yeast. Intriguingly, this is not the case (66). This suggests the possibility that some pre-tRNAs must be processed by RNase Z in the absence of La and if so, La is not required for 3′ endonucleolytic cleavage of some pre-tRNAs (16). However, another interpretation is considered below.
S. cerevisiae tRNase ZL (scTRZ1) and human ELAC2 were shown to function in S. pombe for 3′ processing of nuclear encoded pre-tRNAs (66). It is therefore intriguing that scTRZ1 and ELAC2 complement the temperature sensitive growth defect of sptrz1–1, but not the sptrz1-null mutant. These data suggest that sptrz1+, scTRZ1 and ELAC2 exhibit a conserved activity for the 3′ processing of those pre-tRNAs that were examined (66) but that sptrz1+ also exerts an additional essential function that is species-specific. It is formally possible that an essential S. pombe pre-tRNA requires sptrz1+ but can not be processed by scTRZ1 or ELAC2. It also seems plausible that the presumed additional essential function of sptrz1+ is independent of tRNA production. Since other ribonuclease activities for RNAse Z have been demonstrated (67, 69, 74), sptrz1+ may be required for the processing of another essential RNA, perhaps guided by a protein or RNA partner that can not interact with scTRZ1 or ELAC2. In any case, the distinctive genetics of the S. pombe tRNA 3′ processing factors may be well suited to uncover the essential function of tRNase ZL and lead to a better understanding of the relationship between La and RNase Z.
Pre-tRNA 3′ exonucleases
As reviewed previously, 3′ end maturation of pre-tRNAs takes place in the nucleus and can occur via multiple mechanisms (75). Although it appears that endonucleolytic cleavage by RNase Z reflects the major pathway, the 3′ exonucleases Rex1p and Rrp6p perform 3′ end processing under some conditions and it is suspected that additional exonucleases may also contribute (14, 15). Rex1p appears to be the major 3′ exonuclease that contributes to 3′ end maturation in yeast cells containing La (14). Substitution mutations to Rex1p residues that are predicted to be important for catalysis indeed inactivate its exoribonuclease activity in vitro and in vivo (15). Some data suggested that Rex1p may specifically process tRNAiMet precursors with long 3′ but not short 3′ trailers (15).
Although deletion of RRP6 from S. cerevisiae appeared to restore the pattern of intron-containing pre-tRNASer intermediates that is otherwise aberrant in a Lhp1-Δ Rex1-Δ double mutant, it is not clear if RRP6 contributes positively to production of mature tRNA (14). Deletion of S. pombe rrp6+ had no negative effect on suppressor tRNA maturation; for suppressor tRNASer alleles with little or no structural impairment, deletion of rrp6+ did not reduce suppressor activity in the presence or absence of sla1+ suggesting that Rrp6p does not make significant contributions to tRNA maturation (24). It would appear for most pre-tRNAs not cleaved by RNase Z, that Rex1p processes their 3′ ends and that although Rrp6p may contribute under some conditions it probably serves more to degrade pre-tRNAs as a component of nuclear surveillance (14).
Pre-tRNA nuclear surveillance competes for 3′ end metabolism
Nuclear surveillance was first proposed as a means by which cells perceive defects in tRNA processing or nuclear export and react by decreasing translation initiation mediated by tRNAiMet (76). It was later found that some mutants that reduce tRNAiMet levels do so by nuclear degradation of hypomodified pre-tRNAiMet caused by lack of modification of A58 of pre-tRNAiMet to m1A58 (77). It was discovered that the degradation of hypomodified pre-tRNAiMet requires Rrp44p, a 3′-5′ exoribonuclease subunit of the exosome complex as well as Trf4p, a poly(A) polymerase (78). The hypomodified pre-tRNAiMet is first polyadenylated on its 3′ end by Trf4, creating a single-stranded landing pad for exonuclease loading, and is then degraded in 3′-5′ direction by the Rrp6p-containing nuclear exosome (78, 79). Trf4p is a member of the TRAMP (Trf4/Air2/Mtr4p Polyadenylation) complex comprised of poly (A) polymerase (Trf4 or Trf5), RNA helicase (Mtr4) and a Zinc knuckle protein (Air1 or Air2) which interacts with the nuclear 3′ exoribonuclease, Rrp6p of the nuclear exosome (80, 81). This intranuclear pathway is also operational for other aberrant RNAs that bear a structural perturbation or result from an error in processing (reviewed in 82). However, the details of how this pathway preferentially targets aberrant RNAs are not clear.
Nuclear surveillance would appear to be in competition with pre-tRNA 3′ end processing. It was reported that while hypomodified pre-tRNAiMet is subjected to degradation by nuclear surveillance, the yeast La homologous protein, Lhp1p could offset this (77, 83). In gcd10/14-mutants defective for m1A58 modification of pre-tRNAiMet, over expression of Lhp1p leads to increased levels of mature tRNAiMet and suppresses their phenotypes (77). Deletion of LHP1 in the gcd10/14- mutants exacerbates pre-tRNAiMet 3′ exonuclease-mediated degradation and the associated phenotypes (77, 83).
The Trf4p poly(A) polymerase component of the TRAMP complex can polyadenylate substrates with single strand 3′ overhangs of no less than 3 nucleotides (84). The endonuclease RNase Z leaves only one unpaired nucleotide at the 3′ end and would not generate good substrates for Trf4p (84). This fits with the idea that 3′ trailers subjected to partial processing by 3′ exonuclease(s) as well as those pre-tRNas with too few 3′ terminal Us for La binding would be preferential substrates of Trf4p. Knowing that La binds and protects pre-tRNA 3′ trailers from 3′ exonuclease, the data suggest that La competes with the TRAMP/nuclear exosome surveillance system for the 3′ end of the hypomodified pre-tRNAiMet.
