Abstract
BRCA1 has been implicated in the DNA damage pathway and regulation of genome stability, however, it does not contain intrinsic catalytic activity to repair the DNA lesions. Thus, a potential activity of BRCA1 is to assemble proteins that sense DNA damage and to transduce checkpoint signals to downstream. We have recently isolated a protein termed BAAT1, which binds to BRCA1, ATM, DNA-PKcs, and SMC1. Phosphorylation of ATM/DNA-PKcs is greatly reduced in BAAT1-knockdown cells, suggesting that sensing of DNA lesions mediated by BRCA1/BAAT1 is critical for activation of these kinases.
Keywords: DNA damage pathway, BRCA1, BAAT1, ATM, DNA-PKcs, SMC1
Introduction: BRCA1 Structure and Interacting Proteins
The BRCA1 gene encodes a nuclear phosphoprotein of 1863 amino acids with a predicted molecular weight of 220 kDa.1 The amino terminus contains a RING domain that has been shown to mediate the interaction with another RING domain–containing protein, BARD1. The BRCA1/BARD1 heterodimer shows ubiquitin ligase activity.2,3 Exon 11 of Brca1 encodes approximately 60% of BRCA1. The central region interacts with the DNA repair protein complex Mre11-Rad50-NBS1, the transcriptional repressor ZBRK1,4,5 and transcription factors p53 and STAT1.6,7 The carboxyl terminus of BRCA1 contains 2 repeats of the BRCA1 C-terminal (BRCT) domain. This region is highly acidic, and early studies indicated that the BRCT domain displays transcriptional activation when fused to the yeast GAL4 DNA binding domain,6,8 establishing a potential role of BRCA1 in regulating gene expression.9,10 Soon after that, this BRCT region has been demonstrated to bind to phosphopeptides,11,12 and their in vivo recognition targets are phosphorylated serines in short linear motifs present in proteins involved in cell cycle checkpoints. Several proteins, including BACH1, CtIP, Acetyl-CoA carboxylase, Abraxas/CCDC98, and RAP80, interact with the BRCT domain of BRCA1 in a phosphorylation-dependent manner.13 Our group performed yeast two-hybrid screen to search the BRCT-interacting proteins and identified several proteins.14 BAAT1, BRCA1, and ATM-associated protein 1 are among them, which we found bind to both BRCA1 and ATM.14
Given this ability of BRCA1 to associate with diverse cellular proteins, BRCA1 maintains genomic stability through gene expression, DNA repair, and cell cycle checkpoint activation by interacting these proteins.
A Novel Protein, BAAT1
BAAT1 was isolated by the yeast 2-hybrid screen using aa1314-1863 of BRCA1 as bait. More detailed biochemical analysis revealed that both aa1600-1749 and aa1749-1863 of BRCA1 bind to BAAT1 but that aa1749-1863 carrying breast cancer–associated M1775R mutation does not bind, suggesting that BRCA1/BAAT1 interaction is important for BRCA1’s tumor-suppressive activity.
The human BAAT1 gene is ~ 17 kb on chromosome 7p22.3 and consists of 14 exons (www.ncrna.org). The full-length BAAT1 protein consists of 821 amino acids of ~ 88 kDa. The genome database at www.ensembl.org (gene name C7ORF27) indicates that a total of 41 orthologs are found in the dog, chimpanzee, mouse, chicken, zebrafish, horse, giant panda, etc. For example, human amino acid sequences share 74% or 99% identity with mouse or chimpanzee sequences, respectively. There are at least 8 forms of human BAAT1 mRNA generated by alternative splicing, and 4 of them encode proteins. As shown in Figure 1, the C-terminal half of the protein contains 2 HEAT (Huntingtin, Elongation factor 3, A subunit of protein phosphatase 2A, and TOR1) repeat domains (aa495-531, aa544-576) and a putative phosphorylation residue of Ser742 by Akt/cdk/MAPK, which is commonly predicted in both www.ensembl.org and GPS2.1 programs (Fig. 1). From the www.ensembl.org database, among total 767 sequence variants identified, human BAAT1 has 52 SNP sites resulting in an amino acid change in the encoded peptide sequence and 5 frameshift mutations in coding sequence resulting in truncation of the protein, although it is not known whether these sequence variations are pathogenic. Interestingly, computational alignment indicates that aa54 to aa95 of BAAT1 are the CIDE-N domain (cell death–inducing DFF45-like effector [CIDE] domain),15 suggesting the possible function of the protein in apoptosis.
Figure 1.
