Abstract
DNA methylation inheritance is the process of copying, via the DNA methyltransferase 1 (Dnmt1), the pre-existing methylation patterns onto the new DNA strand during DNA replication. Experiments of chromatin immunoprecipitation, measurement of maintenance methyltransferase activity, proximity ligation in situ assays (P-LISA, Duolink/Olink), and transcription factor arrays demonstrate that Dnmt1 interacts with transcription factors to promote site-specific DNA methylation inheritance, while the Dnmt1-PCNA-UHRF1 complex promotes the DNA methylation inheritance without site preference. We also show that the Dnmt1-PCNA-UHRF1 and Dnmt1/transcription factor complexes methylate DNA by acting as a single player or in cooperation. Thus, our data establish that the copying of the pre-existing methylation pattern is governed by the orchestration of the untargeted and the targeted mechanisms of DNA methylation inheritance, which are themselves dictated by the partners of Dnmt1.
Keywords: epigenetics, DNA methylation, Dnmt
Introduction
DNA methylation inheritance is a crucial event in epigenetics because it consists of the copying of the pre-existing methylation patterns onto the neosynthesized DNA strand during DNA replication. The DNA methyltransferase 1 (Dnmt1) is the principal enzyme responsible for catalyzing the reaction of maintenance of CpG methylation because this enzyme has an intrinsic affinity for hemimethylated DNA.1 At the molecular level, the correct loading of Dnmt1 onto neosynthesized DNA involves an interaction between Dnmt1 and PCNA, and the preference of Dnmt1 for hemimethylated DNA is reinforced by its interaction with UHRF1, a protein known to recognize and bind hemimethylation sites via its SET and RING-associated (SRA) domain.2-7 Thus, a model has been proposed for DNA methylation inheritance consisting of a complex formed by the Dnmt1, UHRF1, PCNA, Ga9, RB1, and PARP1 proteins. In terms of targets, Sharif et al.7 and Bostick et al.5 observed that the disruption of Dnmt1-UHRF1 complexes via the invalidation of the UHRF1 gene induced the global DNA hypomethylation underlined by the hypomethylation of DNA repeat elements such as LINE-1 and of some genes such as the IGF2 and CDKN1c. These findings suggest that the complex formed by the Dnmt1, UHRF1, PCNA, Ga9, RB1, and PARP1 proteins catalyzes the unspecific DNA methylation inheritance.
In parallel, literature reports have mentioned that Dnmt1 can interact with some transcription factors to induce the recruitment of Dnmt1 on DNA sites usually bound by transcription factors and promote the DNA methylation maintenance of CpG located on or in the vicinity of these sites. For example, Esteve et al. demonstrated that Dnmt1 and p53 cooperate to modulate the expression of p53-repressed promoters.8 After an analysis of the complex formed by Dnmt1, E2F1, Rb, and HDAC1, Robertson et al. showed that Dnmt1 could repress the transcription of E2F-responsive promoters.9 It was also reported that STAT3 and Dnmt1 mediated the epigenetic silencing of the SHP-1 tyrosine phosphatase tumor suppressor gene.10 In other words, these results suggest that Dnmt1 directly interacts with transcription factors to maintain the methylation status of the promoter regions of certain genes.
Thus, the machinery of maintenance DNA methylation can be composed of 2 sides: first, crucial in the inheritance of DNA methylation and unspecific in terms of targets, represented by the “Dnmt1-PCNA-UHRF1” complex; and second, characterized by a targeted maintenance methyltransferase activity, represented by the complexes encompassing the Dnmt1 and a transcription factor. Despite their description, the orchestration between the 2 sides of the machinery of maintenance DNA methylation and the identification of a transcription factor interacting potentially with Dnmt1 have not been elucidated. In the present report, we cured that by identifying 58 transcription factors interacting with recombinant Dnmt1. Moreover, we answer several key questions on the respective roles of the 2 sides of the machinery of maintenance DNA methylation. Indeed, chromatin immunoprecipitation (ChIP) and sequential or Re-ChIP experiments reveal that Dnmt1-PCNA-UHRF1 (i.e., the representative of the untargeted side of the DNA methylation inheritance mechanisms) and Dnmt1/Sp1 (i.e., a representative of the targeted side of the DNA methylation inheritance mechanisms) can maintain the DNA methylation by acting as a single player, or they can cooperate in DNA methylation inheritance.
Results
Dnmt1/Sp1 or Dnmt1-PCNA-UHRF1 acts as a single player to maintain the methylation of the −1180/−1091 region of TNFSF10 and the 1163/1270 region of LINE-1 or the −583/−488 region of HOXA2
In order to describe the mechanisms of DNA methylation inheritance in glioma cells, we decided to identify the mechanisms governing the maintenance of the methylation status of LINE-1 (a DNA repeat element frequently hypomethylated in glioma cells) and on the maintenance of the methylation status of the HOXA2, TNFSF10, and SLIT2 genes, that is, 3 genes epigenetically regulated in glioma. For this purpose, we choose to use the U87 cells because LINE-1, HOXA2, TNFSF10, and SLIT2 are methylated in these cells according to the results obtained via the realization of methylated DNA immunoprecipitation (MeDIP) assays (Fig. 1A).
Figure 1.