Opposite effects of La and nuclear surveillance components were demonstrated for suppressor pre-tRNASerUCA alleles of S. pombe (24). For the subset of alleles that were affected, deletion of rrp6+ increased suppressor activity whereas deletion of sla1+ decreased activity (24). The degree to which rrp6+ and sla1+ exerted their effects was greater for some suppressor tRNASer alleles than for others, in general accordance with the degree of structural impairment, i.e., containing one, two or three base substitutions that weaken basepairing potential by changing a G:C pair to a G:U. The most severely affected of the pre-tRNAsSerUCA were shown to be polyadenylated and deletion of rrp6+ increased the mature suppressor tRNASerUCA levels twelve-fold (24). The activity of the U40 allele, which contains a single G:C to G:U change in the anticodon stem, was unaffected by rrp6+ or sla1+ deletion (24). Activity of another allele, C37:10, was diminished by sla1+ deletion but fully restored by rrp6+ deletion in the double mutant. Since Rrp6p acts to degrade pre-tRNAs after polyadenylation by TRAMP, it would seem that La and TRAMP compete for the 3′ ends of at least some pre-tRNAs although it is also possible that Rrp6p and La protein compete more directly as well.
Wolin and colleagues analyzed a more complete set of factors that may be involved in pre-tRNA 3′ end metabolism and extended the list of individual pre-tRNAs that succumb to nuclear surveillance (14). They also showed that competition with La for pre-tRNA 3′ ends extends to the 3′ exonuclease Rex1p. Moreover, Rex1p-dependent 3′ trimming appears to target some pre-tRNAs for decay by TRAMP (14). Anderson and colleagues found that loss of Rex1p results in polyadenylation of pre-tRNAiMet, suggesting that defects in 3′ end processing can activate the nuclear surveillance pathway. Thus, direct competition between La and Rex1p may determine the subsequent fate of a pre-tRNA. The conclusion that Rex1p contributes to pre-tRNA 3′ end maturation even in cells containing La protein (14) is consistent with the idea that both 5′-before-3′ and 3′-before-‘5’ processing occur simultaneously.
It would seem that while nuclear surveillance targets defective pre-tRNAs for decay, La protein can protect those same pre-tRNAs and promote their processing to functional mature tRNAs. A question that arises is why should there be these opposing activities? It is intriguing that the defects in hypomodified pre-tRNAiMet and impaired suppressor tRNASerUCA appear to subject them to degradation in the nucleus more so than preventing them from functioning at the ribosome in the cytoplasm. Appreciation that RNA degradation may compete with productive RNA processing has arisen in part because of the prevalence and efficiency of RNA degradation. While popular belief once suggested that cells should not waste resources on RNA degradation, a prevailing view that cells make RNA that is targeted for degradation questions this (85-87).
Lsm proteins affect pre-tRNA metabolism
Although a report indicates convincingly that some of the Lsm proteins are required for the normal pathway of early pre-tRNA metabolism in S. cerevisiae and for pre-tRNA association with La-homologous protein Lhp1p, it has not been clear how this requirement fits into the pre-tRNA processing pathway (19). Deletion of Lsm2p to Lsm5p and Lsm8p led to alterations in pre-tRNA intermediates while mature tRNA levels were unaffected, somewhat reminiscent of the effects of sla1+ or LHP1 deletion in S. pombe and S. cerevisiae respectively. Lsm6-Δ and Lsm7-Δ also exhibited a similar pre-tRNA phenotype but in this case only for one of the transcripts examined, pre-tRNAAla (19). Thus, the previously characterized seven subunit Lsm2p-8p complex is presumably involved in pre-tRNA metabolism. The Lsm2p-Lsm8p forms a heptameric ring structure that plays a major role in the metabolism, stabilization, nuclear retention and function of the U6 snRNP (88-91).
The Lsm2p-Lsm8p complex also functions in the degradation of pre-mRNAs in the nucleus (92). By contrast, the Lsm1p-7p complex functions in cytoplasmic mRNA degradation (reviewed in 93). Genetic experiments indicate that the only essential function of LSM8 is related to its role in U6 snRNA metabolism or function (94).
Surprisingly, Lsm1-Δ cells also showed accumulation of pre-tRNAAla (19). Although Lsm1p is cytoplasmic, its dysregulation (overexpression) has been shown to perturb U6 snRNA accumulation presumably by disrupting the normal stoichiometric equilibrium of the other Lsm components of the Lsm2p-8p complex (95).
Characteristics of the relationship between the Lsm2-8p complex and Lhp1p in pre-tRNA metabolism are clearly distinguishable from Lsm2-8p participation with Lhp1p in the metabolism of another pol III transcript, U6 snRNA (88, 94). While La/Lhp1p is transiently associated with a small fraction of U6 snRNA that represents the nascent pol III transcript, the Lsm2-8p complex is associated with mature U6 snRNA as part of the stable U6 snRNP (90, 91, 96). La protein requires a 3′ hydroxyl group on its RNA ligands which is present only on newly synthesized yeast U6 snRNA but not mature U6 snRNA which contains a 3′ phosphate and is bound by Lsm2-8p (94). This order of binding to U6 snRNA, Lhp1p before Lsm2-8p (94) is consistent with the idea that Lhp1p functions to stabilize U6 snRNA for assembly into the U6 snRNP while the Lsm2-8p complex participates in the function of the U6 snRNP (91, 96).
Metabolic pulse-chase experiments indicate that pre-tRNA processing was delayed in cells that previously underwent transcriptional shut off of Lsm3p (19). Steady state levels of pre-tRNAs but not mature tRNAs were altered in lsm-deletion mutants (19). In this case, a significant increase in pre-tRNA levels occurred in lsm-mutants, consistent with delay or obstruction of their processing or degradation. Moreover, while coprecipitation of the pre-tRNAs with Lsm3p suggest a direct role for the Lsm proteins in pre-tRNA metabolism, the relatively low efficiency of coprecipitation suggested a transient association (19).
Aberrant pre-tRNAs with extended 3′ ends also accumulated in the lsm-mutants. These aberrant 3′-extended pre-tRNAs appear to represent transcripts produced by read-through of the normal pol III transcription termination site of the tRNA gene, that would be rapidly degraded in wild-type cells but accumulate in the lsm-mutants (19). However, that the elevated levels of the other pre-tRNAs in the lsm-mutants might represent misfolded or defective pre-tRNAs that would otherwise be targeted for degradation was considered unlikely because it implied a substantial amount of pre-tRNA decay in wild-type cells and because similar accumulation was not observed in exosome-deficient strains (19). On the other hand, the authors did note two-hybrid interactions between Lsm8p and the exosome subunit Mtr3p and between Lsm8p and the 5′ exonuclease Xrn1p, as well as physical association of Lsm8p with another exosome subunit Rrp42p (see citations in 19), consistent with the association of Lsm2p-Lsm8p with the RNA degradation machinery. Subsequent work by this group revealed that the Lsm2p-8p complex indeed targets nuclear RNA, albeit capped pre-mRNA, for degradation (92). These observations suggest that Lsm2-8p involvement in pre-tRNA metabolism may be related to degradation more than productive processing.
As noted, the transient association between the Lsm complex and the pre-tRNAs with the concomitant reduction of Lhp1p binding to its substrate pre-tRNAs demonstrated by Kufel et al., is consistent with the Lsm complex functioning as a pre-tRNA chaperone (92), possibly involved in quality control. The limited available evidence is consistent with a role for the Lsm2-8p complex in either pre-tRNA degradation or processing. The experimental data do not address whether the Lsm proteins involved in pre-tRNA metabolism contributes positively or negatively to the production of mature tRNA. However, this may be addressed using the S. pombe suppressor-tRNA system described above. In this case, increases or decreases in suppressor tRNA levels and with concomitant alteration of suppression activity may be monitored and compared in strains carrying various lsm-alleles, sla1-alleles and suppressor tRNA alleles.
Molecular chaperone and RNA chaperone activities
As referred to in the previous section, the Lsm2-8p complex is believed to possess chaperone activity during U6 snRNP assembly and/or function and may also provide this for pre-tRNAs (92). Multiple sources of evidence of chaperone activity for the La proteins from several species has been accumulating. First, it should be noted that two types of chaperone activity have been distinguished. That La meets the definition of a ‘molecular chaperone’ as a protein that binds and stabilizes a substrate and exhibits controlled binding and release was first documented for U6 snRNA and suggested for nascent pol III transcripts in general (see 88). A second definition, that for ‘RNA chaperone activity’ refers to participation of a given protein in the RNA folding process specifically, often by preventing the RNA from becoming trapped in a misfolded conformer (97). Evidence that La fulfills this activity, that it is required for the efficient folding of a pre-tRNA in vivo is also available (56). In addition, recombinant human La protein has been shown to exhibit RNA chaperone activity in vitro using a self-splicing intron RNA misfolding trap assay (23, 57). This RNA chaperone activity has been mapped to one of the two conserved RNA binding motifs of human La protein, the canonical RNA binding surface of the RNA recognition motif-1 (RRM1) (23). An intact RRM1 binding surface of La protein (both human La and Sla1p) has also been shown to be required for the maturation of a structurally impaired pre-tRNASerUCA (24).
Two activities differentially map to the two conserved RNA binding motifs of La proteins, the La motif (LAM) and RNA recognition motif-1 (RRM1) (23, 24). The principal activity of La, binding to the 3′ oligo(U) tract, which is mediated mostly by its La motif (LAM) and serves to protect pre-tRNAs from exonucleases and as reviewed in an earlier section imposes a 5′-before-3′ end processing pathway. As noted above, this 3′ oligo(U) binding activity can offset the TRAMP & 3′ exonuclease Rrp6p nuclear surveillance system which leads to degradation of aberrant pre-tRNAs.
For the maturation of other, more severely impaired pre-tRNAs, there is a requirement for La's principal activity to be accompanied by its second activity (activity-2), mediated by the La RRM β-sheet surface, that functions to offset pre-tRNA decay that would otherwise result from an as yet uncharacterized surveillance system distinct from Rrp6p (24). It remains unclear if this second activity, as examined for human La and Sla1p in S. pombe, offsets or sterically blocks a nuclease activity that degrades the structurally impaired pre-tRNA or if it helps the pre-tRNA avoid or overcome misfolding (24). In support of the second possibility, La RRM, the same RNA binding motif required for maturation of the structurally impaired pre-tRNA in vivo is also required for RNA chaperone activity in vitro (23, 24). It is tempting to speculate that this activity is also responsible for the requirement of Lhp1p in pre-tRNAArgCCG folding (97).
