The Diels-Alder reaction is a [4+2] cycloaddition reaction in which a cyclohexene ring is formed between a 1,3-diene and an electron-deficient alkene via a single pericyclic transition state.1 This reaction has been proposed as a key transformation in the biosynthesis of many cyclohexene-containing secondary metabolites.2–5 However, only four purified enzymes have thus far been implicated in biotransformations that are consistent with a Diels-Alder reaction, namely solanapyrone synthase,6 LovB,7,8 macrophomate synthase,9,10 and riboflavin synthease11,12. Although the stereochemical outcomes of these reactions indicate that the product formation could be enzyme-guided in each case, these enzymes typically demonstrate more than one catalytic activity leaving their specific influence on the cycloaddition step uncertain. In our studies of the biosynthesis of spinosyn A, a tetracyclic polyketide-derived insecticide from Saccharopolyspora spinosa,13,14 we identified a cyclase, SpnF, that catalyzes a transannular [4+2] cycloaddition to form the cyclohexene ring in spinosyn A. Kinetic analysis demonstrates that SpnF specifically accelerates the ring formation reaction with an estimated 500-fold rate enhancement (kcat,spnF versus knon). A second enzyme, SpnL, was also identified as responsible for the final cross-bridging step that completes the tetracyclic core of spinosyn A in a manner consistent with a Rauhut-Currier reaction15. This work is significant because SpnF represents the first example characterized in vitro of a stand-alone enzyme solely committed to the catalysis of a [4+2] cycloaddition reaction. In addition, the mode of formation of the complex perhydro-as-indacene moiety in spinosyn A is now fully established.
Spinosyn A (1), an active ingredient of several highly effective and environmentally benign commercial insecticides, has a complex aglycone structure comprised of a perhydro-as-indacene moiety fused to a 12-membered macrolactone.13,14 How this tetracyclic ring system is biosynthesized has been a subject of much speculation.16–18 Attention has largely focused on the construction of the cyclohexene ring due to the potential involvement of an enzyme that catalyzes the [4+2] cycloaddition, which if concerted would represent a so-called "Diels-Alderase." Four genes in the spinosyn A biosynthetic gene cluster of S. spinosa – spnF, spnJ, spnL, and spnM – were proposed to convert product (2) of the polyketide synthase (PKS) to the tetracyclic aglycone (4) (see Figure 1).16 The gene product of spnJ, which is a flavin-dependent dehydrogenase, was recently demonstrated to catalyze oxidation of the 15-OH of 2 to form the keto-intermediate 3.19 However, the functions of the enzymes encoded by the remaining three genes, spnF, spnL, and spnM, which show significant sequence similarity to lipases (SpnM) and S-adenosyl-l-methionine (SAM)-dependent methyltransferases (SpnF and SpnL),16 remain elusive.
Figure 1. Proposed spinosyn biosynthetic pathway.
To investigate the functions of SpnF, SpnL, and SpnM, the corresponding genes were heterologously overexpressed in Escherichia coli BL21(DE3)* and their products purified as N-terminal His6-tagged proteins (>95% purity). Each of the purified enzymes was tested for activity with 3. As shown in Figure 2A, neither SpnF nor SpnL processes 3. In contrast, complete conversion of 3 to a new product was observed after a 2-hr incubation with SpnM. NMR and MS analysis of this new product (8) revealed a 15,6,5-tricyclic skeleton. Further investigation of the reaction time course led to the discovery of a transient intermediate (Fig. 2D, orange peak), which was identified by spectral analysis as the monocyclic macrolactone 5.
Figure 2. HPLC analysis showing the reactions catalyzed by SpnF, SpnL, and SpnM.
A. Incubation of 2 mM 3 for 2 hr alone (a), and with either 1.25 µM SpnF (b), 1.25 µM SpnL (c), or 1.25 µM SpnM (d). B. Incubation of 2 mM 8 (generated in situ from 3) for 2 hr either alone (a), with 15 µM SpnL (b), or with 15 µM SpnF (c). Trace (d) is the authentic standard of 4. C. Incubation of 1 mM 8 for 2 hr either alone (a); with both 20 µM SpnG and 1.5 mM 9 (b); or with 15 µM SpnL, 20 µM SpnG, and 1.5 mM 9 (c). Trace (d) is the authentic standard of 13. D. Time course of 2 mM 3 in the presence of 1.25 µM SpnM. E. Time course of 2 mM 3 in the presence of both 1.25 µM SpnM and 10 µM SpnF.
These findings suggested that the SpnM-catalyzed conversion of 3 to 8 might be a two-step process involving 1,4-dehydration of 3 followed by a transannular [4+2] cycloaddition between the Δ11,12-alkene and the conjugated Δ4,5,Δ6,7-diene of intermediate 5 to form the cyclohexene moiety in 8 (Fig. 4). To investigate whether SpnM is indeed a cyclase of dual function catalyzing both the dehydration and cyclization steps, the dependence of the rate of each step on the concentration of SpnM was determined. In this experiment the formation and consumption of 5, which respectively reflect the dehydration and cyclization steps, was monitored by HPLC as a function of time.
Figure 4. The spinosyn aglycone biosynthetic pathway.
SpnM catalyzes a dehydration reaction to convert 3 to 5, and SpnF subsequently catalyzes cyclization of 5 to afford 8. The resulting tricyclic macrolactone 8 is then modified with a rhamnose moiety at the C-9 hydroxyl group by SpnG rather than being converted directly to the aglycone 4 (see Scheme 1) as previously thought. SpnL completes the cross-bridging process by interlinking the C-3 and C-14 carbon centers of 10 to produce the tetracyclic nucleus (13) of spinosyn A.
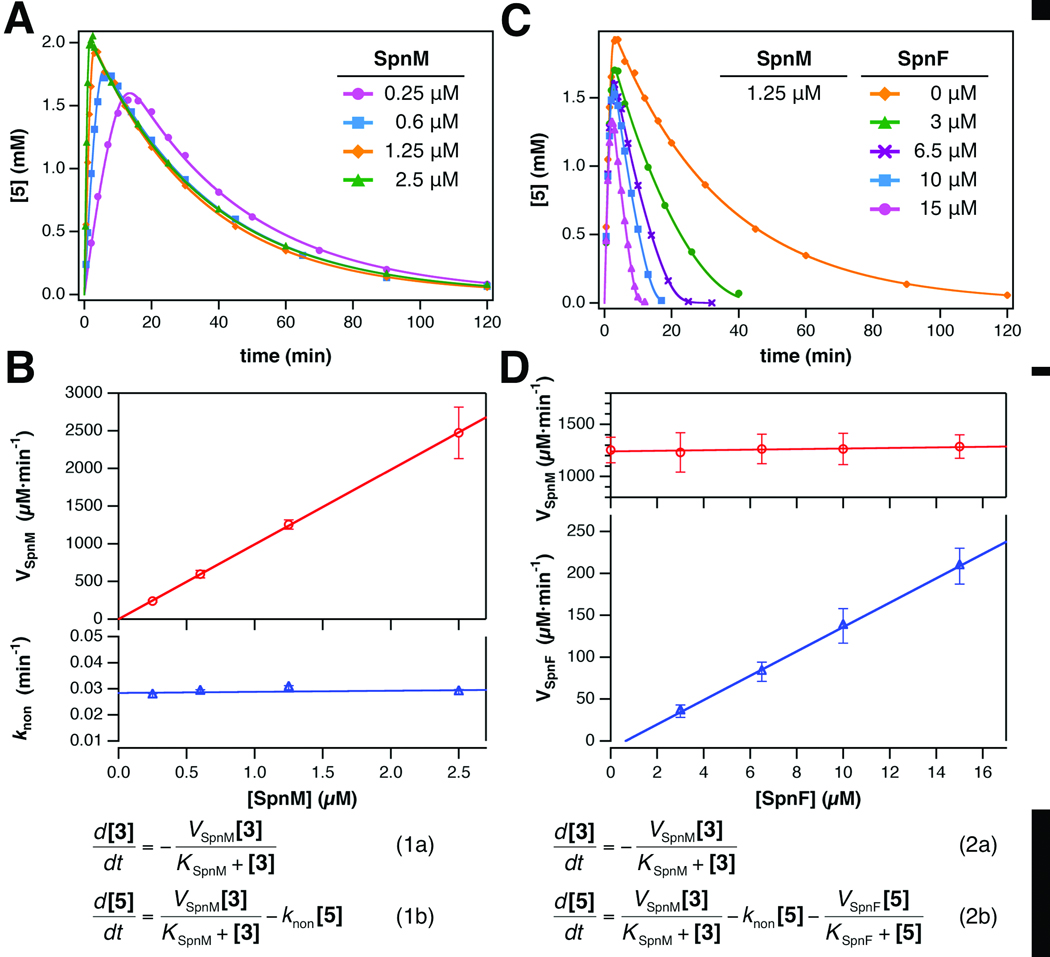
As shown in Figure 3A, rate enhancement of the dehydration step was observed with the increase in SpnM concentration, while the rate of cyclization was unaffected. The full time courses of the formation and decay of 5 were analyzed using numerically integrated coupled rate equations based on different kinetic models (see SI Section S5).20 The observed data was fit best to the integrated rate equations 1a and 1b, which model Michaelis-Menten kinetics for the dehydration step (VSpnM and KSpnM) and first order kinetics (knon) for the cyclization step. Correlation of the fitted parameters versus the concentration of SpnM (Fig. 3B) reveals a significant dependence of VSpnM, but not KSpnM or knon, on SpnM concentration. This analysis indicates that SpnM functions only as a dehydratase, and its product (5) can cyclize nonenzymatically to 8 with a first order rate constant of ~ 0.03 min−1.
Figure 3. Kinetic analysis demonstrating that SpnM and SpnF respectively catalyze the dehydration and cyclization steps of macrolactone 3.
Formation and consumption of 5 was monitored by HPLC and the concentration ([5]) plotted versus time (A & C). Each reaction mixture initially contained 2 mM 3, and the indicated amounts of SpnM and SpnF. A. Variable SpnM with no SpnF and fits based on the rate equations (1). B. Fitted parameters VSpnM and knon versus [SpnM]. The measured turnover number for SpnM, kcat,SpnM, is 1020 ± 57 min−1 while the first order rate constant for nonenzymatic cyclization of 5, knon, is 0.0288 ± 0.00041 min−1. The apparent Michaelis constant for SpnM, KSpnM, is 380 ± 51 µM. C. Variable SpnF at a fixed concentration of SpnM and fits based on rate equations (2). D. Correlation of VSpnM and VSpnF versus [SpnF]. The turnover number for SpnF, kcat,SpnF, is 14 ± 1.6 min−1. The apparent Michaelis constant for SpnF, KSpnF, which is independent of SpnF concentration, is 120 ± 46 µM. All values and error bars are ± s.e.
Having established that SpnM functions as a dehydratase, whereas the [4+2] cycloaddition can proceed nonenzymatically, we next considered the possible involvement of the two remaining gene products, SpnF and SpnL, in the final C–C bond formation between C-3 and C-14. Surprisingly, incubation of SpnF or SpnL with 8, which was generated in situ from 3 in the presence of SpnM, did not produce the aglycone 4 (Fig. 2B). However, when SpnF was added to the assay mixture, a change in the product distribution was noted including rapid disappearance of 5 (Fig. 2B, trace (c)). As shown in Figure 2E, the consumption of 5 with concomitant formation of a product having a retention time consistent with 8 was completed in 20 min in the presence of 10 µM SpnF, instead of requiring more than 2 hr in its absence (Fig. 2D). The structure of this product was established to be 8 by NMR analysis. Additional controls including examining the effects of denatured SpnF as well as mutated SpnF on the rate of cyclization were also performed. In all cases only native (His6-tagged) SpnF lead to appreciably increased rates of cyclization (see SI Section S4.4). These results clearly indicate that SpnF is responsible for the observed rate enhancement of the cyclization of 5 to 8.