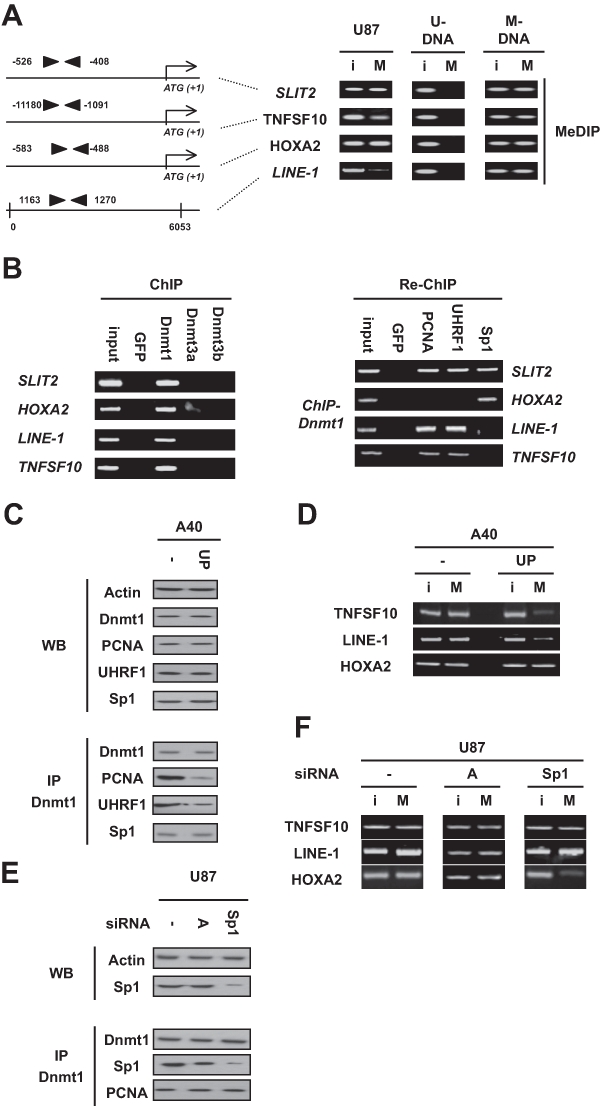
Dnmt1/Sp1 or Dnmt1-PCNA-UHRF1 acts as a single player to maintain the methylation of the −1180/−1091 region of TNFSF10 and the 1163/1270 region of LINE-1 or the −583/−488 region of HOXA2. (A) The methylation status of the SLIT2 (ENSG00000145147), TNFSF10 (ENSG00000121858), and HOXA2 (ENSG00000105996) genes was determined by methylation-specific PCR (MSP). U = unmethylated; M = methylated. The methylation status of LINE-1 (L19092.1) was determined by the methylated DNA immunoprecipitated method (MeDIP). I = input, i.e., no immunoprecipitated DNA; M = MeDIP, i.e., immunoprecipitated DNA. Black triangles represent primers used in MeDIP. U-DNA = CpGenome Universal unmethylated DNA set and M-DNA/CpGenome Universal methylated DNA set (Chemicon, Millipore, Molsheim, France). (B) In U87 cells, chromatin immunoprecipitation (ChIP) experiments were performed to determine the presence of Dnmt1, Dnmt3a, or Dnmt3b on indicated genes. Re-chromatin immunoprecipitation (Re-ChIP) experiments were performed to determine the presence of the considered proteins on indicated genes. (C, D) Impact of the disruption of the Dnmt1-PCNA-UHRF1 interaction on the methylation status of SLIT2, TNFSF10, HOXA2, and LINE-1. The disruption of the Dnmt1-PCNA-UHRF1 interactions via the nucleofection of the Astro40 cells with the pUP plasmid (UP) or plasmid control was monitored by the immunoprecipitation of the Dnmt1 (IP-Dnmt1). Western blot illustrates the expression of Dnmt1, PCNA, UHRF1, and Sp1 in indicated cells. The methylation status of SLIT2, TNFSF10, HOXA2, and LINE-1 was assessed by the methylated DNA immunoprecipitated method (MeDIP). I = input, i.e., no immunoprecipitated DNA; M = MeDIP, i.e., immunoprecipitated DNA. (E, F) Impact of the disruption of the Dnmt1/Sp1 interaction on the methylation status of SLIT2, TNFSF10, HOXA2, and LINE-1. The disruption of the Dnmt1/Sp1 interactions via the Sp1 down-regulation in Astro40 cells by siRNA-Sp1 (sc#29487; Tebu-Bio, Le-Perray-en-Yvelines, France) was monitored by the immunoprecipitation of Dnmt1 (IP-Dnmt1). siRNA-A (sc#37007; Tebu-Bio) is used as control. The methylation status of SLIT2, TNFSF10, HOXA2, and LINE-1 was assessed by the methylated DNA immunoprecipitated method (MeDIP). I = input, i.e., no immunoprecipitated DNA; M = MeDIP, i.e., immunoprecipitated DNA.
To identify the actors and more specially the DNA methyltransferase(s) (Dnmt) implicated in the methylation of LINE-1, HOXA2, TNFSF10, and SLIT2, ChIP analyses were firstly performed with antibodies direct against Dnmt1, Dnmt3a, and Dnmt3b. As illustrated in Figure 1B, we noted that Dnmt1 is the only Dnmt recruited to the −583/−488 region of HOXA2, the −526/−408 region of SLIT2, the −1180/−1091 region of TNFSF10, and the 1163/1270 region of LINE-1.