Distinguishable chaperone activities for pre-tRNAs
Thus, it appears that La can facilitate tRNA processing by two mechanisms: simple 3′ end binding with consequent 3′ end protection, and by promoting folding and/or the structural integrity of a tRNA. Discerning these as separate activities has been a multifaceted process. By screening for and characterizing mutations that confer yeast synthetic lethality in the absence of La, substitutions were found that disrupt basepairing in the anticodon stem of the single copy gene essential for tRNASerCGA (16). It was later reported that substitutions in a suppressor-tRNA in S. pombe confer dependence on La for functional maturation (22). Yeast La indeed promotes proper folding of a mutation-containing pre-tRNAArgCCG with a demonstrable propensity for misfolding (56). By genetic and biochemical dissection it was formally shown that La confers two distinct activities in tRNA maturation, mediated by separate RNA binding surfaces on the two highly conserved motifs of the La protein (24).
The degree to which a pre-tRNA requires or engages La for its principal and secondary activities can vary (24). Some structurally impaired suppressor pre-tRNA alleles trigger decay more than others and are differentially sensitive to Rrp6p and rescue by La. For S. pombe pre-tRNASerUCA, a G:CΠG:U basepair change in the variable arm causes it to be much more highly dependent on La for maturation than basepair disruptions elsewhere in the pre-tRNA, even in the absence of rrp6+ (24). It is important to note for the purpose of this review that while these two activities of La protein are distinguishable, both require intact oligo(U) 3′ end binding activity. Thus, while activity 2 is distinguishable from activity 1 in pre-tRNA metabolism, it is dependent on activity 1 (23). This is in agreement with a two binding site model for La interaction with pre-tRNA in which the La motif binds 3′ oligo(U) and allows the adjacent RRM RNA binding site to mediate activity 2 (23).
We suggest that the ‘molecular chaperone’ and ‘RNA chaperone’ activities of La are represented by distinct activities that differentially map to the two conserved RNA binding motifs of La proteins, the La motif (LAM) and RNA recognition motif-1 (RRM1) respectively (23, 24). The molecular chaperone activity as initially characterized for U6 snRNA and involving Lsm2-8p appears to be related to UUU-3′ end protection and stability during U6 snRNP assembly (94). This activity appears to apply to pre-tRNAs as La can protect their 3′ ends from 3′ exonucleases and other competing activities such as nuclear surveillance as reviewed above, and keep them on a productive maturation pathway. The in vitro RNA chaperone activity and the in vivo activity required for the maturation of structurally-impaired pre-tRNA requires an intact RRM1 β-sheet surface. By contrast an intact RRM1 β-sheet surface is not required for the maturation of other, less impaired pre-tRNAs that succumb to nuclear surveillance that do require an intact LAM but not RRM1 β-sheet surface (23, 24).
It is noteworthy that processing of the apparently nonimpaired pre-tRNALysCUU was also affected in La activity-2 mutants (24). This is consistent with the observation that the RNA binding surface of RRM1 involved in activity-2 participates in, but is not required for, the metabolism of normal pre-tRNA. In this case, La uses both of its conserved RNA binding surfaces, the LAM and RRM, to distinguish pre-tRNAs from 3′ processed tRNAs (23). Both surfaces engage different parts of a contiguous pre-tRNA for high affinity stable binding whereas after 3′ cleavage, each site exhibits a faster off rate and more readily releases its RNA (23).
Acknowledgments
We thank members of the Maraia lab for comments and discussion as well as J. Iben, A. Russo and V. Cherkasova for editing. This work was supported by the Intramural Research Program of the NICHD, NIH. R.J.M. is a Commissioned Officer in the U.S. Public Health Service.
Abbreviations
- pre-tRNA
precursor tRNA
- pol III
RNA polymerase III
- nt
nucleotide
References
- 1.Hopper AK, Pai DA, Engelke DR. Cellular dynamics of tRNAs and their genes. FEBS Lett. 2010;584:310–317. doi: 10.1016/j.febslet.2009.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hopper AK, Phizicky EM. tRNA transfers to the limelight. Genes Dev. 2003;17:162–180. doi: 10.1101/gad.1049103. [DOI] [PubMed] [Google Scholar]
- 3.Phizicky EM, Alfonzo JD. Do all modifications benefit all tRNAs? FEBS Lett. 2010;584:265–271. doi: 10.1016/j.febslet.2009.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirsebom LA. RNase P RNA mediated cleavage: substrate recognition and catalysis. Biochimie. 2007;89:1183–1194. doi: 10.1016/j.biochi.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 5.Lai LB, Vioque A, Kirsebom LA, Gopalan V. Unexpected diversity of RNase P, an ancient tRNA processing enzyme: challenges and prospects. FEBS Lett. 2010;584:287–296. doi: 10.1016/j.febslet.2009.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McClain WH, Lai LB, Gopalan V. Trials, Travails and Triumphs: An Account of RNA Catalysis in RNase P. J Mol Biol. 2010 doi: 10.1016/j.jmb.2010.01.038. [DOI] [PubMed] [Google Scholar]