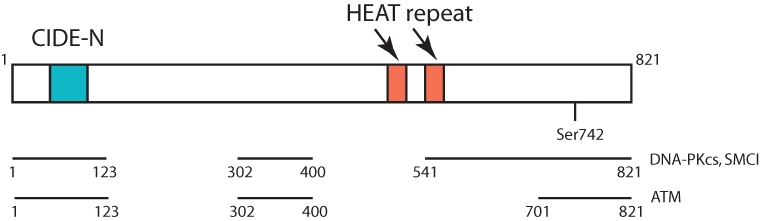
Structure and interaction interface of BAAT1. The N-terminal CIDE-N domain (light green) and 2 HEAT repeats (light red) are indicated. Ser742 is also indicated. BAAT1’s binding regions for DNA-PKcs/SMC1 or ATM are demonstrated in Figure 2.
When BAAT1 is knocked down in mouse embryonic fibroblasts (MEFs) and human osteosarcoma cell line U2OS, constitutive levels of apoptosis are increased, however they are not significantly augmented when these cells are exposed to IR.14 Such results support a notion that BAAT1 may regulate the apoptosis pathway, although the mechanism of induction of apoptosis in BAAT1-knockdown cells is not clear.
BAAT1 mRNA is ubiquitously expressed,although levels in the testis, pancreas, prostate, and salivary gland are higher than the other tissues examined (http://biogps.gnf.org/#goto=genereport&id=221927; http://www.ncbi.nlm.nih.gov).14Detailed immunoblot analysis using mouse brain tissues displayed more interesting distribution of the protein; BAAT1 is highly expressed in the cortex compared to the hippocampus and cerebellum,16 suggesting that BAAT1 is involved in neuronal function, although it has not been illustrated yet.
Roles of BAAT1 in ATM Pathway
BAAT1 was originally isolated as a BRCA1 interacting protein; however, subsequent studies showed that it also binds to ATM.14 When cells are exposed to ionizing radiation (IR), levels of endogenous BAAT1 increase, and fractions of BAAT1 that co-immunoprecipitates with ATM also increase. γH2AX, an indicator of the localization of DNA double-strand breaks (DSBs),17,18 co-localizes with BAAT1 under conditions of IR treatment, with all these results supporting an idea that BAAT1 participates in the DNA damage pathway. Phosphorylation of ATM at Ser1981 has been implicated in activation of ATM in response to cell stress19; however, it is abolished when BAAT1 was transiently knocked down by siRNA. Phosphorylation of histone H2AX and NBS1 that are both well-characterized ATM substrates is significantly decreased in BAAT1 knockdown cells, although phosphorylation of Chk2 at Thr68 is weakly attenuated.14 More recently, the mutant mouse ATM protein deficient for phosphorylation of Ser1987 (equivalent to human ATM Ser1981) does not exhibit dominant-negative interfering activity when expressed physiologically or overexpressed in the context of ATM heterozygous mice. These results suggest an alternative mode for stimulation of ATM by DSBs in which ATM autophosphorylation at Ser 1987 is a consequence rather than a cause of ATM activation.20 Further, A. Nuzzenzweig’s laboratory generated and characterized a triple-negative mutant of mouse ATM in which 3 phosphorylation sites (Ser367, Ser1899, and Ser1987) are simultaneously mutated to Alanine, and found that ATM autophosphorylation correlates with the DNA damage-induced activation of the kinase but is not required for ATM function in vivo.21 Of note, phosphorylation of ATM at Ser1981 similarly occurs in normal mammary epithelial cells, BRCA1-mutant HCC1937 breast cancer cell line, and BRCA1-mutant SNU251 ovarian cancer cell line.14 On the basis of these results, BAAT1 is crucial for ATM’s autophosphorylation, rather than activation.
The mechanism of ATM regulation by BAAT1 is largely unknown. Lowered levels of ATM phosphorylation at Ser1981 in BAAT1 knockdown cells under conditions of IR treatment can be recovered when these cells are simultaneously treated with okadaic acid, a phosphatase inhibitor, suggesting that a phosphatase that may be involved in ATM dephosphorylation is activated when BAAT1 is transiently knocked down. Biochemical studies have demonstrated that IR-induced ATM phosphorylation at Ser1981 is dephosphorylated by incubating with purified PP2A phosphatase, but this dephosphorylation is inhibited by co-incubating with baculovirus-produced BAAT1.14 On the other hand, overexpression of BAAT1 does not enhance phosphorylation of ATM after IR (not published). Although these results are obtained from in vitro assay, it is suggested that, when levels of BAAT1 are reduced, a phosphatase(s) that dephosphorylates ATM Ser1981 could access ATM more efficiently, providing a model that BAAT1 modulates ATM phosphorylation after IR stress rather than directly inducing ATM’s autophosphorylation (Fig. 2).
Figure 2.
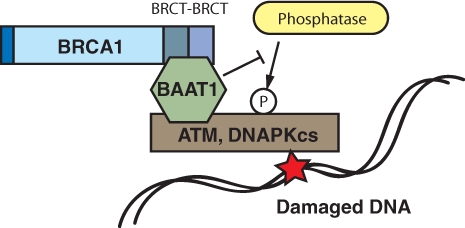
Schematic model of BAAT1 function in regulating ATM/DNA-PKcs phosphorylation. BAAT1 constitutively binds to BRCT domains of BRCA1 and stress kinases such as ATM/DNA-PKcs. Phosphorylation of ATM at Ser1981 and DNA-PKcs at Ser2956 occurs under conditions of DNA stress. In this model, BAAT1 potentially protects these residues from phosphatases that dephosphorylate these residues.
