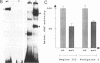Abstract
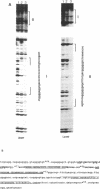
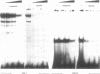
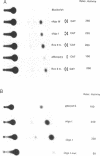
The replacement histone H1(0) of the H1 group, known to interact with general transcription factors, has been found associated with transcriptionally repressed chromatin. Transcription of the gene in F9 stem cells is low but can be stimulated by treating the cells with retinoic acid. Using mutant deletions, we now demonstrate that basal level transcription in F9 cells is mediated by an 80 bp DNA fragment, located 430 bp upstream of the TATA box, which does not include the retinoic acid responsive element (RARE) known to bind retinoic acid receptors and stimulate transcription from an heterologous promoter after retinoic acid treatment. By footprinting, DMS interference, site-directed mutagenesis and UV-cross linking techniques we demonstrate that at least two nuclear factors, with MW of 90,000 and 30,000, bind to the 80 bp fragment and that this binding is necessary for transcription. Furthermore, positioning of this fragment upstream of the HSV-tk gene promoter stimulates transcription 2-3 times over control values, far less than the activity observed for this fragment in the homologous promoter, indicating that full activity of this fragment requires sequences located in the proximal part of the promoter.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alonso A., Breuer B., Bouterfa H., Doenecke D. Early increase in histone H1(0) mRNA during differentiation of F9 cells to parietal endoderm. EMBO J. 1988 Oct;7(10):3003–3008. doi: 10.1002/j.1460-2075.1988.tb03163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A., Jorcano J. L., Beck E., Hovemann B., Schmidt T. Drosophila melanogaster U1 snRNA genes. J Mol Biol. 1984 Dec 25;180(4):825–836. doi: 10.1016/0022-2836(84)90259-6. [DOI] [PubMed] [Google Scholar]
- Aubert D., Garcia M., Benchaibi M., Poncet D., Chebloune Y., Verdier G., Nigon V., Samarut J., Mura C. V. Inhibition of proliferation of primary avian fibroblasts through expression of histone H5 depends on the degree of phosphorylation of the protein. J Cell Biol. 1991 May;113(3):497–506. doi: 10.1083/jcb.113.3.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer B., Fischer J., Alonso A. Cloning and characterization of the mouse histone H1(0) promoter region. Gene. 1989 Sep 30;81(2):307–314. doi: 10.1016/0378-1119(89)90191-1. [DOI] [PubMed] [Google Scholar]
- Croston G. E., Kerrigan L. A., Lira L. M., Marshak D. R., Kadonaga J. T. Sequence-specific antirepression of histone H1-mediated inhibition of basal RNA polymerase II transcription. Science. 1991 Feb 8;251(4994):643–649. doi: 10.1126/science.1899487. [DOI] [PubMed] [Google Scholar]
- Dailey L., Hanly S. M., Roeder R. G., Heintz N. Distinct transcription factors bind specifically to two regions of the human histone H4 promoter. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7241–7245. doi: 10.1073/pnas.83.19.7241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey L., Roberts S. B., Heintz N. Purification of the human histone H4 gene-specific transcription factors H4TF-1 and H4TF-2. Genes Dev. 1988 Dec;2(12B):1700–1712. doi: 10.1101/gad.2.12b.1700. [DOI] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992 Jan 16;355(6357):219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- Glass C. K., DiRenzo J., Kurokawa R., Han Z. H. Regulation of gene expression by retinoic acid receptors. DNA Cell Biol. 1991 Nov;10(9):623–638. doi: 10.1089/dna.1991.10.623. [DOI] [PubMed] [Google Scholar]
- Helms S. R., van Wijnen A. J., Kroeger P., Shiels A., Stewart C., Hirshman J., Stein J. L., Stein G. S. Identification of an enhancer-like element upstream from a cell cycle dependent human H4 histone gene. J Cell Physiol. 1987 Sep;132(3):552–558. doi: 10.1002/jcp.1041320319. [DOI] [PubMed] [Google Scholar]