- 7.Reiner R, Ben-Asouli Y, Krilovetzky I, Jarrous N. A role for the catalytic ribonucleoprotein RNase P in RNA polymerase III transcription. Genes Dev. 2006;20:1621–1635. doi: 10.1101/gad.386706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fairley JA, Kantidakis T, Kenneth NS, Intine RV, Maraia RJ, White RJ. Human La is Found at RNA Polymerase III-Transcribed Genes In Vivo. Proc Nat Acad Sci, USA. 2005;102:18350–18355. doi: 10.1073/pnas.0506415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stefano JE. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell. 1984;36:145–154. doi: 10.1016/0092-8674(84)90083-7. [DOI] [PubMed] [Google Scholar]
- 10.Melton DA, De Robertis EM, Cortese R. Order and intracellular location of the events involved in the maturation of a spliced tRNA. Nature. 1980;284:143–148. doi: 10.1038/284143a0. [DOI] [PubMed] [Google Scholar]
- 11.Engelke DR, Gegenheimer P, Abelson J. Nucleolytic processing of a tRNAArg-tRNAAsp dimeric precursor by a homologous component from Saccharomyces cerevisiae. J Biol Chem. 1985;260:1271–1279. [PubMed] [Google Scholar]
- 12.O'Connor JP, Peebles CL. In vivo pre-tRNA processing in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:425–439. doi: 10.1128/mcb.11.1.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kufel J, Tollervey D. 3′-processing of yeast tRNATrp precedes 5′-processing. RNA. 2003;9:202–208. doi: 10.1261/rna.2145103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Copela LA, Fernandez CF, Sherrer RL, Wolin SL. Competition between the Rex1 exonuclease and the La protein affects both Trf4p-mediated RNA quality control and pre-tRNA maturation. RNA. 2008;14:1214–1227. doi: 10.1261/rna.1050408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ozanick SG, Wang X, Costanzo M, Brost RL, Boone C, Anderson JT. Rex1p deficiency leads to accumulation of precursor initiator tRNAMet and polyadenylation of substrate RNAs in Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:298–308. doi: 10.1093/nar/gkn925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoo CJ, Wolin SL. The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell. 1997;89:393–402. doi: 10.1016/s0092-8674(00)80220-2. [DOI] [PubMed] [Google Scholar]
- 17.Teplova M, Yuan YR, Ilin S, Malinina L, Phan AT, Teplov A, Patel DJ. Structural basis for recognition and sequestration of UUU-OH 3′-termini of nascent RNA pol III transcripts by La, a rheumatic disease autoantigen. Mol Cell. 2006;21:75–85. doi: 10.1016/j.molcel.2005.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Horn DJ, Yoo CJ, Xue D, Shi H, Wolin SL. The La protein in Schizosaccharomyces pombe: a conserved yet dispensable phosphoprotein that functions in tRNA maturation. RNA. 1997;3:1434–1443. [PMC free article] [PubMed] [Google Scholar]
- 19.Kufel J, Allmang C, Verdone L, Beggs JD, Tollervey D. Lsm proteins are required for normal processing of pre-tRNAs and their efficient association with La-homologous protein Lhp1p. Mol Cell Biol. 2002;22:5248–5256. doi: 10.1128/MCB.22.14.5248-5256.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Y, Intine RV, Mozlin A, Hasson S, Maraia RJ. Mutations in the RNA Polymerase III Subunit Rpc11p That Decrease RNA 3′ Cleavage Activity Increase 3′-Terminal Oligo(U) Length and La-Dependent tRNA Processing. Mol Cell Biol. 2005;25:621–636. doi: 10.1128/MCB.25.2.621-636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohli J, Munz P, Soll D. In: Molecular Biology of the Fission Yeast. Nasim A, Young P, Johnson BF, editors. Academic Press Inc.; San Diego: 1989. pp. 75–96. [Google Scholar]
- 22.Intine RVA, Sakulich AL, Koduru SB, Huang Y, Pierstorrf E, Goodier JL, Phan L, Maraia RJ. Control of transfer RNA maturation by phosphorylation of the human La antigen on serine 366. Mol Cell. 2000;6:339–348. doi: 10.1016/s1097-2765(00)00034-4. [DOI] [PubMed] [Google Scholar]
- 23.Bayfield MA, Maraia RJ. Precursor-product discrimination by La protein during tRNA metabolism. Nat Struct & Mol Biol. 2009;16:430–437. doi: 10.1038/nsmb.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang Y, Bayfield MA, Intine RV, Maraia RJ. Separate RNA-binding surfaces on the multifunctional La protein mediate distinguishable activities in tRNA maturation. Nat Struct Mol Biol. 2006;13:611–618. doi: 10.1038/nsmb1110. [DOI] [PubMed] [Google Scholar]
- 25.Hamada M, Sakulich AL, Koduru SB, Maraia R. Transcription termination by RNA polymerase III in fission yeast: A genetic and biochemical model system. J Biol Chem. 2000;275:29076–29081. doi: 10.1074/jbc.M003980200. [DOI] [PubMed] [Google Scholar]
- 26.Bogenhagen DF, Brown DD. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell. 1981;24:261–270. doi: 10.1016/0092-8674(81)90522-5. [DOI] [PubMed] [Google Scholar]
- 27.Braglia P, Percudani R, Dieci G. Sequence context effects on oligo(dT) termination signal recognition by Saccharomyces cerevisiae RNA polymerase III. J Biol Chem. 2005;280:19551–19562. doi: 10.1074/jbc.M412238200. [DOI] [PubMed] [Google Scholar]