Roles of BAAT1 in DNA-PK and SMC1 Pathway
Extensive studies of biochemical analysis of BAAT1 illustrated that this protein shows functional interaction with not only ATM but also a catalytic subunit of DNA-dependent kinase (DNA-PKcs) and SMC1 (Fig. 1).16 GST pulldown analysis revealed that both DNA-PKcs and SMC1 bind to fragment #1 (aa1-123), #3 (aa302-400), #5 (aa541-700), and #6 (aa701-821) before NCS treatment, but interaction with #1 was decreased after NCS treatment. ATM binds to #1, #3, and #6 of BAAT1, although interaction with BAAT1 #1 was much weaker than the others. After NCS1 treatment, ATM binds primarily to the BAAT1 #6 fragment, although BAAT1 #3 showed weak interaction. Interaction between ATM and BAAT1 #1 was not detected after NCS treatment. These results suggest that protein modification and/or allosteric change of the conformation occurs in BAAT1, DNA-PKcs, ATM, and/or SMC1. Physiological roles of SNPs in the BAAT1 gene in these protein-protein interactions still remain to be elucidated.
Similar to the results of inhibition of ATM phosphorylation observed in BAAT1 knockdown cells, autophosphorylation of DNA-PKcs at Ser2056 is also attenuated when BAAT1 is depleted.16 It has been well established that catalytic activity of DNA-PKcs is regulated primarily by its physical interaction with Ku70/80 subunits and DNA fragment22; however, we could not detect direct interaction between BAAT1 and Ku70/80 subunits, suggesting that the mechanism of regulation of DNA-PKcs by BAAT1 perhaps does not involve Ku subunits. More recently, studies from Lees-Miller’s laboratory demonstrated that DNA-PKcs interacts with protein phosphatase 2A and 6.23 Taken together, these studies suggest that BAAT1 is potentially involved in protection of phosphorylated residues of the kinase (ATM and DNA-PKcs) rather than directly involved in process of their activation.
SMC1 proteins play a pivotal role in sister chromatid cohesion, chromosome condensation, sex chromosome dosage compensation, and DNA recombination and repair.24-27 Heterodimers of the SMC1 and SMC3 proteins have been implicated specifically in both sister chromatid cohesion and DNA recombination. ATM phosphorylates SMC1 protein at Ser957 and Ser966 after ionizing irradiation, and expression of a phospho-deficient form of SMC1 protein mutated at these phosphorylation sites abrogates the ionizing irradiation-induced S phase cell cycle checkpoint.28,29 When cells were treated with NCS, phosphorylation of SMC1 at Ser966 was significantly increased, but this phosphorylation was reduced in BAAT1 knockdown cells. It remains to be elucidated how SMC1 and its phosphorylation is important to regulate S phase checkpoint. Although there is not evidence that BAAT1 functions in S phase checkpoint, these results suggest that S phase checkpoint is abrogated in BAAT1 knockdown cells due to lowered phosphorylation of SMC1.
Regulation of ATM/DNA-PKcs Activity by BAAT1
As described here, BAAT1 is essential for the activation of ATM and DNA-PKcs. Phosphorylation of these kinases has been used as readout of their activation. Inhibition or decreased phosphorylation of these kinases in BAAT1 knockdown cells suggests BAAT’s potential function in the pathway, not only of phosphatases but also of some other possibilities. For example, in BAAT1-depleted cells, 1) ATM/DNA-PKcs cannot relocalize to the sites of DNA lesions; 2) kinase activities are inhibited, although relocalization to DNA lesions is fine; 3) phosphatase activity is increased, leading to lower autophosphorylation; and 4) phosphatase activity is not significantly increased, although access of phosphatase to kinase is increased. Our preliminary data suggest a certain interaction between BAAT1 and a phosphatase(s); however, other possibilities raised here also need to be examined.
Acknowledgments
This review article is dedicated to Dr. Saburo Hanafusa, who died in Osaka on March 15, 2009. He has been a mentor to a number of cancer biologists since he has been at The Rockefeller University in New York. Both authors of this article participated in Saburo’s laboratory in the early 1990s. We are grateful for his legacy and his sincere attitude as a sensei (“teacher” in Japanese).
Footnotes
The author(s) declared no potential conflicts of interest with respect to the authorship and/or publication of this article.
This work was supported by the National Institutes of Health/National Cancer Institute [grant numbers R01CA79892, R01CA90631 (to T.O.)]; the Susan G. Komen Foundation [Breast Cancer Research Grant]; and the AVON Breast Cancer Research Foundation.
References
- 1. Miki Y, