Abstract
Emerging data suggest that SSeCKS/Gravin/AKAP12 (“AKAP12”), originally identified as an autoantigen in cases of myasthenia gravis, controls multiple biological processes through its ability to scaffold key signaling proteins such as protein kinase (PK) C and A, calmodulin, cyclins, phosphoinositides, “long” β-1,4 galactosyltransferase (GalTase) isoform, Src, as well as the actin cytoskeleton in a spatiotemporal manner. Specialized functions attributed to AKAP12 include the suppression of cancer malignancy, especially aspects of metastatic progression, regulation of blood-brain and blood-retina barrier formation, and resensitization of β2-adrenergic pain receptors. Recent data identify a direct role for AKAP12 in cytokinesis completion, further suggesting a function as a negative regulator of cell senescence. The current review will discuss the emerging knowledge base of AKAP12-related biological roles and how the factors that affect AKAP12 expression or that interact with AKAP12 at the protein level control cancer progression and blood-tissue barrier formation.
Keywords: SSeCKS/Gravin/AKAP12, PKA, PKC, cyclin D, VEGF, metastasis, Src, Ras, prostate cancer, cell-cell barriers, G1→S progression, cytokinesis, cell motility, tumor invasiveness, neovascularization
Introduction
What was originally identified as a minor autoantigen correlating with poor prognosis in myasthenia gravis, and thus called Gravin,6,7 was subsequently shown to be orthologous to a rodent protein, SSeCKS (pronounced essex), a major PKC substrate and binding protein whose expression is suppressed in Src- and Ras-transformed cells.10 The demonstration that Gravin and SSeCKS also bind RII isoforms of PKA11 led to the renaming of these orthologs to A kinase anchoring protein (AKAP)-12. Importantly, AKAP12 orthologs have been identified in all vertebrate species analyzed including Danio and Fugu, but no credible orthologs have been found in insects, fungi, or worms. Whereas mouse and rat SSeCKS are >90% identical at the protein level, rodent SSeCKS is 83% identical to human AKAP12 over the N-terminal (approximately 1,000 a.a.) and 40% identical to the Xenopus ortholog, Xgl, over the N-terminal (800 a.a.).14 Vertebrates encode only a single copy of their AKAP12 ortholog, and whole genome sequencing has failed to identify homologous gene family members within a given species. In humans, AKAP12 maps to 6q24-25.2, a deletion hotspot in cases of advanced prostate, breast, and ovarian cancer.14 This region is syntenic with the telomeric end of mouse chromosome 10, and indeed, mouse AKAP12 maps to this region (www.ensembl.org).
In humans and rodents, 2 major AKAP12 transcripts, α and β, are expressed from independent promoters spaced roughly 35 Kb apart; yet, they splice to a common large exon that results in their sharing >93% amino acid identity.19 Whereas α and β AKAP12 protein isoforms are expressed ubiquitously in the embryo and the adult as 305- or 290-kDa products, respectively (290 or 280 kDa in rodents), there is high expression in some epithelial populations such as in the prostatic luminal cells, in some specialized cells such as glomerular mesangial cells and podocytes, and in Purkinje cells in the brain. The highest organ-specific expression is detected in the testes, ovary, brain, lung, prostate, and cardiac muscle. In contrast, a smaller testes-specific γAKAP12 isoform shares roughly 85% of the common C-terminal α and β residues.20 AKAP12-null mice, deficient in all 3 major isoforms, are viable, although they suffer from delayed fertility and prostatic hyperplasia.21 In contrast, the loss of AKAP12 causes selective gastrulation defects in Danio embryos,22 although it is unclear whether these are sufficient to prevent development of adult fish.
AKAP12 Scaffolding Functions
Scaffolding proteins are defined by their ability to bind signaling and/or cytoskeletal proteins in a spatiotemporal manner, typically involving scaffolding protein multimerization and the ability to associate with one or more specific cellular sites. Association with plasma and vesicle membrane sites by AKAP12 is facilitated by N-terminal myristylation, found only in the α isoform,1 and by 3 so-called polybasic effector domains13 that likely bind phosphoinositides and other phospholipids based on their homology to a membrane-binding domain in the MARCKS protein.23 Indeed, mouse AKAP12 was identified independently as a phosphatidylserine-binding protein,16,24 a function that maps to the effector domains and that facilitates association with plasma membranes and PKC.13 The 3 effector domains are required for AKAP12 to mediate cell flattening.25
Although AKAP12 is a weak F-actin cross-linking protein in vitro, it co-stains with F-actin, and this association can be antagonized by AKAP12 tyrosine phosphorylation via an EGF- or PDGF-induced, FAK-dependent pathway.4 Recent data suggest that AKAP12 may also associate with the actin or tubulin cytoskeleton, respectively, via direct or indirect association to profilin or dynein (below). Evidence that AKAP12 multimerizes comes from the Scatchard analysis of AKAP12/F-actin in vitro binding4 and from protein-protein interaction databases such as MINT (http://mint.bio.uniroma2.it/mint/).
AKAP12 binding partners have been identified by homology searching, yeast 2-hybrid analysis, and mass spectrometry of coimmunoprecipitating proteins. To date, protein binding domains have been identified that bind PKC, PKA-RII, cyclins, calmodulin, β1,4-galactosyltransferase-polypeptide 1 (B4galT1), Src, β2-adrenergic receptor,26 F-actin, and cAMP-specific 3′,5′-cyclic phosphodiesterase 4D27 (Fig. 1). Recent large-scale protein-protein interaction screens identified associations with profilin (PFN1), pyruvate kinase M2 fragment (PKM2), premature ovarian failure protein (POF1B), peptidyl prolyl isomerase (PPIA; cyclophilin-A), C1 segment protein (SC-9C5.12), small proline-rich protein II (SPRR2E), and transferring (TF) and valosin-containing protein (VCP), although specific binding sites on AKAP12 have not yet been characterized (http://www.mitocheck.org/cgi-bin/mtc?query=MCG_0000007). Association with SC-9C5.12, which binds dynein, would facilitate interaction between AKAP12 and the tubulin cytoskeleton, whereas association with VCP, which binds Cdc42, might facilitate AKAP12 association with the leading edge of motile cells. Finally, the ability of AKAP12 to associate with and control the cytokinesis apparatus might be facilitated by its interaction with transferrin, which in turn binds a MIS12/Bub1 complex. Importantly, there is evidence that phosphorylation of AKAP12 by mitogen-induced kinases antagonizes AKAP12 scaffolding activity, whereas phosphorylation by kinases associated with differentiation, such as PKA, might enhance AKAP12 scaffolding activity. Both the serine and tyrosine phosphorylation of AKAP12 increases during G1→S progression and then decreases precipitously in G2/M,4,28 even though there is only a small decrease in total AKAP12 protein levels during these phases.28 Thus, prephosphorylation of AKAP12 by PKC decreases its ability to bind PKC, calmodulin, and cyclins in vitro and in cellulo.8,15 In contrast, PKA can phosphorylate AKAP12 and enhance association with a Src SH3 domain, thereby facilitating β2-adrenergic receptor complex resensitization.29-32 Whereas AKAP12 binding to PKC inhibits its kinase activity1,2,11 and sequesters PKC isozymes at plasma membrane sites, AKAP12 regulates cyclin D, in part, by sequestering cyclin D pools in the cytoplasm.8 Interestingly, SSeCKS phosphorylation by PKC in neuronal cells causes a decrease in AKAP12-PKC scaffolding but no change in AKAP12-PKA binding33 or agonist-induced PKA activation,11 suggesting that AKAP12 controls the well-recognized, mutually exclusive activation cross-talk between PKC and PKA. The differential PKC-PKA control likely relates to overlap between PKC binding and phosphorylation sites mapping to the N-terminus of AKAP12, in contrast to the PKA-RII binding site mapping to a C-terminal region lacking phosphorylation sites (Fig. 1). Indeed, the C-terminal AKAP site is required for AKAP12 to target PKA-RII to the cell periphery,34 and PKC activation by phorbol esters causes the AKAP12/PKA-RII complex to translocate to the perinucleus.13,33 Lastly, in round spermatids, AKAP12 co-stains with calcineurin.35
Figure 1.