- 28.Gottlieb E, Steitz JA. The RNA binding protein La influences both the accuracy and the efficiency of RNA polymerase III transcription in vitro. EMBO J. 1989;8:841–850. doi: 10.1002/j.1460-2075.1989.tb03445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathews MB, Francoeur AM. La antigen recognizes and binds to the 3′-oligouridylate tail of a small RNA. Mol Cell Biol. 1984;4:1134–1140. doi: 10.1128/mcb.4.6.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mazabraud A, Scherly D, Muller F, Rungger D, Clarkson SG. Structure and transcription termination of a lysine tRNA gene from Xenopus laevis. J Mol Biol. 1987;195:835–845. doi: 10.1016/0022-2836(87)90488-8. [DOI] [PubMed] [Google Scholar]
- 31.Reddy R, Henning D, Tan E, Busch H. Identification of a La protein binding site in a RNA polymerase III transcript (4.5 I RNA) J Biol Chem. 1983;258:8352–8356. [PubMed] [Google Scholar]
- 32.Rinke J, Steitz JA. Precursor molecules of both human 5S ribosomal RNA and transfer RNAs are bound by a cellular protein reactive with anti-La lupus antibodies. Cell. 1982;29:149–159. doi: 10.1016/0092-8674(82)90099-x. [DOI] [PubMed] [Google Scholar]
- 33.Rinke J, Steitz JA. Association of the lupus antigen La with a subset of U6 snRNA molecules. Nucleic Acids Res. 1985;13:2617–2629. doi: 10.1093/nar/13.7.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zasloff M, Santos T, Romeo P, Rosenberg M. Transcription and precursor processing of normal and mutant human tRNAiMet genes in a homologous cell-free system. J Biol Chem. 1982;257:7857–7863. [PubMed] [Google Scholar]
- 35.Campbell FE, Setzer DR. Transcription termination by RNA polymerase III: uncoupling of polymerase release from termination signal recognition. Mol Cell Biol. 1992;12:2260–2272. doi: 10.1128/mcb.12.5.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cozzarelli NR, Gerrard SP, Schlissel M, Brown DD, Bogenhagen DF. Purified RNA polymerase III accurately and efficiently terminates transcription of 5S RNA genes. Cell. 1983;34:829–835. doi: 10.1016/0092-8674(83)90540-8. [DOI] [PubMed] [Google Scholar]
- 37.Matsuzaki H, Kassavetis GA, Geiduschek EP. Analysis of RNA chain elongation and termination by Saccharomyces cerevisiae RNA polymerase III. J Mol Biol. 1994;235:1173–1192. doi: 10.1006/jmbi.1994.1072. [DOI] [PubMed] [Google Scholar]
- 38.Shaaban SA, Bobkova EV, Chudzik DM, Hall BD. In vitro analysis of elongation and termination by mutant RNA polymerases with altered termination behavior. Mol Cell Biol. 1996;16:6468–6476. doi: 10.1128/mcb.16.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitehall SK, Bardeleben C, Kassavetis GA. Hydrolytic cleavage of nascent RNA in RNA polymerase III ternary transcription complexes. J Biol Chem. 1994;269:2299–2306. [PubMed] [Google Scholar]
- 40.Shaaban SA, Krupp BM, Hall BD. Termination-altering mutations in the second-largest subunit of yeast RNA polymerase III. Mol Cell Biol. 1995;15:14671478. doi: 10.1128/mcb.15.3.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bobkova EV, Hall BD. Substrate specificity of the RNase activity of yeast RNA polymerase III. J Biol Chem. 1997;272:22832–22839. doi: 10.1074/jbc.272.36.22832. [DOI] [PubMed] [Google Scholar]
- 42.Chedin S, Riva M, Schultz P, Sentenac A, Carles C. The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination. Genes Dev. 1998;12:3857–3871. doi: 10.1101/gad.12.24.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bayfield MA, Yang R, Maraia RJ. Conserved and divergent features of the structure and function of La and La-related proteins (LARPs) Biochim Biophys Acta. 2010 doi: 10.1016/j.bbagrm.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maraia RJ, Bayfield MA. The La protein-RNA complex surfaces. Mol Cell. 2006;21:149–152. doi: 10.1016/j.molcel.2006.01.004. review. [DOI] [PubMed] [Google Scholar]
- 45.Wolin SL, Cedervall T. The La protein. Annu Rev Biochem. 2002;71:375–403. doi: 10.1146/annurev.biochem.71.090501.150003. [DOI] [PubMed] [Google Scholar]
- 46.Celma ML, Pan J, Weissman SM. Studies of low molecular weight RNA from cells infected with adenovirus 2. I. The sequences at the 3′ end of VA-RNA I. J Biol Chem. 1977;252:9032–9042. [PubMed] [Google Scholar]
- 47.Maraia RJ, Chang DY, Wolffe AP, Vorce RL, Hsu K. The RNA polymerase III terminator used by a B1-Alu element can modulate 3′ processing of the intermediate RNA product. Mol Cell Biol. 1992;12:1500–1506. doi: 10.1128/mcb.12.4.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maraia RJ, Kenan DJ, Keene JD. Eukaryotic transcription termination factor La mediates transcript release and facilitates reinitiation by RNA polymerase III. Mol Cell Biol. 1994;14:2147–2158. doi: 10.1128/mcb.14.3.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.French SL, Osheim YN, Schneider DA, Sikes ML, Fernandez CF, Copela LA, Misra VA, Nomura M, Wolin SL, Beyer AL. Visual analysis of the yeast 5S rRNA gene transcriptome: regulation and role of La protein. Mol Cell Biol. 2008;28:4576–4587. doi: 10.1128/MCB.00127-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jarrous N, Reiner R. Human RNase P: a tRNA-processing enzyme and transcription factor. Nucleic Acids Res. 2007;35:3519–3524. doi: 10.1093/nar/gkm071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fan H, Sakulich AL, Goodier JL, Zhang X, Qin J, Maraia RJ. Phosphorylation of the human La antigen on serine 366 can regulate recycling of RNA polymerase III transcription complexes. Cell. 1997;88:707–715. doi: 10.1016/s0092-8674(00)81913-3. [DOI] [PubMed] [Google Scholar]