We then complemented the identification of the machinery of DNA methylation inheritance of LINE-1/TNFSF10 and HOXA2 and SLIT2 by performing re-ChIP experiments using antibodies directed against PCNA and UHRF1 or Sp1 because 1) the Dnmt1-PCNA-UHRF1 multiprotein complex has been described as being the major actor of DNA methylation inheritance; 2) Sp1 boxes are included in the considerate regions of the 3 genes of interest (according to the Patch program); and 3) the literature describes a Dnmt1/Sp1 complex as an epigenetic regulator of gene expression.5,7,11 We observed that the Dnmt1-PCNA-UHRF1 complex was corecruited to the −526/−408 region of SLIT2, the −1180/−1091 region of TNFSF10, and the 1163/1270 region of LINE-1 but not to the −583/−488 region of HOXA2 (Fig. 1B). Re-ChIP experiments also revealed that Dnmt1/Sp1 was corecruited to the −526/−408 region of SLIT2 and to the −583/−488 region of HOXA2. Together, these results indicate that Dnmt1-PCNA-UHRF1 governs the methylation of the 1163/1270 region of LINE-1 and the −1180/−1091 region of TNFSF10, while Dnmt1/Sp1 controls the methylation of the −583/−488 region of HOXA2, and Dnmt1-PCNA-UHRF1 and Dnmt1/Sp1 cooperate to methylate the −526/−408 region of SLIT2.
To reinforce the demonstration that Dnmt1-PCNA-UHRF1 governs the methylation of the −1180/−1091 region of TNFSF10 and the 1163/1270 region of LINE-1, we used the Astro40/Astro40-UP cells (referred to as A40) because of the abrogation of the formation of the Dnmt1-PCNA-UHRF1 interaction via the expression of the “UP” peptide as described previously (Fig. 1C).12 MeDIP analyses showed that the disruption of the Dnmt1-PCNA-UHRF1 interaction in Astro40-UP cells was associated with a decrease in the methylation level in the −1180/−1091 region of TNFSF10 and the 1163/1270 region of LINE-1 (Fig. 1D). As expected, we noted that the methylation level of the −583/−488 region of HOXA2 was unaffected by the disruption of the Dnmt1-PCNA-UHRF1 interaction in Astro40-UP cells. To confirm that Dnmt1/Sp1 controls the methylation in the −583/−488 region of HOXA2, we next decided to decrease the expression using Sp1 by an RNA interference approach. Western blot and immunoprecipitation showed that the partial decrease of expression of Sp1 was associated with the significant decrease of the Dnmt1/Sp1 interactions (Fig. 1E). MeDIP experiments showed that the methylation level in the −583/−488 region of HOXA2 decreased under these conditions, while the methylation level of the −1180/−1091 region of TNFSF10 and the 1163/1270 region of LINE-1 was not affected by the decrease of the Dnmt1/Sp1 interaction (Fig. 1F). Moreover, bisulfite sequencing analysis confirmed this point by showing that the CG dinucleotides included in the Sp1 boxes located between the −553/−441 region are unmethylated when U87 cells were treated with siRNA-Sp1 (Suppl. Fig. S1).
Dnmt1/Sp1 complements the DNA methylation inheritance of the SLIT2 gene initiated by Dnmt1-PCNA-UHRF1
We next decided to dissect the cooperation existing between Dnmt1-PCNA-UHRF1 and Dnmt1/Sp1 to methylate the −526/−408 region of SLIT2. For this purpose, we performed a kinetic ChIP assay on G0/G1 synchronized cells (72 hours after serum deprivation). Indeed, cell cycle phase analysis indicated that, after this treatment, 94.7% of the cells were in G0/G1, 2.6% in S, and 2.6% in G2/M (NucleoCounter NC-3000 Kit; Chemometec, Allerød, Denmark). As illustrated in Figure 2A, it appears that the coengagement of Dnmt1 and UHRF1 on the −526/−408 region of SLIT2 precedes the corecruitment of Dnmt1 and Sp1. Thus, these results could suggest that the Dnmt1/Sp1 engagement on the −526/−408 region of SLIT2 could complement the methylation of the −526/−408 region of SLIT2 initiated by the Dnmt1-PCNA-UHRF1 complex. To demonstrate this point, we evaluated the methylation status of the different CG dinucleotides included in the −526/−408 region of SLIT2 at t = 0 minutes, t = 65 minutes, and t = 105 minutes, that is, 15 minutes after the peak of the Dnmt1/UHRF1 and Dnmt1/Sp1 recruitments on the −526/−408 region of SLIT2. For this purpose, we performed MSP analyses: MSPa targeting the CG#1-2-3, MSPb targeting the CG#4, and MSPc targeting the CG#5-6 (Fig. 2B). We noted that the DNA methylation of CG#1-2-3 and CG#5-6 was complete at t = 65 minutes, while the methylation of CG#4 was not performed at t = 65 minutes and was almost complete at t = 105 minutes. According to the kinetics of recruitment of Dnmt1/UHRF1 and Dnmt1/Sp1, our data indicate that Dnmt1/UHRF1 performed the DNA methylation inheritance of CG#1-2-3 and CG#5-6, while the methylation of CG#4 is performed by Dnmt1/Sp1. The necessity of recruited Dnmt1/Sp1 to maintain the methylation of CG#4 is also supported by the fact that the disruption of the Dnmt1/Sp1 interaction (in U87 cells treated with siRNA-Sp1) promoted the loss of methylation of this CG dinucleotide, while the methylation of CG#1-2 is unaffected (Fig. 2C). Indeed, the observation of similar quantities of PCR product in the methylation sensitive restriction assay (MSRA) performed using digested and undigested DNA extracted from U87 cells treated or not with siRNA-Sp1 indicated that BstUI did not cleave DNA. With DNA methylation blocking the cleavage by BstUI, it appears that CG#1-2 was methylated in U87 cells, independently of the presence of Dnmt1/Sp1 interaction. A contrario, less PCR product was obtained when DNA was digested by DpnI and when U87 cells were treated with siRNA-Sp1. Thus, with methylation blocking the DpnI cleavage, these data indicate that CG#4 is mainly unmethylated in U87 cells treated with siRNA-Sp1. Taken together, these results strongly demonstrate the idea that the DNA methylation inheritance of the SLIT2 gene is performed via the cooperation between the Dnmt1-PCNA-UHRF1 and Dnmt1/Sp1 complexes.