AKAP12 scaffolding domains. Known PKC phosphorylation sites1,2 are identified at top as Ser507/515, 599, and 748 (based on the rat protein sequence: NP_476444.2) as well as an implied EGF- or PDGF-induced, FAK-dependent tyrosine phosphorylation site at Y835.4 A Src SH3 binding site (P15xxP).5 AKAP12 also encodes at least 4 nuclear localization signals (NLS) homologous to those from SV40 Tag, a cyclin binding domain containing 2 CY motifs,8 an acidic/basic region (KR- and E/D-rich), an A kinase anchoring protein (AKAP) domain for PKA RII isoform binding,11 a nuclear exclusion domain,12 3 polybasic domains involved in membrane association,13 at least 4 calmodulin binding domains of the so-called 1-5-10 motif,15 a phosphatidylserine (PS) binding domain16 that facilitates association with PKC isoforms,17 2 distinct binding domains for β1,4-galactosyltransferase (B4GALT1),18 and an F-actin binding domain.4
AKAP12 protein levels increase >10-fold during contact inhibition,8,28,36 and the relative low basal and mitogen-inducible phosphorylation of AKAP12 under this condition4,28 suggests that its scaffolding activity for signaling proteins, and thus its activity as a negative mitogenic regulator (below), is highest during contact inhibition.
AKAP12 Functions in Normal Cell Cycle: Regulator of Mitogenesis and Cytokinesis
Several lines of evidence suggest that AKAP12 functions in normal cells to 1) prevent inappropriate cell cycle progression and 2) facilitate completion of cytokinesis and mitosis. Early attempts to constitutively express AKAP1210 led to the finding that its overexpression in untransformed fibroblasts8 and epithelial cells36 caused G1 arrest due to the suppression of serum-induced, MEK/ERK-dependent cyclin D expression.8 Interestingly, AKAP12 encodes 2 so-called cyclin binding (CY) motifs, which can also cause G1 arrest by sequestering pools of cyclin D in the cytoplasm during contact-inhibited growth.8 PKC-induced phosphorylation of AKAP12 at sites within the CY domains antagonizes AKAP12-cyclin binding,15 resulting in the nuclear translocation of cyclin D even in contact-inhibited cultures, followed by one additional round of DNA replication.8 Recent data indicate that compared to WT mouse embryo fibroblasts (MEF), AKAP12-null MEF have higher basal- and serum-induced MEK/ERK activation, rates of serum-induced proliferation, saturation densities, and autocrine growth associated with increased basal cyclin D levels (Akakura S, Nochajski P, Gao L, Sotomayor P, Matsui S, Gelman IH. Rb-dependent cellular senescence, multinucleation and susceptibility to oncogenic transformation through PKC scaffolding by SSeCKS/AKAP12. Cell Cycle: in press.) (Gao and Gelman, unpublished data).
A pool of AKAP12 may help facilitate completion of cytokinesis during normal cell cycle progression. Recent data indicate that although AKAP12-null MEF initially proliferate faster than WT MEF, they suffer from premature senescence, exhibiting binucleation or polyploidy. This senescence is Rb but not p53 dependent, and it correlates with increased PKCα and δ activity due to the loss of AKAP12 scaffolding. Activated PKCδ causes the downregulation of Lats1, a mitotic exit network kinase required for completion of cytokinesis.37 In parallel, higher PKCα kinase levels induce p16Ink4a expression and Rb activation by increasing MEK kinase activity, which in turn downregulates expression of Id1, a negative regulator of p16Ink4a expression (Akakura S, Nochajski P, Gao L, Sotomayor P, Matsui S, Gelman IH. Rb-dependent cellular senescence, multinucleation and susceptibility to oncogenic transformation through PKC scaffolding by SSeCKS/AKAP12. Cell Cycle: in press.). Indeed, higher AKAP12 protein levels are found in senescent human diploid fibroblasts and in aging rat and human keratinocytes,38 and the siRNA-mediated knockdown of AKAP12 is sufficient to induce senescence associated with binucleation (Akakura S, Nochajski P, Gao L, Sotomayor P, Matsui S, Gelman IH. Rb-dependent cellular senescence, multinucleation and susceptibility to oncogenic transformation through PKC scaffolding by SSeCKS/AKAP12. Cell Cycle: in press.). Moreover, AKAP12 may directly regulate cytokinesis based on the demonstration of a pool staining in the anaphase abscission furrow,39,40 and that loss of AKAP12 leads to multinucleation.39,41 The furrow contains the actin-myosin ring whose PKC-RhoGTPase–dependent contraction helps complete daughter chromosome separation,42 and thus, it is conceivable that AKAP12 might regulate cytokinesis via its ability to scaffold PKC and actin and to attenuate RhoGTPase activity.43 Strengthening this notion, a recent systems biology analysis predicts the existence of a mitotic protein complex containing AKAP12, phosphodiesterase 4D, ATM, Polo-like kinase 4, APC, dynein, and profilin (complex MCC_0000069; http://www.mitocheck.org/cgi-bin/mtc?query=MCG_0000007).
AKAP12 and Cell Motility
Overexpression and knockdown experiments indicate that AKAP12 likely does not control generic cell motility but that it might attenuate specialized motility in response to specific chemoattractants. For example, AKAP12-re-expressing MAT-LyLu prostate cancer cells have normal short- and long-term motility in monolayer wound-healing assays.36 In contrast, AKAP12 inhibits chemotaxis in these same cells due to the inhibition of a PKC-Raf/MEK/ERK pathway,44 and similarly, AKAP12-null MEF display higher chemotaxis towards serum or PDGF-BB than passage-matched WT MEF (Hyun-Kyung Ko and Irwin Gelman, unpublished data). A recent study by Busch et al.45 strongly suggests that upregulated AKAP12 facilitates HGF-induced, c-Met–dependent cell motility through the upregulation of PKA activity and PKA-induced genes, presumably through AKAP12’s scaffolding function. Taken together, these data strengthen the notion that AKAP12 facilitates the differential activation of PKC and PKA in processes such as cell motility.
Consistent with AKAP12 being a cytoskeletal protein that can regulate the remodeling of the actin cytoskeleton,46 either AKAP12 overexpression or deficiency in untransformed cells causes cell flattening3,8,9,46 (Fig. 2). However, whereas overexpression antagonizes stress fiber formation and leads to the production of long filamentous projections, AKAP12 deficiency results in the production of thickened, longitudinal stress fibers and increased numbers of focal adhesion plaques. Indeed, in the absence of FAK, AKAP12 co-stains with stress fibers, whereas in EGF- or PDGF-stimulated FAK+/+ cells, AKAP12 stains at the plasma membrane and along a cortical cytoskeleton.4 It is reasonable to speculate that the dynamic exchange of AKAP12 between filamentous and cortical cytoskeletal networks plays a role in controlling specialized cell motility. Taken with the ability of AKAP12 to normalize actin cytoskeletal structures in cancer cells (below), these data suggest that AKAP12 functions as a morphostat, that is, a protein that regulates oncogenic and specialized motility pathways through the normalization of cytoskeletal architecture.
Figure 2.

Control of cell shape and cytoskeletal dynamics by AKAP12. The downregulation of AKAP12 in v-Src–transformed NIH3T3 murine fibroblasts correlates with a transition from the polygonal morphology of parental NIH3T3 (top panel, middle) to a so-called fusiform morphology (top panel, left). In contrast, overexpression of AKAP12 via a Tet-OFF vector system3 causes cell flattening, the thinning of stress fibers, and the production of long filamentous (F) projections (top panel, right). The staining intensities in the top panel reflect relative levels of AKAP12. AKAP12 loss in MEF (bottom left) or in stellate mesangial cells due to treatment with antisense (ASN) AKAP12 oligonucleotides (bottom right) results in cell flattening marked by thickened, longitudinal stress fibers (“F-actin”) and increased numbers of focal adhesion plaques (“vinculin”).9
AKAP12 as a Tumor/Metastasis Suppressor
Evidence that AKAP12 might be a tumor and/or metastasis suppressor comes from its mapping to 6q24-25.2, a known deletion hotspot in many cancers,36 its downregulation by specific oncogenes and in cancer cell lines and human cancer tissues compared to normal controls, its upregulation by treatments that suppress oncogenic growth, by direct demonstrations showing that re-expression suppresses in vitro and in vivo oncogenic growth, especially metastasis formation, or that its loss produces a tumor- or metastasis-prone condition.
An early identification of AKAP12 was based on its severe downregulation in Src-transformed fibroblasts1 and the finding that its re-expression could suppress in vitro parameters of oncogenic growth.3,36 Later studies showed 5- to 15-fold AKAP12 downregulation in fibroblasts and epithelial cells transformed by oncogenic versions of Ras,1,47 Myc,1,48-50 Jun,51 Fos and Dnmt1,47 and Wnt I,52 but not by oncogenic Raf, Mos, or Neu.10 This indicates that AKAP12 loss is not a generic effect of oncogenic transformation but due to specific oncogenic pathways. Moreover, AKAP12 levels track with transformation status; namely, they are downregulated following Ras transformation, they rise to normal levels in revertants, and then they are downregulated in retransformants, correlating especially with anchorage-independent growth.3 Whereas WT MEF require two or more oncogenes for efficient transformation,53 AKAP12-null MEF can be transformed efficiently by single oncogenes such as v-Src or Ras, strengthening the idea that AKAP12 functions in cell culture systems as a tumor suppressor gene (Akakura S, Nochajski P, Gao L, Sotomayor P, Matsui S, Gelman IH. Rb-dependent cellular senescence, multinucleation and susceptibility to oncogenic transformation through PKC scaffolding by SSeCKS/AKAP12. Cell Cycle: in press.). A recent study showed that downregulation of the αAKAP12 promoter by Src requires a short proximal sequence that binds USF1, Sp1, Sp3, HDAC1,54 plus a form of TFII-I that is tyrosine phosphorylated by activated Src.55
AKAP12 expression is also induced by genes or treatments that antagonize oncogenic transformation. For example, AKAP12 is upregulated by re-expression of the p53 tumor suppressor,56 Smad-4–dependent, TGF-β–induced cell cycle arrest,57-59 by antagonizing STAT3β-dependent oncogenic signaling,60 and by differentiating agents that suppress tumor growth such as vitamin D3 analogs61,62 and retinoids63,64 (Sunamoto and Gelman, unpublished observations; Streb JW, Long X, Lee T-H, Sun Q, Kitchen CM, Georger MA, Metlay LA, Blaner WS, Carr DW, Gelman IH, Miano JM. Retinoid-induced expression of an immediate early tumor suppressor gene in vascular smooth muscle cells in vitro and in vivo. Submitted for publication.). C/EBPα-induced growth arrest in BCR-ABL–positive KCL22 cells results in a 15-fold increase in AKAP12 expression.65 AKAP12 expression is induced by androgen levels that cause growth arrest in untransformed human prostate epithelial cells and in androgen-dependent LNCaP cells,66,67 whereas selection of androgen-independent LNCaP cells correlates with AKAP12 downregulation.68 AKAP12 expression can be stimulated by the forced expression of the GATA-3 transcription factor, which normally facilitates breast development but which is downregulated during progression to malignancy.69
Re-expression of AKAP12 suppresses many parameters of oncogenic growth in cell culture assays. For example, AKAP12 re-expression suppressed v-Src-, Jun- or Ras-induced anchorage- and growth factor–independent growth, focus formation, Matrigel invasiveness, and podosome formation, while re-establishing normalized cytoskeletal structures such as stress fibers and mature focal adhesion plaques.28,36,43,51 AKAP12 inhibits neither Src autophosphorylation activity during this suppression3,43 nor Src’s ability to phosphorylate signaling proteins such as Shc or invasion-related proteins such as Tks5.43,44 However, AKAP12 does inhibit invasiveness and chemotaxis through disengaging Src from activating downstream PKC-Raf/MEK/ERK pathways controlling cytoskeletal remodeling, podosome formation, and MMP-2/9 expression.44 Choi et al.70 were also able to reverse several in vitro oncogenic growth parameters of AGS gastric cancer cells by the re-expression of human AKAP12.
AKAP12 can be defined as a metastasis suppressor based on the currently accepted definition of a gene that fails to significantly affect primary tumor growth but which suppresses one or more parameters of metastasis.71 Thus, the tetracycline-regulated re-expression of AKAP12 in MAT-LyLu rat prostate cancer cells causes a small decrease in primary subcutaneous tumors yet severely suppresses the formation of macroscopic lung metastases.36 Interestingly, AKAP12 re-expression did not alter lung colonization activity, but it did result in relatively avascular micrometastases.72 The finding that astrocyte-encoded AKAP12 can suppress JNK-dependent VEGF expression and angiogenesis during the establishment of the blood-brain barrier right after birth57 led to our follow-up study, which showed that AKAP12 could suppress the expression of VEGF and other proangiogenic genes in tumor and mural cells and that the forced expression of VEGF-A (165 or 121 isoforms) in the AKAP12-re-expressing MAT-LyLu cells could partially rescue formation of lung macrometastases.72 AKAP12’s role as a metastasis suppressor is strengthened by multiple gene expression array studies (www.oncomine.org) showing decreased levels of AKAP12 transcripts in metastases versus primary-site colon, endocrine, and breast cancers (Fig. 3) or versus primary prostate cancers (Ray M, Zheng S, Nochajski P, Davis W, Mohler JL, Marshall J, Gelman IH. Loss of SSeCKS/Gravin/AKAP12 expression in prostate cancer tissues correlates with disease progression. Submitted for publication.) or in predicting metastasis of bladder cancer after 1 year (Fig. 3).
Figure 3.

Decreased AKAP12 expression in metastatic progression. Oncomine study data showing statistically lower AKAP12 RNA expression levels in metastases compared to primary-site colon, endocrine, or breast cancers, as well as lower AKAP12 RNA levels correlating with incidence of metastasis formation at 1 year after primary diagnosis (left).