To quantify the effect of SpnF on the rate of the cyclization step, the production and consumption of 5 was again monitored by HPLC, this time as a function of SpnF concentration while keeping that of SpnM fixed. As shown in Figure 3C, the rate of conversion of 5 to 8 is clearly enhanced as the concentration of SpnF increases. The data was fit best using the coupled rate equations 2a and 2b, where the cyclization event is modeled as the sum of a first order and a Michaelis-Menten process. In these fits all parameters including the initial concentration of 3 were allowed to float except for knon, which was fixed at the fitted value obtained in the absence of SpnF. Correlation of the fitted parameters with SpnF concentration indicates a significant dependence of VSpnF on SpnF concentration (Fig. 3D), which is not true for the VSpnM parameter. These results firmly establish that SpnF is a cyclase catalyzing the conversion of 5 to 8 with an apparent kcat of 14 ± 1.6 min−1 (± s.e.) for an estimated rate enhancement (kcat,spnF versus knon) of ~ 500-fold.
This result left spnL as the only gene without an assigned function, and it was hypothesized that SpnL plays a role in the transannular cyclization reaction between C-3 and C-14. However, the observed inability of SpnL to convert 8 to the anticipated product (4) (Fig. 2B, trace (b)) prompted us to reconsider the sequence of events of the proposed biosynthetic pathway in Figure 1. Previously, glycosylations had been thought to occur on the tetracyclic aglycone (4), because transfer of the rhamnose moiety from TDP-β-l-rhamnose (9) to 9-OH of 4 by the rhamnosyltransferase, SpnG, had been demonstrated.21,22 However, this observation did not necessarily rule out an alternative pathway in which rhamnosylation of 8 precedes C-3/C-14 cross-bridging, because glycosyltransferases involved in the biosynthesis of secondary metabolites are known to be promiscuous with regard to their substrate specificity.23,24
To test this possibility, the ability of SpnG to accept 8 as a substrate was investigated. As shown in Figure 2C (trace b), incubation of 8 and 9 with SpnG resulted in the disappearance of 8 with the concomitant appearance of a new peak (retention time of 17 min). This new product was identified as 10 by NMR and MS analysis (Fig. 4). Upon addition of SpnL to this reaction mixture, the peak at 17 min disappeared and a new peak appeared at 27 min (Fig. 2C, trace c). Both transformations were highly efficient. Identification of the new product as 13 was confirmed by high-resolution MS analysis and HPLC co-elution with an authentic sample of 13. These results strongly suggest that 10 rather than 8 is the biological substrate for SpnL, which catalyzes the final cyclization step to generate the perhydro-as-indacene core.
The mechanisms by which SpnF and SpnL catalyze their respective cyclization reactions are a point of interest. The SpnF-catalyzed endo-mode syn-addition of an alkenyl to a dienyl functionality appears consistent with a Diels-Alder reaction; however, confirmation of this hypothesis will require demonstrating that the reaction progresses through a single pericyclic transition state such as 6. Therefore, a stepwise [4+2] cycloaddition mechanism, e.g., one involving a dipolar intermediate such as 7, cannot at present be ruled out. In contrast, the C–C bond formation catalyzed by SpnL may involve a Rauhut-Currier mechanism15 consistent with the observation that 10 is susceptible toward nucleophilic addition by a thiol (see SI Section S3.10) forming a covalent adduct that may be structurally analogous to 11 or 12 (see Figure 4), though the specific site of attack remains unknown. While these mechanistic proposals are at present speculative, it is worth noting that Roush and coworkers were able to accomplish their non-enzymatic total synthesis of spinosyn A by exploiting both the transannular Diels-Alder and Rauhut-Currier reactions in an analogous fashion.25 This precedent in chemical synthesis certainly substantiates the feasibility of the mechanisms proposed for the SpnF- and SpnL-catalyzed reactions.