Figure 2.
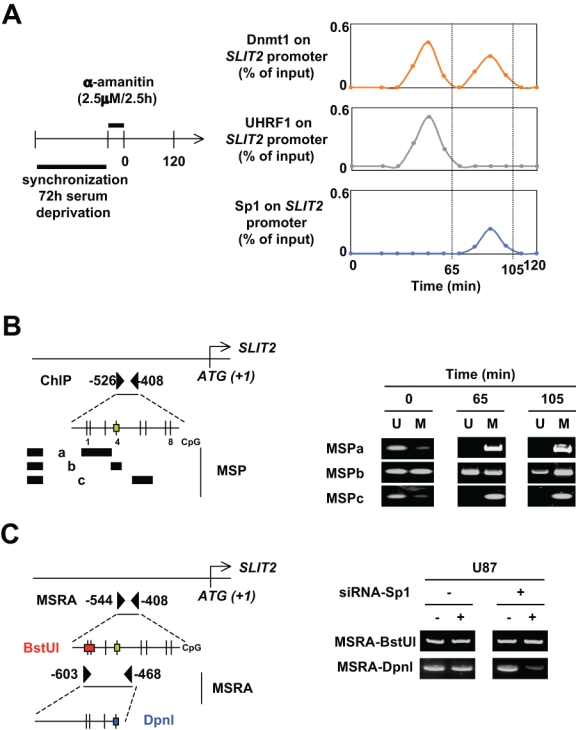
Description of the cooperation between Dnmt1/Sp1 and Dnmt1-PCNA-UHRF1 to maintain the DNA methylation. (A) The kinetic of ChIP experiments was realized at an indicated time after cell (U87) synchronization (72 hours of serum deprivation) and α-amanitin treatment. ChIP was performed with the EZ ChIP kit (Millipore, Molsheim, France) and with indicated antibodies. The amount of immunoprecipitated promoter was quantified by semiquantitative PCR and normalized to input. (B) Identification by methylation-specific PCR (MSP) of the methylation status of CG dinucleotides localized into the −526/−408 region of the SLIT2 gene in U87 cells. MSPa was designed to investigate the methylation status of CG dinucleotides #1, #2, and #3; MSPb was designed to investigate the methylation status of CG dinucleotide #4; and MSPc was designed to investigate the methylation status of CG dinucleotides #5 and #6. The green box indicates the presence of Sp1 binding site according to the use of the Patch program. U = unmethylated; M = methylated. Black boxes represent primers. (C) Impact of siRNA-induced decrease expression of Sp1 on the methylation status of specific CG dinucloetides of the SLIT2 genes. Methylation status of specific CG dicnuleotides was determined by methylation sensitive restriction assay (MSRA). DNA methylation blocks the cleavage of BstUI and DpnI when the CGCG and GATCg sequences are methylated, respectively. Thus, when DNA is initially unmethylated, no or weak DNA is amplified by PCR. Black triangles represent primers.
Comparison of the maintenance methyltransferase activity of the Dnmt1-PCNA-UHRF1 and Dnmt1/Sp1 complexes
Based on our previous results, we then hypothesized that the 2 sides can compose the machinery of maintenance DNA methylation: one unspecific in terms of DNA target and represented by the Dnmt1-PCNA-UHRF1 complex and the other specific/targeted symbolized by the Dnmt1/Sp1 complex, that is, by a complex including Dnmt1 and a transcription factor. To validate this idea, we focused on the Dnmt1/Sp1 interaction and compared the maintenance methyltransferase (mMTase) activity of Dnmt1 with the mMTase activities of Dnmt1-PCNA-UHRF1 and Dnmt1/Sp1 towards 8 hemimethylated double-stranded DNA probes. Dnmt–magnetic beads (DMB) assay revealed that the coincubation of PCNA and UHRF1 with Dnmt1 increased the 3H-incorporation (symbolizing the mMTase activity) with all probes used in our assay, while PCNA and/or UHRF1 are devoid of mMTase activity (Fig. 3A and Suppl. Fig. S2). The coincubation of Sp1 with Dnmt1 increased the 3H-incorporation in probe#4 only, that is, in the probe designed akin to the Sp1 box. The specificity of the mMTase activity was validated by the presence of antibody directed against Sp1, which blocked the binding of Sp1 to DNA and abrogated the peak of the 3H-incorporation on probe#4 (Fig. 3A). The fact that the 3H-incorporation was similar for all probes used in our tests suggested that the mMTase activity catalyzed by Dnmt1-PCNA-UHRF1 was unspecific in terms of targets. The absence of a specific site for Dnmt1-PCNA-UHRF1–induced inheritance of methylation is also confirmed by the absence of specificity to the consensus sequence designed from probes used in this study (Fig. 3B). All these data provide support for the idea that Dnmt1-PCNA-UHRF1 maintains DNA methylation in an unspecific manner in terms of DNA target, while the interaction of Dnmt1 with Sp1 promoted the targeted DNA methylation inheritance in DNA regions including Sp1 boxes.
Figure 3.
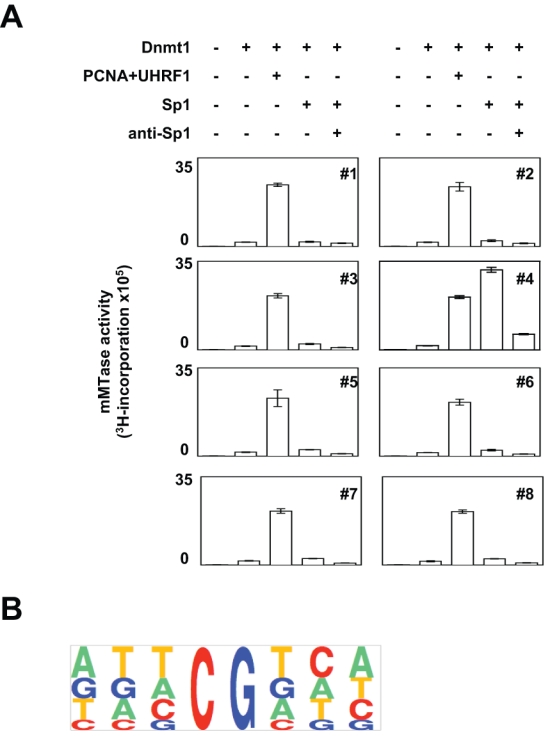
Comparison of maintenance methyltransferase activities (mMTase) of Dnmt1 in the presence of PCNA/UHRF1 or Sp1. (A) mMTase activities were assessed by Dnmt–magnetic beads (DMB) assays and the measurement of the 3H-incorporation in 8 double-stranded hemimethylated probes. (B) Consensus sequence for DNA methylation by Dnmt1-PCNA-UHRF1 obtained by using the Pictogram program, available at http://genes.mit.edu/pictogram.html.
Description of transcription factors interacting with Dnmt1
To identify transcription factors interacting with Dnmt1, we monitored the binding of recombinant Dnmt1 (Dnmt1R) in transcription factors arrays. In this acellular assay, Dnmt1R interacted with 58 transcription factors, that is, 56% of transcription factors spotted on the arrays (Fig. 4A and Suppl. Fig. S4). Thus, this analysis confirmed several interactions already described in the literature, such as the Dnmt1/Sp1 or Dnmt1/p53 interactions, and identified a majority of potential interactions not yet described to date. To cure that, we attempted to determine the existence of some interaction in cells. By performing proximity ligation in situ assay (P-LISA) via Olink/Duolink experiments and immunoprecipitation experiments, we confirmed that Dnmt1 interacts with Sp1, p53, C-EBPα, and YY1 but not with Sp4 or NFκB-p50 (Fig. 4 B and C). Moreover, the specificity of the interaction of Dnmt1R in this analysis is also reinforced by the fact that we noted homologies and differences between the transcription factor interacting with Dnmt1, Dnmt3a, and Dnmt3b.13 For example, we noted that ATF1 interacted with Dnmt1 but not with Dnmt3a and Dnmt3b. Pax9 is devoid of interaction in our system with Dnmt1, Dnmt3a, and Dnmt3b, while GATA1 interacted with all 3 Dnmts.
Figure 4.

Identification of transcription factors interacting with Dnmt1. (A) List of transcription factors interacting with Dnmt1 according to the results obtained from the hybridation of 5 µg of Dnmt1R on transcription factors arrays (Ozyme, Saint-Quentin-en-Yvelines, France). (B) Validation of the Dnmt1/transcription factors interactions in U251 glioma cells using the P-LISA–Olink/Duolink method. Nucleus/DNA was stained with DAPI (blue), and interactions were visualized by red fluorescence according to the P-LISA–Olink/Duolink method according to a previous report.13 (C) Validation of the Dnmt1/transcription factors interactions in U251 glioma cells using the immunoprecipitation method. Immunoprecipitations were realized using the Catch and Release v2.0 Reversible Immunoprecipitation System (Millipore, Molsheim, France). Nuclear extract is obtained using the EpiQuik Nuclear Extraction Kit I (Euromedex, Souffelweyersheim, France). (D) Model of DNA methylation inheritance machinery. Two sides compose the DNA methylation inheritance machinery: one untargeted, represented by the Dnmt1-PCNA-UHRF1 complex, and a second targeted, represented by the Dnmt1/transcription factor interactions. These 2 sides can work in solo or in duet to maintain the DNA methylation level of genes or DNA repeat elements.