Downregulation of AKAP12 in Human Cancers
A large corpus of data indicates that AKAP12 expression is downregulated in many solid and liquid cancers types, either associated with gene deletion or epigenetic downregulation due to promoter hypermethylation or changes in chromatinization. For example, AKAP12 staining is consistently lost in prostate cancers with Gleason sum >636 (Ray M, Zheng S, Nochajski P, Davis W, Mohler JL, Marshall J, Gelman IH. Loss of SSeCKS/Gravin/AKAP12 expression in prostate cancer tissues correlates with disease progression. Submitted for publication.).
Roughly 40% of the Gleason sum 7 to 10 lesions that were deficient AKAP12 staining also exhibited gene deletion, as determined by laser capture microdissection followed by PCR analysis (AKAP12 and GAPDH control primers) (Gelman, unpublished data). Reports showing cancer-related loss of AKAP12 expression include human and rat prostate cancer cell lines,36 pulmonary adenocarcinomas,73 leiomymoma,74 chronic and acute myeloid leukemias and myelodysplastic syndromes,75-77 multiple myelomas,78 papillary thyroid carcinoma,79 pediatric acute lymphoblastic leukemia,80 gastric cancer,70 non-small cell lung carcinoma,81 osteosarcomas,82,83 melanomas,84,85 retinoblastomas,86 colon cancer,87,88 fibrosarcomas,89 and squamous cell lung carcinoma.90,91 In all these cases, AKAP12 transcript levels are suppressed 5- to 15-fold compared to matched controls. Many microarray-based studies demonstrate 3- to 10-fold reduction in relative AKAP12 mRNA levels in breast, prostate, lung, and ovarian cancers and in gliomas,77,92-100 and others have been cited in Entrez GEO (Gene Expression Omnibus; www.ncbi.nlm.nih.gov/geo) or Oncomine (www.oncomine.org) linking AKAP12 expression with tumor suppression. In silico analyses at the Cancer Genome Anatomy Project SAGE (Serial Analysis of Gene Expression) site (http://cgap.nci.nih.gov/SAGE) identify AKAP12 downregulation in human thyroid, lung, and liver cancer tissue and in human ovarian cancer cell lines. The majority of cancers suffering from epigenetic AKAP12 silencing seem to be regulated via promoter hypermethylation,70,78,85,88,99,100 although instances exist where AKAP12 silencing is through changes in histone acetylation.78 Indeed, the silencing of AKAP12 via promoter hypermethylation101,102 has been shown to function as a neoplastic progression biomarker in Barrett’s esophagus.103
In contrast to these examples of AKAP12 downregulation in cancer, AKAP12 expression is moderately induced in several cancer cell lines and human cancers, likely reflecting the varying biologies and/or activated signaling pathways of these cancer types. These include chronic myelogenous leukemia,104 the pancreatic cancer cell lines PANC-1 and SU8686 versus normal mucosa,105 muscle-invasive versus superficial bladder cancer,106 giant cell granuloma of the jaw,107 and high-grade follicular lymphoma.108
Control of Neovascularization and Barriergenesis
AKAP12 can attenuate neovascularization and barrier formation, such as the blood-brain (BBB) or blood-retina (BRB) barriers, through the downregulation of proangiogenic genes such as those encoding HIF-1α or VEGF. The first such study identified AKAP12 upregulation during normoxic transition of the mouse embryo right at birth and showed that AKAP12 was responsible for suppressing angiogenesis through a JNK-dependent downregulation of VEGF and for inducing formation of the BBB after birth.57 Indeed, chemotherapies that paradoxically induce angiogenesis result in increased VEGF and decreased AKAP12 mRNA levels,109 strengthening the idea that AKAP12 is an antagonist of VEGF. Our data showing that AKAP12 re-expression leads to the suppression of neovascularization at metastatic sites through the downregulation of VEGF72 further strengthens this concept. AKAP12 seems to strengthen BRB formation by upregulating tight junction proteins and by decreasing VEGF expression through the enhancement of HIF-1α/VHL binding.86 Interestingly, the studies above show that AKAP12 can suppress BBB, BRB, and metastasis-related angiogenesis both when expressed ectopically in barrier-forming endothelial cells or through the induction of secreted factors after AKAP12 expression in astrocytes or stromal cells. Moreover, the ability of AKAP12 to secrete antiangiogenic and probarriergenic factors requires the suppression of PKC and RhoGTPase/Rho kinase activity,110 presumably through a direct scaffolding of PKC by AKAP12. Consistent with the notion that PKC-mediated phosphorylation of AKAP12 antagonizes its scaffolding activity, You et al. showed that the ability of IL-17F, IL-1β, or TNFα to induce permeability of microvascular endothelial layers correlated with the activation of PKC and the hyperphosphorylation/downregulation of AKAP12.111,112
Conclusion
AKAP12 likely regulates cell cycle progression, specialized cell motility, and angiogenesis by controlling actin cytoskeletal remodeling and signaling pathways through its multiple scaffolding domains (Fig. 4). (Similar functions are thought to be involved with AKAP12 regulation of β2-adrenergic receptor resensitization.32,113) Current data indicate that AKAP12 can regulate Raf/MEK/ERK pathways through direct scaffolding functioning upstream of ERK, JNK, and PKC signaling. Suppression of oncogenic proliferation, invasiveness, chemotaxis, neovascularization, and cell senescence all likely involve attenuation of PKC activation through AKAP12’s spatiotemporal scaffolding functions. Some of these may also involve the downstream attenuation of Raf/MEK/ERK signaling, such as in specialized motility or MMP, HIF-1α or VEGF expression. Other AKAP12-regulated parameters, such as podosome formation, are downstream of PKC and RhoGTPases but independent of MEK, and yet, others involve PKC- and MEK-independent JNK pathways, such as VEGF and MMP secretion.
Figure 4.
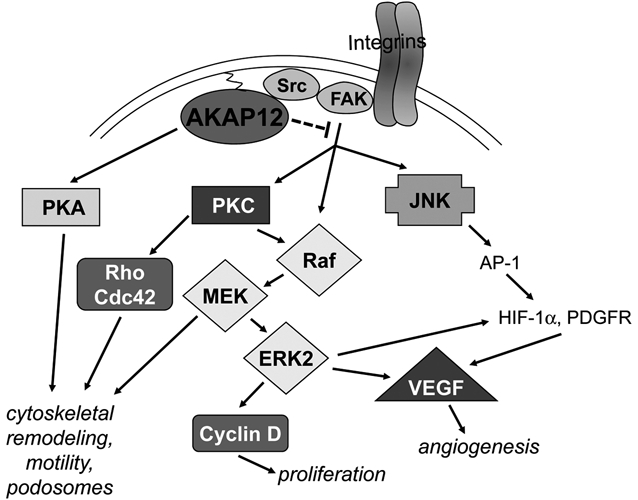
Pathways controlled by AKAP12 in cancer. AKAP12 suppresses cancer progression by disengaging adhesion- and growth factor–induced activation of Src-FAK complexes from transducing Raf/MEK/ERK- and JNK-mediated proliferation, angiogenesis, and cytoskeletal remodeling signals, possibly through a direct binding of AKAP12 to the Src-SH3 domain, resulting in the altering of Src-FAK signaling complexes. AKAP12 also directly inhibits PKC activation by a direct scaffolding function. In contrast, AKAP12 scaffolding of PKA facilitates cAMP-induced cytoskeletal remodeling.