- 52.Goodier JL, Fan H, Maraia RJ. A carboxy-terminal basic region controls RNA polymerase III transcription factor activity of human La protein. Mol Cell Biol. 1997;17:5823–5832. doi: 10.1128/mcb.17.10.5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gottlieb E, Steitz JA. Function of the mammalian La protein: evidence for its action in transcription termination by RNA polymerase III. EMBO J. 1989;8:851–861. doi: 10.1002/j.1460-2075.1989.tb03446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maraia RJ. Transcription termination factor La is also an initiation factor for RNA polymerase III. Proc Natl Acad Sci USA. 1996;93:3383–3387. doi: 10.1073/pnas.93.8.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Z, Luo T, Roeder RG. Identification of an autonomously initiating RNA polymerase III holoenzyme containing a novel factor that is selectively inactivated during protein synthesis inhibition. Genes Dev. 1997;11:2371–2382. doi: 10.1101/gad.11.18.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chakshusmathi G, Kim SD, Rubinson DA, Wolin SL. A La protein requirement for efficient pre-tRNA folding. EMBO J. 2003;22:6562–6572. doi: 10.1093/emboj/cdg625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Belisova A, Semrad K, Mayer O, Kocian G, Waigmann E, Schroeder R, Steiner G. RNA chaperone activity of protein components of human Ro RNPs. RNA. 2005;11:1084–1094. doi: 10.1261/rna.7263905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee Y, Kindelberger DW, Lee JY, McClennen S, Chamberlain J, Engelke DR. Nuclear pre-tRNA terminal structure and RNase P recognition. RNA. 1997;3:175–185. [PMC free article] [PubMed] [Google Scholar]
- 59.Leontis N, DaLio A, Strobel M, Engelke D. Effects of tRNA-intron structure on cleavage of precursor tRNAs by RNase P from Saccharomyces cerevisiae. Nucleic Acids Res. 1988;16:2537–2552. doi: 10.1093/nar/16.6.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Willis I, Frendewey D, Nichols M, Hottinger-Werlen A, Schaack J, Soll D. A single base change in the intron of a serine tRNA affects the rate of RNase P cleavage in vitro and suppressor activity in vivo in Saccharomyces cerevisiae. J Biol Chem. 1986;261:5878–5885. [PubMed] [Google Scholar]
- 61.Fan H, Goodier JL, Chamberlain J, Engelke DR, Maraia RJ. 5′ Processing of tRNA precursors can be modulated by the human La antigen phosphoprotein. Mol Cell Biol. 1998;18:3201–3211. doi: 10.1128/mcb.18.6.3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Intine RV, Tenenbaum SA, Sakulich AS, Keene JD, Maraia RJ. Differential phosphorylation and subcellular localization of La RNPs associated with precursor tRNAs and translation-related mRNAs. Molecular Cell. 2003;12:1301–1307. doi: 10.1016/s1097-2765(03)00429-5. [DOI] [PubMed] [Google Scholar]
- 63.Bayfield MA, Kaiser TE, Intine RV, Maraia RJ. Conservation of a masked nuclear export activity of La proteins and its effects on tRNA maturation. Mol Cell Biol. 2007;27:3303–3312. doi: 10.1128/MCB.00026-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Intine RV, Dundr M, Misteli T, Maraia RJ. Aberrant nuclear trafficking of La protein leads to disordered processing of associated precursor tRNAs. Mol Cell. 2002;9:1113–1123. doi: 10.1016/s1097-2765(02)00533-6. [DOI] [PubMed] [Google Scholar]
- 65.Nashimoto M, Nashimoto C, Tamura M, Kaspar RL, Ochi K. The inhibitory effect of the autoantigen La on in vitro 3′ processing of mammalian precursor tRNAs. J Mol Biol. 2001;312:975–984. doi: 10.1006/jmbi.2001.5026. [DOI] [PubMed] [Google Scholar]
- 66.Zhao Z, Su W, Yuan S, Huang Y. Functional conservation of tRNase ZL among Saccharomyces cerevisiae, Schizosaccharomyces pombe and humans. Biochem J. 2009;422:483–492. doi: 10.1042/BJ20090743. [DOI] [PubMed] [Google Scholar]
- 67.Takaku H, Minagawa A, Takagi M, Nashimoto M. The N-terminal half-domain of the long form of tRNase Z is required for the RNase 65 activity. Nucleic Acids Res. 2004;32:4429–4438. doi: 10.1093/nar/gkh774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Elbarbary RA, Takaku H, Uchiumi N, Tamiya H, Abe M, Nishida H, Nashimoto M. Human cytosolic tRNase ZL can downregulate gene expression through miRNA. FEBS Lett. 2009;583:3241–3246. doi: 10.1016/j.febslet.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 69.Elbarbary RA, Takaku H, Uchiumi N, Tamiya H, Abe M, Takahashi M, Nishida H, Nashimoto M. Modulation of gene expression by human cytosolic tRNase Z(L) through 5′-half-tRNA. PLoS One. 2009;4:e5908. doi: 10.1371/journal.pone.0005908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nakashima A, Takaku H, Shibata HS, Negishi Y, Takagi M, Tamura M, Nashimoto M. Gene silencing by the tRNA maturase tRNase ZL under the direction of small-guide RNA. Gene therapy. 2007;14:78–85. doi: 10.1038/sj.gt.3302841. [DOI] [PubMed] [Google Scholar]