In summary, the biosynthetic pathway for spinosyn A is now fully established (Fig. 4), with the specific functions of SpnM as a dehydratase and SpnF as well as SpnL as the two cyclases in the cross-bridging steps biochemically determined. SpnF represents the first enzyme for which specific acceleration of a [4+2] cycloaddition reaction has been experimentally verified as its only known function. It stands in contrast to macrophomate synthase, for which evidence has been provided suggesting a tandem Michael-Aldol reaction mechanism,26,27 as well as the multifunctional solanapyrone synthase, LovB, and riboflavin synthase, which respectively participate in hydroxyl oxidation,5,6 polyketide synthesis,7,8 and hydride transfer,12 in addition to the [4+2] cycloaddition reactions, the concertedness of which have yet to be verified. For this reason, the SpnF reaction provides a unique system for detailed mechanistic investigation of enzyme-catalyzed [4+2] cycloadditions and the existence of a bona fide Diels-Alderase.
Method Summary
All proteins used in this work were overexpressed in E. coli BL21(DE3)* (Invitrogen, Carlsbad, CA) and purified by Ni-NTA (Qiagen, Valencia, CA) affinity chromatography. Specifically, SpnF was co-overexpressed with the chaperone protein pair, GroEL/ES, to improve the solubility. Because overexpression of the protein encoded by the originally assigned spnM gene16 failed to afford an active soluble protein, the gene sequence was re-examined and revised to include 204 additional base pairs (Supplementary Fig. 2). Overexpression of the revised spnM gene produced an active enzyme with significantly improved protein yield. All enzyme reaction products (5, 8, 10, and 13) were extracted with ethyl acetate or chloroform and purified using silica gel column chromatography or HPLC. Their structures were characterized by 1D- and 2D-NMR spectroscopy and/or high-resolution mass spectrometry. In particular, the stereochemistry of 8 was assigned based on its 1H-1H NOESY spectrum. All substrate specificity and time course assays were run in 50 mM Tris∙HCl buffer (pH 8) at 30 °C. Reaction aliquots were quenched with an excess volume of ethanol after a given incubation time and centrifuged to remove protein. The supernatant was then analyzed by reverse phase HPLC with detection by UV absorbance at 254 nm (Figure 2) or 280 nm (Figure 3). Time course assays also included p-methoxyacetophenone as an internal standard to normalize the peak areas corresponding to 5. Numerical integration of equations (1) and (2) employed the fourth order Runge-Kutta algorithm (Supplementary Ref. 9) following non-dimensionalization of substrate concentration. The resulting simulated progress curves were fit using steepest descent (Supplementary Ref. 10) directly to full time courses of normalized substrate concentration obtained via the HPLC discontinuous assay to provide the kinetic parameters and a concentration normalization factor. Further detail regarding experimental procedures as well as data fitting and analysis is described in the Supplementary Information.
Supplementary Material
Acknowledgements
We thank Dr. Christian Whitman for review of the manuscript, Dr. Lin Hong for carrying out the early cloning work, Dr. Ben Shoulders and Steve Sorey for assistance with the interpretation of the NMR spectra, and Dr. Steven Mansoorabadi and Eta Isiorho for discussions on the reaction mechanisms and structural modelling of SpnF. This work is supported in part by grants from the National Institutes of Health (GM035906, F32AI082906), the Texas Higher Education Coordination Board (ARP-003658-0093-2007), and the Welch Foundation (F-1511).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author contributions H.-w.L. provided the scientific direction and the overall experimental design for the studies. H.J.K designed and performed most of the experiments. S.H.C participated in the chemical synthesis of the substrate for SpnJ (2) and the characterization of the structures of the enzyme reaction products. M.W.R analyzed the kinetic experiment data. Y.-n.L. carried out the mutation studies of SpnF. H.J.K, M.W.R and H.-w.L. wrote the manuscript.
Author information Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Huisgen R. Cycloadditions – definition, classification, and characterization. Angew. Chem. Int. Ed. 1968;7:321–328. [Google Scholar]
- 2.Stocking EM, Williams RM. Chemistry and biology of biosynthetic Diels-Alder reactions. Angew. Chem. Int. Ed. 2003;42:3078–3115. doi: 10.1002/anie.200200534. [DOI] [PubMed] [Google Scholar]
- 3.Oikawa H. Involvement of the Diels-Alderases in the biosynthesis of natural products. Bull. Chem. Soc. Jpn. 2005;78:537–554. [Google Scholar]
- 4.Kelly WL. Intramolecular cyclizations of polyketide biosynthesis: mining for a "Diels-Alderase?". Org. Biomol. Chem. 2008;6:4483–4493. doi: 10.1039/b814552k. [DOI] [PubMed] [Google Scholar]
- 5.Oikawa H. Diels-Alderases. In: Mander L, Liu H-w, editors. Comprehensive Natural Products II, Chemistry and Biology. Vol. 8. Amsterdam: Elsevier; 2010. pp. 277–314. [Google Scholar]
- 6.Oikawa H, Katayama K, Suzuki Y, Ichihara A. Enzymatic-activity catalyzing exo-selective Diels-Alder reaction in solanapyrone biosynthesis. J. Chem. Soc. Chem. Comm. 1995:1321–1322. [Google Scholar]
- 7.Auclair K, Sutherland A, Kennedy J, Witter DJ, Van den Heever JP, Hutchinson CR, Vederas JC. Lovastatin nonaketide synthase catalyzes an intramolecular Diels-Alder reaction of a substrate analogue. J. Am. Chem. Soc. 2000;122:11519–11520. [Google Scholar]
- 8.Ma SM, Li JW-H, Choi JW, Zhou H, Lee KKM, Moorthie VA, Xie X, Kealey JT, Da Silva NA, Vederas JC, Tang Y. Complete Reconstitution of a highly reducing iterative polyketide synthase. Science. 2009;326:589–592. doi: 10.1126/science.1175602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watanabe K, Mie T, Ichihara A, Oikawa H, Honma M. Detailed reaction mechanism of macrophomate synthase. J. Biol. Chem. 2000;275:38393–38401. doi: 10.1074/jbc.M003119200. [DOI] [PubMed] [Google Scholar]
- 10.Ose T, Watanabe K, Mie T, Honma M, Watanabe H, Yao M, Oikawa H, Tanaka I. Insight into a natural Diels-Alder reaction from the structure of macrophomate synthase. Nature. 2003;422:185–189. doi: 10.1038/nature01454. [DOI] [PubMed] [Google Scholar]
- 11.Eberhardt S, Zingler N, Kemter K, Richter G, Cushman M, Bacher A. Domain structure of riboflavin synthase. Eur. J. Biochem. 2001;268:4315–4323. doi: 10.1046/j.1432-1327.2001.02351.x. [DOI] [PubMed] [Google Scholar]
- 12.Kim R-R, Illarionov B, Joshi M, Cushman M, Lee CY, Eisenreich W, Fischer M, Bacher A. Mechanistic insights on riboflavin synthase inspired by selective binding of the 6,7-dimethyl-8-ribityllumazine exomethylene anion. J. Am. Chem. Soc. 2010;132:2983–2990. doi: 10.1021/ja908395r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirst HA, Michel KH, Martin JW, Creemer LC, Chio EH, Yao RC, Nakatsukasa WM, Boeck L, Occolowitz JL, Paschal JW, Deeter JB, Jones ND, Thompson GD. A83543A-D, Unique fermentation-derived tetracyclic macrolides. Tetrahedron Lett. 1991;32:4839–4842. [Google Scholar]
- 14.Kirst HA. The spinosyn family of insecticides: realizing the potential of natural products research. J. Antibiot. 2010;63:101–111. doi: 10.1038/ja.2010.5. [DOI] [PubMed] [Google Scholar]