Discussion
The understanding of molecular actors governing the mechanisms of maintenance of DNA methylation patterns is essential because this epigenetic regulation of DNA is implicated in the regulation of gene expression, silencing of parasitic DNA elements, genomic imprinting, and embryogenesis.14-16
Based on the description of molecular actors of the DNA methylation inheritance of CG dinucleotides included in certain regions of LINE-1, SLIT2, TNFSF10, and HOXA2, our data indicate that the machinery of DNA methylation inheritance is composed of at least 2 arms (Fig. 4D). The first arm is represented in our study, by the Dnmt1-PCNA-UHRF1 complex, and is unspecific in terms of DNA targeting. Indeed, no consensus sequence for Dnmt1-PCNA-UHRF1 was observed with the 8 probes used in our DMB assays. A contrario, the second arm represented in our paper by the Dnmt1/Sp1 complex is specific in terms of DNA targeting. Besides, this idea of specific DNA methylation inheritance has been already introduced by others via the description of some interactions between Dnmt1 and transcription factors.8-11 Concerning the Dnmt1/Sp1 interaction, we complemented our data by observing that only 12% of Sp1 is engaged with Dnmt1 in a multiprotein complex because we counted, on average, 75 dots of Sp1 per nucleus and 9 dots of Dnmt1/Sp1 per nucleus (Suppl. Fig. S5). These last results suggest that a small (but a considerable) quantity of Sp1 is engaged in the specific anchorage of Dnmt1 on DNA. Thus, the DNA methylation inheritance by the multiprotein complexes including Dnmt1 and transcription factors appears as an important alternative mechanism of DNA methylation, probably quite important to the selective anchoring of DNA methyltransferases 3A and 3B to nucleosomes containing methylated DNA or against the loading of Dnmt3a/b or Dnmt1 on DNA via its interactions with UHRF1 (also named Np95) or PCNA.2,5,7,17,18
Our results also indicate that these 2 sides can promote the DNA methylation inheritance by acting in “solo” or in “duet.” Interestingly, the fact that the 2 arms of DNA methylation inheritance can act in solo suggests the existence of an absence of a redundant system to maintain the level of DNA methylation of some genes or DNA repeat elements. This point can also suggest that the inhibition or the dysfunction of the machinery of DNA methylation inheritance acting in solo can be a source of passive DNA demethylation promoting the DNA hypomethylation of genes or DNA repeat elements. Indeed, we here underline that the methylation status of specific regions of LINE-1 (a DNA repeat element) and TNFSF10 is governed in solo by Dnmt1-PCNA-UHRF1. We have reported that this complex is disrupted during the initiation and the progression of gliomagenesis, and several articles have reported that the DNA repeat elements are generally hypomethylated in cancer (specially in glioma) and that the TNFSF10 gene is one of the genes hypomethylated in more than 20% of GBM.12,19 Thus, based on this work and on some of our previous results, it appears that the disruption of Dnmt1-PCNA-UHRF1 is a source of hypomethylation of certain regions of DNA repeat elements and genes and by ricochet of global DNA hypomethylation. The presence of Dnmt1/transcription factor interactions can be a crucial actor in the promotion of the persistence of local (hyper)methylation of certain genes in tumors. Indeed, in addition to the −583/−488 region of the HOXA2 gene, we also observed that the methylation of CG dinucleotides located in the −801/−693 region of the ZNF215 gene is dependent on the corecruitment of Dnmt1/Sp1 but not of Dnmt1-PCNA-UHRF1, Dnmt3a, Dnmt3b, or other Dnmt1/transcription factor interactions tested (Suppl. Fig. S6). Martinez et al. have reported that the ZNF215 gene is one of the genes hypermethylated in more than 20% of gliomas.19 Thus, the presence in gliomas of the Dnmt1/Sp1 interaction could constitute a rational molecular explanation to the (hyper)methylation of the ZNF215 gene. The description of the operating process in duet between the targeted and untargeted DNA methylation inheritance machineries observed for the SLIT2 gene, is, to date, the first of a cooperation between the 2 arms (targeted and untargeted) of DNA methylation inheritance machineries. Although implicating the Dnmt1-PCNA-UHRF1 complex on the DNA methylation inheritance of DNA repeat elements, Bostick et al. and Sharif et al. do not mention the existence of different sides of DNA methylation inheritance machineries.5,7 Inversely, the papers describing the targeted DNA methylation inheritance as a result of interactions between Dnmt1 and transcription factors, such as E2F1, p53, or Stat3, did not mention the existence of the cooperation of these mechanisms of DNA methylation inheritance with others.8-10 Several papers have only reported that different Dnmts can cooperate to methylate DNA. Thus, the cooperation of Dnmt1 and Dnmt3b has been already described.20
The use of kinetic ChIP experiments reinforces the existence of the cooperation between 2 arms of DNA methylation inheritance machineries and indicates that this cooperation, to maintain the methylation of the −526/−408 region of the SLIT2 gene, is sequential. Indeed, our data strongly demonstrated that the corecruitment of Dnmt1/Sp1 after the corecruitment of Dnmt1-PCNA-UHRF1 is indispensable to promote the complete methylation inheritance of the CG dinucleotides included within the −526/−408 region of the SLIT2 gene. The sequential recruitment of Dnmts on a gene to promote its methylation has been also reported in the context of cyclical DNA methylation.21,22 However, neither the presence of a DNA demethylation phase nor the sequential recruitment of Dnmt3a and Dnmt3b was observed in the present paper. Thus, our data underline that the sequential recruitment of Dnmt can also occur during an operating process of DNA methylation inheritance.
The description of a targeted DNA methylation inheritance machinery echoes the description and identification of the role played by the Dnmt3a/Dnmt3b and transcription factors interactions in the targeted de novo DNA methylation process.13 Moreover, the description of a targeted DNA methylation inheritance machinery gives birth to the idea to identify the potential partners of Dnmt1 and more specially the identity of transcription factors able to interact with Dnmt1. The use of transcription factor arrays allowed for the identification of 58 transcription factors able to interact with the recombinant Dnmt1 protein in an acellular system. As already described about the Dnmt3/transcription factor interactions, we noted that all members of a transcription factor family did not interact with Dnmt1: MEF2A and MEF2B interact with Dnmt1 but not MEF2C and MEF2D, for example. The fact that Dnmt1 could potentially interact with 58 transcription factors, that is, with 56% of transcription factors spotted on arrays, suggests that the role played by the multiprotein complexes including Dnmt1 and a transcription factor (to form the targeted DNA methylation inheritance machinery) is not negligible in the maintenance of the global level of DNA methylation. However, despite the presence of transcription factor binding sites on several DNA repeat elements, our ChIP analyses revealed the absence, in the majority of cases, of the corecruitment of Dnmt1 and transcription factors on the considered regions of several DNA repeat elements, while Dnmt1-PCNA-UHRF1 was corecruited on these regions (Suppl. Fig. S7). Thus, it seems that the methylation of the considered regions of several DNA repeat elements is not governed, in general, by the targeted DNA methylation inheritance machinery in spite of the fact that this machinery is composed by an important number of complexes including the direct interaction of Dnmt1 and transcription factors.
To summarize, the description of the Dnmt1/transcription factor interactions as a second actor in the mechanism of DNA methylation inheritance reinforces the knowledge of a molecular mechanism governing the DNA methylation by 1) introducing a new mechanism of DNA methylation inheritance working in parallel and/or in concert with the untargeted machinery of DNA methylation inheritance represented by the Dnmt1-PCNA-UHRF1 complex, and 2) echoing the description of the Dnmt3/transcription factor interactions as crucial players in target DNA (hyper)methylation.
Materials and Methods
Analyses of DNA methylation
DNA was extracted by using the QiaAmp DNA mini Kit (Qiagen, Courtaboeuf, France). Four micrograms of genomic DNA were treated with the EZ DNA methylation Gold kit (Proteigene, Zymo Research, Saint Marcel, France) according to the manufacturer’s instructions. Five microliters of bisulfite DNA were used for methylation-specific PCR analysis. Methylated DNA immunoprecipitation experiments were realized by using a MeDIP kit (Diagenode, Liège, Belgium).
Methylation sensitive restriction assay (MSRA) was performed by digesting 1 µg of genomic DNA with BstUI (Ozyme, Saint-Quentin-en-Yvelines, France) or DpnI (Fermentas, St. Rémy Les Chevreuse, France) for 16 hours. BstUI and DpnI digest only unmethylated DNA and leave intact methylated DNA. One microliter of digested and undigested DNA was then used for PCR.
ChIP and Re-ChIP experiments
Briefly, chromatin was purified from cells after cross-linking with 1% formaldehyde for 10 minutes at room temperature. ChIP and Re-ChIP assays were performed with the EZ ChIP kit (Millipore, Molsheim, France) with indicated antibodies and primers.
Measure of de novo methyltransferase activity via DMB assay
Dnmts–magnetic beads (DMB) assays were performed as described by Yokochi and Robertson.23 Briefly, a typical methylation reaction required 30 nM recombinant Dnmt1 protein (Methylation Ltd., Port Orange, FL), 125 nM DNA oligonucleotides (probes), and 900 nM tritium-labeled AdoMet (1 mCi/mL; Amersham Bioscience, GE Healthcare, Piscataway, NJ), in reaction buffer (50 mM Tris, pH 8.0, 5 mM EDTA, 10% glycerol, 0.5 mM phenylmethylsulfonyl fluoride). Coincubations of PCNA/UHRF1 or Sp1 were performed with 30 nM protein (Tebu-Bio, Le Perray-en-Yvelines, France). After incubation at 37°C for 1 hour, reactions were quenched with an equal volume of magnetic beads suspension and incubated for 15 minutes at room temperature. Next, the beads were magnetically isolated from the reaction mix, and tritium incorporation was measured by scintillation counting.
Western blot
In brief, proteins were size fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Proteins were transferred onto nitrocellulose or PVDF membrane. Saturation and blotting were realized by using the SNAP i.d Protein Detection System (Millipore). The detection of proteins was performed using ECL (Amersham Biosciences, GE Healthcare) and/or SuperSignal West Femto Maximum Sensitivity (Pierce, Thermo Fisher Scientific, Brebières, France) chemiluminescence reagents. Bands were quantified using Quantity One quantification software (Bio-Rad Laboratories, Hercules, CA).