How these pathways are regulated by AKAP12 in the context of metastasis remains unclear. However, the growing evidence implicating AKAP12 in the regulation of specialized biologies such as chemotaxis, invasiveness, and neovascularization at metastatic sites, possibly through antagonizing required Src-signaling pathways,114,115 strongly suggests that AKAP12 regulatory functions are heavily influenced by specific microenvironmental conditions. Thus, although AKAP12 expression is high in many tissues, the loss of its expression in transgenic mice results in hyperplasia and/or dysplasia in only some sites.21 Similarly, this may be why AKAP12 downregulation plays a direct role in metastasis suppression in specific tumor sites such as the prostate or pancreas.116
Acknowledgments
The author thanks Andrei Bakin for critical reading of this article. This paper is dedicated to the memory of my postdoctoral mentor, Dr. Hidesaburo Hanafusa, who taught me that scientific advancement required a keen sense of observation, faithfulness to detail, dogged perseverance, and the humility to withstand strong critical review from the outside as well as even stronger criticism from within.
Footnotes
The author(s) declared no potential conflicts of interest with respect to the authorship and/or publication of this article.
This work was supported by the National institutes of Health [grant number CA94108]; Department of Defense [grant numbers PC074228, PC061246, BC086529]; and the Roswell Park Alliance Foundation.
References
- 1. Ln X, Tombler E, Nelson PJ, Ross M, Gelman IH. A novel src- and ras-suppressed protein kinase C substrate associated with cytoskeletal architecture. J Biol Chem. 1996;271:28, 430,-28, 438 [DOI] [PubMed] [Google Scholar]
- 2. Chapline C, Cottom J, Tobin H, Hulmes J, Crabb J, Jaken S. A major, transformation-sensitive PKC-binding protein is also a PKC substrate involved in cytoskeletal remodeling. J Biol Chem. 1998;273:19482-9 [DOI] [PubMed] [Google Scholar]
- 3. Lin X, Gelman IH. Re-expression of the major protein kinase C substrate, SSeCKS, suppresses v-src-induced morphological transformation and tumorigenesis. Cancer Res. 1997;57:2304-12 [PubMed] [Google Scholar]
- 4. Xia W, Gelman IH. Mitogen- and FAK-regulated tyrosine phosphorylation of the SSeCKS scaffolding protein modulates its actin-binding properties. Exp Cell Res. 2002;277:139-51 [DOI] [PubMed] [Google Scholar]
- 5. Tao J, Wang HY, Malbon C. Src docks to AKAP Gravin, regulating beta-adrenergic receptor resensitization and recycling. J Biol Chem. 2007;282:6597-608 [DOI] [PubMed] [Google Scholar]
- 6. Gordon T, Grove B, Loftus JC, et al. Molecular cloning and prelimnary characteriztion of a novel cytoplasmic antigen recognized by myasthenia gravis sera. J Clin Invest. 1992;90:992-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sasaki H, Kunimatsu M, Funii Y, et al. Autoantibody to gravin is expressed more strongly in younger and nonthymomatous patients with myasthenia gravis. Surgery Today. 2001;31:1036-7 [DOI] [PubMed] [Google Scholar]
- 8. Lin X, Nelson P, Gelman IH. Regulation of G–>S progression by the SSeCKS tumor suppressor: control of cyclin D expression and cellular compartmentalization. Mol Cell Biol. 2000;20:7259-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nelson PJ, Moissoglu K, Vargas JJ, Klotman PE, Gelman IH. Involvement of the protein kinase C substrate, SSeCKS, in the actin-based stellate morphology of mesangial cells. J Cell Sci. 1999;112:361-70 [DOI] [PubMed] [Google Scholar]
- 10. Lin X, Nelson PJ, Frankfort B, Tombler E, Johnson R, Gelman IH. Isolation and characterization of a novel mitogenic regulatory gene, 322, which is transcriptionally suppressed in cells transformed by src and ras. Mol Cell Biol. 1995;15:2754-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nauert J, Klauck T, Langeberg LK, Scott JD. Gravin, an autoantigen recognized by serum from myasthenia gravis patients, is a kinase scaffolding protein. Curr Biol. 1997;7:52-62 [DOI] [PubMed] [Google Scholar]
- 12. Streb JW, Miano JM. Cross-species sequence analysis reveals multiple charged residue-rich domains that regulate nuclear/cytoplasmic partitioning and membrane localization of a kinase anchoring protein 12 (SSeCKS/Gravin). J Biol Chem. 2005;280:28007-14 [DOI] [PubMed] [Google Scholar]
- 13. Yan X, Walkiewicz M, Carlson J, Leiphon L, Grove B. Gravin dynamics regulates the subcellular distribution of PKA. Exp Cell Res. 2009;315:1247-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gelman IH. The role of the SSeCKS/Gravin/AKAP12 scaffolding proteins in the spaciotemporal control of signaling pathways in oncogenesis and development. Front Biosci. 2002;7:d1782-97 [DOI] [PubMed] [Google Scholar]
- 15. Lin X, Gelman IH. Calmodulin and cyclin D anchoring sites on the Src-suppressed C kinase substrate, SSeCKS. Biochem Biophys Res Commun. 2002;290:1368-75 [DOI] [PubMed] [Google Scholar]
- 16. Caberoy NB, Zhou Y, Alvarado G, Fan X, Li W. Efficient identification of phosphatidylserine-binding proteins by ORF phage display. Biochem Biophys Res Commun. 2009;386:197-201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hyatt SL, Liao L, Aderem A, Nairn AC, Jaken S. Correlation between protein kinase C binding proteins and substrates in REF52 cells. Cell Growth Differ. 1994;5:495-502 [PubMed] [Google Scholar]
- 18. Wassler MJ, Foote CI, Gelman IH, Shur BD. Functional interaction between the SSeCKS scaffolding protein and the cytoplasmic domain of beta 1,4-galactosyltransferase. J Cell Sci. 2001;114:2291-300 [DOI] [PubMed] [Google Scholar]
- 19. Gelman IH. Metastasis suppression by SSeCKS/Gravin/AKAP12 through the spatiotemporal control of oncogenic signaling mediators. In: Georgescu Maria-Magdalena, editor. Adaptor proteins and cancer. Kerala, India: Transworld Research Network; 2008. p. 83-101 [Google Scholar]
- 20. Streb JW, Kitchen CM, Gelman IH, Miano JM. Multiple promoters direct expression of three AKAP12 isoforms with distinct subcellular and tissue distribution profiles. J Biol Chem. 2004;279:56014-23 [DOI] [PubMed] [Google Scholar]
- 21. Akakura S, Huang C, Nelson PJ, Foster B, Gelman IH. Loss of the SSeCKS/Gravin/AKAP12 gene results in prostatic hyperplasia. Cancer Res. 2008;68:5096-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Weiser DC, Pyati UJ, Kimelman D. Gravin regulates mesodermal cell behavior changes required for axis elongation during zebrafish gastrulation. Genes Dev. 2007;21:1559-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Victor K, Jacob J, Cafiso DS. Interactions controlling the membrane binding of basic protein domains: phenylalanine and the attachment of the myristoylated alanine-rich C-kinase substrate protein to interfaces. Biochemistry. 1999;38:12527-36 [DOI] [PubMed] [Google Scholar]