- 71.Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tavtigian SV, Simard J, Teng DH, Abtin V, Baumgard M, Beck A, Camp NJ, Carillo AR, Chen Y, Dayananth P, et al. A candidate prostate cancer susceptibility gene at chromosome 17p. Nat Genet. 2001;27:172–180. doi: 10.1038/84808. [DOI] [PubMed] [Google Scholar]
- 73.Fujiwara H, Emi M, Nagai H, Nishimura T, Konishi N, Kubota Y, Ichikawa T, Takahashi S, Shuin T, Habuchi T, et al. Association of common missense changes in ELAC2 (HPC2) with prostate cancer in a Japanese case-control series. Journal of human genetics. 2002;47:641–648. doi: 10.1007/s100380200099. [DOI] [PubMed] [Google Scholar]
- 74.Nashimoto M. Anomalous RNA substrates for mammalian tRNA 3′ processing endoribonuclease. FEBS Lett. 2000;472:179–186. doi: 10.1016/s0014-5793(00)01462-9. [DOI] [PubMed] [Google Scholar]
- 75.Morl M, Marchfelder A. The final cut. The importance of tRNA 3′-processing. EMBO Rep. 2001;2:17–20. doi: 10.1093/embo-reports/kve006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Qiu H, Hu C, Anderson J, Björk G, Sarkar S, Hopper A, Hinnebusch AG. Defects in tRNA Processing and Nuclear Export Induce GCN4 Translation Independently of Phosphorylation of the Alpha Subunit of eIF2. Mol Cell Biol. 2000;20:2505–2516. doi: 10.1128/mcb.20.7.2505-2516.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Anderson J, Phan L, Cuesta R, Carlson BA, Pak M, Asano K, Bjork GR, Tamame M, Hinnebusch AG. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 1998;12:3650–3662. doi: 10.1101/gad.12.23.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kadaba S, Wang X, Anderson JT. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA. 2006 doi: 10.1261/rna.2305406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 81.Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. A New Yeast Poly(A) Polymerase Complex Involved in RNA Quality Control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Anderson JT. RNA turnover: unexpected consequences of being tailed. Curr Biol. 2005;15:R635–638. doi: 10.1016/j.cub.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 83.Calvo O, Cuesta R, Anderson J, Gutierrez N, Garcia-Barrio MT, Hinnebusch AG, Tamame M. GCD14p, a repressor of GCN4 translation, cooperates with Gcd10p and Lhp1p in the maturation of initiator methionyl-tRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:4167–4181. doi: 10.1128/mcb.19.6.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hamill S, Wolin SL, Reinisch KM. Structure and function of the polymerase core of TRAMP, a RNA surveillance complex. PNAS. 2010;34:15045–15050. doi: 10.1073/pnas.1003505107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 86.McPheeters DS, Cremona N, Sunder S, Chen HM, Averbeck N, Leatherwood J, Wise JA. A complex gene regulatory mechanism that operates at the nexus of multiple RNA processing decisions. Nat Struct Mol Biol. 2009;16:255–264. doi: 10.1038/nsmb.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 88.Pannone B, Xue D, Wolin SL. A role for the yeast La protein in U6 snRNP assembly: evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J. 1998;17:7442–7453. doi: 10.1093/emboj/17.24.7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wilusz CJ, Wilusz J. Eukaryotic Lsm proteins: lessons from bacteria. Nat Struct Mol Biol. 2005;12:1031–1036. doi: 10.1038/nsmb1037. [DOI] [PubMed] [Google Scholar]
- 90.Spiller MP, Boon KL, Reijns MA, Beggs JD. The Lsm2-8 complex determines nuclear localization of the spliceosomal U6 snRNA. Nucleic Acids Res. 2007;35:923–929. doi: 10.1093/nar/gkl1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Karaduman R, Dube P, Stark H, Fabrizio P, Kastner B, Luhrmann R. Structure of yeast U6 snRNPs: arrangement of Prp24p and the LSm complex as revealed by electron microscopy. RNA. 2008;14:2528–2537. doi: 10.1261/rna.1369808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kufel J, Bousquet-Antonelli C, Beggs JD, Tollervey D. Nuclear pre-mRNA decapping and 5′ degradation in yeast require the Lsm2-8p complex. Mol Cell Biol. 2004;24:9646–9657. doi: 10.1128/MCB.24.21.9646-9657.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tharun S. Roles of eukaryotic Lsm proteins in the regulation of mRNA function. International review of cell and molecular biology. 2009;272:149–189. doi: 10.1016/S1937-6448(08)01604-3. [DOI] [PubMed] [Google Scholar]
- 94.Pannone BK, Kim SD, Noe DA, Wolin SL. Multiple functional interactions between components of the Lsm2-Lsm8 complex, U6 snRNA and the yeast La protein. Genetics. 2001;158:187–196. doi: 10.1093/genetics/158.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Luhtala N, Parker R. LSM1 over-expression in Saccharomyces cerevisiae depletes U6 snRNA levels. Nucleic Acids Res. 2009;37:5529–5536. doi: 10.1093/nar/gkp572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Licht K, Medenbach J, Luhrmann R, Kambach C, Bindereif A. 3′-cyclic phosphorylation of U6 snRNA leads to recruitment of recycling factor p110 through LSm proteins. RNA. 2008;14:1532–1538. doi: 10.1261/rna.1129608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, Jantsch MF, Konrat R, Blasi U, Schroeder R. RNA chaperones, RNA annealers and RNA helicases. RNA biology. 2007;4:118–130. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]