- 15.Aroyan CE, Dermenci A, Miller SJ. The Rauhut-Currier reaction: a history and its synthetic application. Tetrahedron. 2009;65:4069–4084. [Google Scholar]
- 16.Waldron C, Matsushima P, Rosteck PR, Broughton MC, Turner J, Madduri K, Crawford KP, Merlo DJ, Baltz RH. Cloning and analysis of the spinosad biosynthetic gene cluster of Saccharopolyspora spinosa. Chem. Biol. 2001;8:487–499. doi: 10.1016/s1074-5521(01)00029-1. [DOI] [PubMed] [Google Scholar]
- 17.Martin CJ, Timoney MC, Sheridan RM, Kendrew SG, Wilkinson B, Staunton JC, Leadlay PF. Heterologous expression in Saccharopolyspora erythraea of a pentaketide synthase derived from the spinosyn polyketide synthase. Org. Biomol. Chem. 2003;1:4144–4147. doi: 10.1039/b310740j. [DOI] [PubMed] [Google Scholar]
- 18.Oikawa H. Biosynthesis of structurally unique fungal metabolite GKK1032A(2): indication of novel carbocyclic formation mechanism in polyketide biosynthesis. J. Org. Chem. 2003;68:3552–3557. doi: 10.1021/jo0267596. [DOI] [PubMed] [Google Scholar]
- 19.Kim HJ, Pongdee R, Wu Q, Hong L, Liu H-w. The biosynthesis of spinosyn in Saccharopolyspora spinosa: synthesis of the cross-bridging precursor and identification of the function of SpnJ. J. Am. Chem. Soc. 2007;129:14582–14584. doi: 10.1021/ja076580i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frieden C. Analysis of kinetic data: practical applications of computer simulation and fitting programs. Method. Enzymol. 1994;240:311–322. doi: 10.1016/s0076-6879(94)40053-9. [DOI] [PubMed] [Google Scholar]
- 21.Chen Y, Lin Y, Tsai K, Chiu H. Functional characterization and substrate specificity of spinosyn rhamnosyltransferase by in vitro reconstitution of spinosyn biosynthetic enzymes. J. Biol. Chem. 2009;284:7352–7363. doi: 10.1074/jbc.M808441200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim HJ, White-Phillip JA, Ogasawara Y, Shin N, Isiorho EA, Liu H-w. Biosynthesis of spinosyn in Saccharopolyspora spinosa: Synthesis of permethylated rhamnose and characterization of the functions of SpnH, SpnI, and SpnK. J. Am. Chem. Soc. 2010;132:2901–2903. doi: 10.1021/ja910223x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thibodeaux CJ, Melançon CE, Liu H-w. Unusual sugar biosynthesis and natural product glycodiversification. Nature. 2007;446:1008–1016. doi: 10.1038/nature05814. [DOI] [PubMed] [Google Scholar]
- 24.Thibodeaux CJ, Melançon CE, Liu H-w. Natural-product sugar biosynthesis and enzymatic glycodiversification. Angew. Chem. Int. Ed. 2008;47:9814–9859. doi: 10.1002/anie.200801204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mergott DJ, Frank SA, Roush WR. Total synthesis of (−)-spinosyn A. Proc. Natl. Acad. Sci. U.S.A. 2004;101:11955–11959. doi: 10.1073/pnas.0401247101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guimarães CRW, Udier-Blagović M, Jørgensen WL. Macrophomate synthase: QM/MM simulations address the Diels-Alder versus Michael-Aldol reaction mechanism. J. Am. Chem. Soc. 2005;127:3577–3588. doi: 10.1021/ja043905b. [DOI] [PubMed] [Google Scholar]
- 27.Serafimov JM, Gillingham D, Kuster S, Hilvert D. The putative Diels-Alderase macrophomate synthase is an efficient aldolase. J. Am. Chem. Soc. 2008;130:7798–7799. doi: 10.1021/ja8017994. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.