Transcription factors arrays
Panomics TranSignal TF protein arrays I, II, and IV (Ozyme) were performed according to the manufacturer’s instructions and by using 5 µg of recombinant Dnmt1 (Methylation Ltd.). Blotting of membranes was realized by using the SNAP i.d Protein Detection System (Millipore) and antibodies directed against Dnmt1. The detection of proteins was performed using ECL (Amersham Biosciences, GE Healthcare). All the results shown in this article are representative of 3 independent experiments.
Supplementary Material
Acknowledgments
The authors thank Philippe Hulin for technical assistance (microPICell, Plateau technique d’imagerie cellulaire) (Centre de Recherche en Cancérologie Nantes-Angers, INSERM U892). Thanks also to Lisa Oliver for helpful discussion.
Footnotes
The author(s) declared no potential conflicts of interest with respect to the authorship and/or publication of this article.
This work was supported by grants from the Association pour la Recherche contre le Cancer (ARC#1020). E.H. was supported by a fellowship from INCa (Région Grand Ouest).
Supplementary material for this article is available on the Genes & Cancer website at http://ganc.sagepub.com/supplemental.
References
- 1. Fatemi M, Hermann A, Pradhan S, Jeltsch A. The activity of the murine DNA methyltransferase Dnmt1 is controlled by interaction of the catalytic domain with the N-terminal part of the enzyme leading to an allosteric activation of the enzyme after binding to methylated DNA. J Mol Biol 2001;309:1189-99 [DOI] [PubMed] [Google Scholar]
- 2. Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, Li BF. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science 1997;277:1996-2000 [DOI] [PubMed] [Google Scholar]
- 3. Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature 2008;455:818-21 [DOI] [PubMed] [Google Scholar]
- 4. Avvakumov G, Walker J, Xue S, et al. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature 2008;455:822-5 [DOI] [PubMed] [Google Scholar]
- 5. Bostick M, Kim J, Estève P, Clark A, Pradhan S, Jacobsen S. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 2007;27:2187-97 [DOI] [PubMed] [Google Scholar]
- 6. Hashimoto H, Horton J, Zhang X, Bostick M, Jacobsen S, Cheng X. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature 2008;455:826-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sharif J, Muto M, Takebayashi S, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 2007;450:908-12 [DOI] [PubMed] [Google Scholar]
- 8. Estève P, Chin H, Pradhan S. Human maintenance DNA (cytosine-5)-methyltransferase and p53 modulate expression of p53-repressed promoters. Proc Natl Acad Sci U S A 2005;102:1000-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet 2000;25:338-42 [DOI] [PubMed] [Google Scholar]
- 10. Zhang Q, Wang H, Marzec M, Raghunath P, Nagasawa T, Wasik M. STAT3- and DNA methyltransferase 1-mediated epigenetic silencing of SHP-1 tyrosine phosphatase tumor suppressor gene in malignant T lymphocytes. Proc Natl Acad Sci U S A 2005;102:6948-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Estève P, Chin H, Pradhan S. Molecular mechanisms of transactivation and doxorubicin-mediated repression of survivin gene in cancer cells. J Biol Chem 2007;282:2616-25 [DOI] [PubMed] [Google Scholar]
- 12. Hervouet E, Lalier L, Debien E. Tumor induction by disruption of the Dnmt1, PCNA and UHRF1 interactions. Nature Precedings 2008. http://hdl.handle.net/10101/npre.2008.2509.1
- 13. Hervouet E, Vallette F, Cartron P. Dnmt3/transcription factor interactions as crucial players in targeted DNA methylation. Epigenetics 2009;4:487-99 [DOI] [PubMed] [Google Scholar]
- 14. Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 1992;69:915-26 [DOI] [PubMed] [Google Scholar]
- 15. Li E, Beard C, Forster A, Bestor T, Jaenisch R. DNA methylation, genomic imprinting, and mammalian development. Cold Spring Harb Symp Quant Biol 1993;58:297-305 [DOI] [PubMed] [Google Scholar]
- 16. Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet 1998;20:116-7 [DOI] [PubMed] [Google Scholar]
- 17. Jeong S, Liang G, Sharma S, et al. Selective anchoring of DNA methyltransferases 3A and 3B to nucleosomes containing methylated DNA. Mol Cell Biol 2009;29:5366-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Meilinger D, Fellinger K, Bultmann S, et al. Np95 interacts with de novo DNA methyltransferases, Dnmt3a and Dnmt3b, and mediates epigenetic silencing of the viral CMV promoter in embryonic stem cells. EMBO Rep 2009;10:1259-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martinez R, Martin-Subero J, Rohde V, Kirsch M, Alaminos M, Fernandez A, et al. A microarray-based DNA methylation study of glioblastoma multiforme. Epigenetics 2009;4:255-64 [DOI] [PubMed] [Google Scholar]
- 20. Rhee I, Bachman KE, Park BH, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 2002;416:552-6 [DOI] [PubMed] [Google Scholar]
- 21. Kangaspeska S, Stride B, Métivier R, et al. Transient cyclical methylation of promoter DNA. Nature 2008;452:112-5 [DOI] [PubMed] [Google Scholar]
- 22. Métivier R, Gallais R, Tiffoche C, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature 2008;452:45-50 [DOI] [PubMed] [Google Scholar]
- 23. Yokochi T, Robertson KD. DMB (DNMT-magnetic beads) assay: measuring DNA methyltransferase activity in vitro. Methods Mol Biol 2004;287:285-96 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