- Kamakaka R. T., Thomas J. O. Chromatin structure of transcriptionally competent and repressed genes. EMBO J. 1990 Dec;9(12):3997–4006. doi: 10.1002/j.1460-2075.1990.tb07621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer E. J., Kistler W. S. Analysis of the promoter for the gene encoding the testis-specific histone H1t in a somatic cell line: evidence for cell-cycle regulation and modulation by distant upstream sequences. Gene. 1992 Jan 15;110(2):167–173. doi: 10.1016/0378-1119(92)90644-5. [DOI] [PubMed] [Google Scholar]
- Laybourn P. J., Kadonaga J. T. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science. 1991 Oct 11;254(5029):238–245. doi: 10.1126/science.254.5029.238. [DOI] [PubMed] [Google Scholar]
- Lee I. J., Tung L., Bumcrot D. A., Weinberg E. S. UHF-1, a factor required for maximal transcription of early and late sea urchin histone H4 genes: analysis of promoter-binding sites. Mol Cell Biol. 1991 Feb;11(2):1048–1061. doi: 10.1128/mcb.11.2.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckow B., Schütz G. CAT constructions with multiple unique restriction sites for the functional analysis of eukaryotic promoters and regulatory elements. Nucleic Acids Res. 1987 Jul 10;15(13):5490–5490. doi: 10.1093/nar/15.13.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelson E., Landsman D., Druckmann S., Bustin M. Immunofractionation of chromatin regions associated with histone H1o. Eur J Biochem. 1986 Oct 15;160(2):253–260. doi: 10.1111/j.1432-1033.1986.tb09964.x. [DOI] [PubMed] [Google Scholar]
- Nacheva G. A., Guschin D. Y., Preobrazhenskaya O. V., Karpov V. L., Ebralidse K. K., Mirzabekov A. D. Change in the pattern of histone binding to DNA upon transcriptional activation. Cell. 1989 Jul 14;58(1):27–36. doi: 10.1016/0092-8674(89)90399-1. [DOI] [PubMed] [Google Scholar]
- Osborne H. B., Chabanas A. Kinetics of histone H10 accumulation and commitment to differentiation in murine erythroleukemia cells. Exp Cell Res. 1984 Jun;152(2):449–458. doi: 10.1016/0014-4827(84)90646-3. [DOI] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988 Oct 20;335(6192):683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Roche J., Gorka C., Goeltz P., Lawrence J. J. Association of histone H1(0) with a gene repressed during liver development. Nature. 1985 Mar 14;314(6007):197–198. doi: 10.1038/314197a0. [DOI] [PubMed] [Google Scholar]
- Rousseau D., Khochbin S., Gorka C., Lawrence J. J. Regulation of histone H1(0) accumulation during induced differentiation of murine erythroleukemia cells. J Mol Biol. 1991 Jan 5;217(1):85–92. doi: 10.1016/0022-2836(91)90613-b. [DOI] [PubMed] [Google Scholar]
- Rousseau S., Renaud J., Ruiz-Carrillo A. Basal expression of the histone H5 gene is controlled by positive and negative cis-acting sequences. Nucleic Acids Res. 1989 Sep 25;17(18):7495–7511. doi: 10.1093/nar/17.18.7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J. M., Wiaderkiewicz R., Ruiz-Carrillo A. Histone H5 in the control of DNA synthesis and cell proliferation. Science. 1989 Jul 7;245(4913):68–71. doi: 10.1126/science.2740916. [DOI] [PubMed] [Google Scholar]
- Tung L., Lee I. J., Rice H. L., Weinberg E. S. Positive and negative transcriptional regulatory elements in the early H4 histone gene of the sea urchin, Strongylocentrotus purpuratus. Nucleic Acids Res. 1990 Dec 25;18(24):7339–7348. doi: 10.1093/nar/18.24.7339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster N., Jin J. R., Green S., Hollis M., Chambon P. The yeast UASG is a transcriptional enhancer in human HeLa cells in the presence of the GAL4 trans-activator. Cell. 1988 Jan 29;52(2):169–178. doi: 10.1016/0092-8674(88)90505-3. [DOI] [PubMed] [Google Scholar]