- 24. Chapline C, Mousseau B, Ramsay K, et al. Identification of a major protein kinase C-binding protein and substrate in rat embryo fibroblasts: decreased expression in transformed cells. J Biol Chem. 1996;271:6417-22 [DOI] [PubMed] [Google Scholar]
- 25. Weiser DC, Julien KR, Lang JS, Kimelman D. Cell shape regulation by Gravin requires N-terminal membrane effector domains. Biochem Biophys Res Commun. 2008;375:512-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shih ML, Lin FB, Scott JD, Wang HY, Malbon CC. Dynamic complexes of β(2)-adrenergic receptors with protein kinases and phosphatases and the role of gravin. J Biol Chem. 1999;274:1588-95 [DOI] [PubMed] [Google Scholar]
- 27. Willoughby D, Wong W, Schaack J, Scott JD, Cooper DM. An anchored PKA and PDE4 complex regulates subplasmalemmal cAMP dynamics. EMBO J. 2006;25:2051-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nelson P, Gelman IH. Cell-cycle regulated expression and serine phosphorylation of the myristylated protein kinase C substrate, SSeCKS: correlation with cell confluency, G0 phase and serum response. Molec Cell Biochem. 1997;175:233-41 [DOI] [PubMed] [Google Scholar]
- 29. Tao J, Wang HY, Malbon CC. Src docks to A-kinase anchoring protein gravin, regulating beta2-adrenergic receptor resensitization and recycling. J Biol Chem. 2007;282:6597-608 [DOI] [PubMed] [Google Scholar]
- 30. Tao J, Wang HY, Malbon CC. Protein kinase A regulates AKAP250 (gravin) scaffold binding to the beta2-adrenergic receptor. EMBO J. 2003;22:6419-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tao J, Wang HY, Malbon C. AKAR2-AKAP12 fusion protein “biosenses” dynamic phosphorylation and localization of a GPCR-based scaffold. J Molec Signal. 2010;5:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Malbon CC, Tao JC, Wang HY. AKAPs (A-kinase anchoring proteins) and molecules that compose their G-protein-coupled receptor signalling complexes. Biochem J. 2004;379:1-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Piontek J, Brandt R. Differential and regulated binding of PKA and PKC isoenzymes to gravin in human model neurons: evidence that gravin provides a dynamic platform for the localization of kinases during neuronal development. J Biol Chem. 2003;278:38970-9 [DOI] [PubMed] [Google Scholar]
- 34. Yan XH, Carlson J, Grove BD. Gravin targets PKA RII to the cell periphery in cultured cells. FASEB J. 2004;18:A784 [Google Scholar]
- 35. Martin LJ, Chen H, Liao X, et al. FK506, a calcineurin inhibitor, prevents cadmium-induced testicular toxicity in mice. Toxicol Sci. 2007;100:474-85 [DOI] [PubMed] [Google Scholar]
- 36. Xia W, Unger P, Miller L, Nelson J, Gelman IH. The Src-suppressed C kinase substrate, SSeCKS, is a potential metastasis inhibitor in prostate cancer. Cancer Res. 2001;61:5644-51 [PubMed] [Google Scholar]
- 37. Ohtani N, Yamakoshi K, Takahashi A, Hara E. The p16INK4a-RB pathway: molecular link between cellular senescence and tumor suppression. J Med Invest. 2004;51:146-53 [DOI] [PubMed] [Google Scholar]
- 38. Rhim JH, Jang IS, Yeo EJ, Song KY, Park SC. Role of protein kinase C-dependent A-kinase anchoring proteins in lysophosphatidic acid-induced cAMP signaling in human diploid fibroblasts. Aging Cell. 2006;5:451-61 [DOI] [PubMed] [Google Scholar]
- 39. Choi MC, Lee YU, Kim SH, et al. A-kinase anchoring protein 12 regulates the completion of cytokinesis. Biochem Biophys Res Commun. 2008;373:85-9 [DOI] [PubMed] [Google Scholar]
- 40. Hutchins JRA, Toyoda Y, Hegemann B, et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328:593-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kittler R, Pelletier L, Heninger AK, et al. Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat Cell Biol. 2007;9:1401-12 [DOI] [PubMed] [Google Scholar]
- 42. Werner M, Glotzer M. Control of cortical contractility during cytokinesis. Biochem Soc Trans. 2008;36:371-7 [DOI] [PubMed] [Google Scholar]
- 43. Gelman IH, Gao L. The SSeCKS/Gravin/AKAP12 metastasis suppressor inhibits podosome formation via RhoA- and Cdc42-dependent pathways. Molec Cancer Res. 2006;4:151-8 [DOI] [PubMed] [Google Scholar]
- 44. Su B, Bu Y, Engelberg D, Gelman IH. SSeCKS/Gravin/AKAP12 inhibits cancer cell invasiveness and chemotaxis by suppressing a PKC-RAF/MEK/ERK pathway. J Biol Chem. 2010;285:4578-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Busch H, Camacho-Trullio D, Rogon Z, et al. Gene network dynamics controlling keratinocyte migration. Mol Syst Biol. 2008;4:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gelman IH, Lee K, Tombler E, Gordon R, Lin X. Control of cytoskeletal architecture by the src-suppressed C kinase substrate, SSeCKS. Cell Motil Cytoskel. 1998;41:1-17 [DOI] [PubMed] [Google Scholar]
- 47. Ordway JM, Williams K, Curran T. Transcription repression in oncogenic transformation: common targets of epigenetic repression in cells transformed by Fos, Ras or Dnmt1. Oncogene. 2004;23:3737-48 [DOI] [PubMed] [Google Scholar]
- 48. Schlosser I, Holzel M, Murnseer M, Burtscher H, Weidle UH, Eick D. A role for c-Myc in the regulation of ribosomal RNA processing. Nucleic Acids Res. 2003;31:6148-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. O’Connell BC, Cheung AF, Simkevich CP, et al. A large scale genetic analysis of c-Myc-regulated gene expression patterns. J Biol Chem. 2003;278:12563-73 [DOI] [PubMed] [Google Scholar]
- 50. Coller HA, Grandori C, Tamayo P, et al. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci U S A. 2000;97:3260-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cohen SB, Waha A, Gelman IH, Vogt PK. Expression of a down-regulated target, SSeCKS, reverses v-Jun-induced transformation of 10T1/2 murine fibroblasts. Oncogene. 2001;20:141-6 [DOI] [PubMed] [Google Scholar]
- 52. Yen A, Lin DM, Lamkin TJ, Varvayanis S. retinoic acid, bromodeoxyuridine, and the Delta 205 mutant polyoma virus middle T antigen regulate expression levels of a common ensemble of proteins associated with early stages of inducing HL-60 leukemic cell differentiation. In Vitro Cell Dev Biol Anim. 2004;40:216-41 [DOI] [PubMed] [Google Scholar]
- 53. Weinberg RA. Oncogenes, antioncogenes, and the molecular bases of multistep carcinogenesis. Cancer Res. 1989;49:3713-21 [PubMed] [Google Scholar]
- 54. Bu Y, Gelman IH. v-Src-mediated down-regulation of SSeCKS metastasis suppressor gene promoter by the recruitment of HDAC1 into a USF1-Sp1-Sp3 complex. J Biol Chem. 2007;282:26725-39 [DOI] [PubMed] [Google Scholar]
- 55. Bu Y, Gao L, Gelman IH. Role for transcription factor TFII-I in the suppression of SSeCKS/Gravin/Akap12 transcription by Src. Int J Cancer. Epub 2010. June 21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Daoud SS, Munson PJ, Reinhold W, et al. Impact of p53 knockout and topotecan treatment on gene expression profiles in human colon carcinoma cells: a pharmacogenomic study. Cancer Res. 2003;63:2782-93 [PubMed] [Google Scholar]
- 57. Lee SW, Kim WJ, Choi YK, et al. SSeCKS regulates angiogenesis and tight junction formation in blood-brain barrier. Nat Med. 2003;9:900-6 [DOI] [PubMed] [Google Scholar]
- 58. Zhang X, Ma L, Enkemann SA, Pledger WJ. Role of Gadd45alpha in the density-dependent G1 arrest induced by p27(Kip1). Oncogene. 2003;22:4166-74 [DOI] [PubMed] [Google Scholar]
- 59. Ali NA, McKay MJ, Molloy MP. Proteomics of Smad4 regulated transforming growth factor-beta signalling in colon cancer cells. Mol Bio-Syst. 2010;6:2332-8 [DOI] [PubMed] [Google Scholar]
- 60. Yoo JY, Huso DL, Nathans D, Desiderio S. Specific ablation of Stat3beta distorts the pattern of Stat3-responsive gene expression and impairs recovery from endotoxic shock. Cell. 2002;108:331-44 [DOI] [PubMed] [Google Scholar]
- 61. Palmer HG, Sanchez-Carbayo M, Ordonez-Moran P, Larriba MJ, Cordon-Cardo C, Munoz A. Genetic signatures of differentiation induced by 1alpha,25-dihydroxyvitamin D3 in human colon cancer cells. Cancer Res. 2003;63:7799-806 [PubMed] [Google Scholar]
- 62. Kovalenko PL, Zhang Z, Cui M, Clinton SK, Fleet JC. 1,25 dihydroxyvitamin D-mediated orchestration of anticancer, transcript-level effects in the immortalized, non-transformed prostate epithelial cell line, RWPE1. BMC Genomics. 2010;11:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chen J, Maltby KM, Miano JM. A novel retinoid-response gene set in vascular smooth muscle cells. Biochem Biophys Res Commun. 2001;281:475-82 [DOI] [PubMed] [Google Scholar]
- 64. Shin I, Kim S, Song H, Kim HR, Moon A. H-Ras-specific activation of Rac-MKK3/6-p38 pathway: its critical role in invasion and migration of breast epithelial cells. J Biol Chem. 2005;280:14675-83 [DOI] [PubMed] [Google Scholar]
- 65. Tavor S, Park DJ, Gery S, Vuong PT, Gombart AF, Koeffler HP. Restoration of C/EBPalpha expression in a BCR-ABL+ cell line induces terminal granulocytic differentiation. J Biol Chem. 2003;278:52651-9 [DOI] [PubMed] [Google Scholar]
- 66. Nelson PS, Clegg N, Arnold H, et al. The program of androgen-responsive genes in neoplastic prostate epithelium. Proc Natl Acad Sci U S A. 2002;99:11890-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shao C, Wang Y, Yue HH, et al. Biphasic effect of androgens on prostate cancer cells and its correlation with androgen receptor coactivator dopa decarboxylase. J Androl. 2007;28:804-12 [DOI] [PubMed] [Google Scholar]
- 68. Singh AP, Bafna S, Chaudhary K, et al. Genome-wide expression profiling reveals transcriptomic variation and perturbed gene networks in androgen-dependent and androgen-independent prostate cancer cells. Cancer Lett. 2008;259:28-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dydensborg AB, Rose AA, Wilson BJ, et al. GATA3 inhibits breast cancer growth and pulmonary breast cancer metastasis. Oncogene. 2009;28:2634-42 [DOI] [PubMed] [Google Scholar]
- 70. Choi MC, Jong HS, Kim TY, et al. AKAP12/Gravin is inactivated by epigenetic mechanism in human gastric carcinoma and shows growth suppressor activity. Oncogene. 2004;23:7095-103 [DOI] [PubMed] [Google Scholar]
- 71. Rinker-Schaeffer CW, O’Keefe JP, Welch DR, Theodorescu D. Metastasis suppressor proteins: discovery, molecular mechanisms, and clinical application. Clin Cancer Res. 2006;12:3882-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Su B, Zheng Q, Vaughan MM, Bu Y, Gelman IH. SSeCKS metastasis-suppressing activity in MatLyLu prostate cancer cells correlates with VEGF inhibition. Cancer Res. 2006;66:5599-607 [DOI] [PubMed] [Google Scholar]
- 73. Wikman H, Kettunen E, Seppanen JK, et al. Identification of differentially expressed genes in pulmonary adenocarcinoma by using cDNA array. Oncogene. 2002;21:5804-13 [DOI] [PubMed] [Google Scholar]
- 74. Chegini N, Verala J, Luo X, Xu J, Williams RS. Gene expression profile of leiomyoma and myometrium and the effect of gonadotropin releasing hormone analogue therapy. J Soc Gynecol Investig. 2003;10:161-71 [DOI] [PubMed] [Google Scholar]
- 75. Boultwood J, Pellagatti A, Watkins F, et al. Low expression of the putative tumour suppressor gene gravin in chronic myeloid leukaemia, myelodysplastic syndromes and acute myeloid leukaemia. Br J Haematol. 2004;126:508-11 [DOI] [PubMed] [Google Scholar]
- 76. Pellagatti A, Cazzola M, Giagounidis A, et al. Deregulated gene expression pathways in myelodysplastic syndrome hematopoietic stem cells. Leukemia. 2010;24:756-64 [DOI] [PubMed] [Google Scholar]
- 77. Yildirim M, Paydas S, Tanriverdi K, Seydaoglu G, Disel U, Yavuz S. Gravin gene expression in acute leukaemias: clinical importance and review of the literature. Leuk Lymphoma. 2007;48:1167-72 [DOI] [PubMed] [Google Scholar]
- 78. Heller G, Schmidt WM, Ziegler B, et al. Genome-wide transcriptional response to 5-aza-2′-deoxycytidine and trichostatin a in multiple myeloma cells. Cancer Res. 2008;68:44-54 [DOI] [PubMed] [Google Scholar]
- 79. Wasenius VM, Hemmer S, Kettunen E, Knuutila S, Franssila K, Joensuu H. Hepatocyte growth factor receptor, matrix metalloproteinase-11, tissue inhibitor of metalloproteinase-1, and fibronectin are up-regulated in papillary thyroid carcinoma: a cDNA and tissue microarray study. Clin Cancer Res. 2003;9:68-75 [PubMed] [Google Scholar]
- 80. Yeoh EJ, Ross ME, Shurtleff SA, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1:133-43 [DOI] [PubMed] [Google Scholar]
- 81. Lim EH, Aggarwal A, Agasthian T, et al. Feasibility of using low-volume tissue samples for gene expression profiling of advanced non-small cell lung cancers. Clin Cancer Res. 2003;9:5980-7 [PubMed] [Google Scholar]
- 82. Daino K, Ugolin N, Altmeyer-Morel S, Guilly MN, Chevillard S. Gene expression profiling of alpha-radiation-induced rat osteosarcomas: identification of dysregulated genes involved in radiation-induced tumorigenesis of bone. Int J Cancer. 2009;125:612-20 [DOI] [PubMed] [Google Scholar]
- 83. Daino K, Roch-Lefevre S, Ugolin N, Altmeyer-Morel S, Guilly MN, Chevillard S. Silencing of Cited2 and Akap12 genes in radiation-induced rat osteosarcomas. Biochem Biophys Res Commun. 2009;390:654-8 [DOI] [PubMed] [Google Scholar]
- 84. Hacker E, Muller K, Whiteman DC, Pavey S, Hayward N, Walker G. Reduced expression of IL-18 is a marker of ultraviolet radiation-induced melanomas. Int J Cancer. 2008;123:227-31 [DOI] [PubMed] [Google Scholar]
- 85. Bonazzi VF, Irwin D, Hayward NK. Identification of candidate tumor suppressor genes inactivated by promoter methylation in melanoma. Genes Chrom Cancer. 2009;48:10-21 [DOI] [PubMed] [Google Scholar]
- 86. Choi YK, Kim JH, Kim WJ, et al. AKAP12 regulates human blood-retinal barrier formation by downregulation of hypoxia-inducible factor-1alpha. J Neurosci. 2007;27:4472-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Feng XD, Huang SG, Shou JY, et al. Analysis of pathway activity in primary tumors and NCI60 cell lines using gene expression profiling data. Genom Proteom Bioinform. 2007;5:15-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Mori Y, Cai K, Cheng Y, et al. A genome-wide search identifies epigenetic silencing of somatostatin, tachykinin-1, and 5 other genes in colon cancer. Gastroenterol. 2006;131:797-808 [DOI] [PubMed] [Google Scholar]
- 89. Yoon DK, Jeong CH, Jun HO, et al. AKAP12 induces apoptotic cell death in human fibrosarcoma cells by regulating CDKI-cyclin D1 and caspase-3 activity. Cancer Lett. 2007;254:111-8 [DOI] [PubMed] [Google Scholar]
- 90. Kettunen E, Anttila S, Seppanen JK, et al. Differentially expressed genes in nonsmall cell lung cancer: expression profiling of cancer-related genes in squamous cell lung cancer. Cancer Genet Cytogenet. 2004;149:98-106 [DOI] [PubMed] [Google Scholar]
- 91. Yan B, Yang X, Lee TL, et al. Genome-wide identification of novel expression signatures reveal distinct patterns and prevalence of binding motifs for p53, nuclear factor-kappaB and other signal transcription factors in head and neck squamous cell carcinoma. Genome Biol. 2007;8:R78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Armstrong SA, Staunton JE, Silverman LB, et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41-7 [DOI] [PubMed] [Google Scholar]
- 93. Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747-52 [DOI] [PubMed] [Google Scholar]
- 94. Tonin PN, Hudson TJ, Rodier F, et al. Microarray analysis of gene expression mirrors the biology of an ovarian cancer model. Oncogene. 2001;20:6617-26 [DOI] [PubMed] [Google Scholar]
- 95. Dhanasekaran SM, Barrette TR, Ghosh D, et al. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412:822-6 [DOI] [PubMed] [Google Scholar]
- 96. Welsh JB, Zarrinkar PP, Sapinoso LM, et al. Analysis of gene expression profiles in normal and neoplastic ovarian tissue samples identifies candidate molecular markers of epithelial ovarian cancer. Proc Natl Acad Sci U S A. 2001;98:1176-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Rosenwald A, Alizadeh AA, Widhopf G, et al. Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. J Exp Med. 2001;194:1639-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Garber ME, Troyanskaya OG, Schluens K, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci U S A. 2001;98:13784-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Jo U, Whang YM, Kim HK, Kim YH. AKAP12alpha is associated with promoter methylation in lung cancer. Cancer Res Treat. 2009;38:144-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Tessema M, Willink R, Do K, et al. Promoter methylation of genes in and around the candidate lung cancer susceptibility locus 6q23-25. Cancer Res. 2008;68:1707-14 [DOI] [PubMed] [Google Scholar]
- 101. Wang J, Qin R, Ma Y, et al. Differential gene expression in normal esophagus and Barrett’s esophagus. J Gastroenterol. 2009;44:897-911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Jin Z, Hamilton JP, Yang J, et al. Hypermethylation of the AKAP12 promoter is a biomarker of Barrett’s-associated esophageal neoplastic progression. Cancer Epidemiol Biomarkers Prev. 2008;17:111-7 [DOI] [PubMed] [Google Scholar]
- 103. Jin Z, Cheng Y, Gu W, et al. A multicenter, double-blinded validation study of methylation biomarkers for progression prediction in Barrett’s esophagus. Cancer Res. 2009;69:4112-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Zhu Y, Hollmen J, Raty R, et al. Investigatory and analytical approaches to differential gene expression profiling in mantle cell lymphoma. Br J Haematol. 2002;119:905-15 [DOI] [PubMed] [Google Scholar]
- 105. Yang J, Kiefer S, Rauchman M. Characterization of the gene encoding mouse retinoblastoma binding protein-7, a component of chromatin-remodeling complexes. Genomics. 2002;80:407-15 [DOI] [PubMed] [Google Scholar]
- 106. Tsujimoto K, Ono T, Sato M, Nishida T, Oguma T, Tadakuma T. Regulation of the expression of caspase-9 by the transcription factor activator protein-4 in glucocorticoid-induced apoptosis. J Biol Chem. 2005;280:27638-44 [DOI] [PubMed] [Google Scholar]
- 107. Carinci F, Piattelli A, Martinelli M, et al. Genetic profiling of central giant cell granuloma of the jaws. J Craniofac Surg. 2005;16:399-407 [DOI] [PubMed] [Google Scholar]
- 108. Jiang BH, Agani F, Passaniti A, Semenza GL. V-SRC induces expression of hypoxia-inducible factor 1 (HIF-1) and transcription of genes encoding vascular endothelial growth factor and enolase 1: involvement of HIF-1 in tumor progression. Cancer Res. 1997;57:5328-35 [PubMed] [Google Scholar]
- 109. Lahav R, Suva ML, Rimoldi D, Patterson PH, Stamenkovic I. Endothelin receptor B inhibition triggers apoptosis and enhances angiogenesis in melanomas. Cancer Res. 2004;64:8945-53 [DOI] [PubMed] [Google Scholar]
- 110. Choi YK, Kim KW. AKAP12 in astrocytes induces barrier functions in human endothelial cells through protein kinase Czeta. FEBS J. 2008;275:2338-53 [DOI] [PubMed] [Google Scholar]
- 111. You QH, Sun GY, Wang N, Shen JL, Wang Y. Interleukin-17F-induced pulmonary microvascular endothelial monolayer hyperpermeability via the protein kinase C pathway. J Surg Res. 2009;162:110-21 [DOI] [PubMed] [Google Scholar]
- 112. You QH, Sun GY, Wang N, Chen S, Luo QL. Role of src-suppressed C kinase substrate in rat pulmonary microvascular endothelial hyperpermeability stimulated by inflammatory cytokines. Inflamm Res. 2010;59:949-58 [DOI] [PubMed] [Google Scholar]
- 113. Malbon CC. A-kinase anchoring proteins: trafficking in G-protein-coupled receptors and the proteins that regulate receptor biology. Curr Opin Drug Discov Devel. 2007;10:573-9 [PubMed] [Google Scholar]
- 114. Weis S, Cui J, Barnes L, Cheresh D. Endothelial barrier disruption by VEGF-mediated Src activity potentiates tumor cell extravasation and metastasis. J Cell Biol. 2004;167:223-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Summy JM, Gallick GE. Src family kinases in tumor progression and metastasis. Cancer Metastasis Rev. 2003;22:337-58 [DOI] [PubMed] [Google Scholar]
- 116. Mardin WA, Petrov KO, Enns A, Senninger N, Haier J, Mees ST. SERPINB5 and AKAP12: expression and promoter methylation of metastasis suppressor genes in pancreatic ductal adenocarcinoma. BMC Cancer. 2010;10:549. [DOI] [PMC free article] [PubMed] [Google Scholar]


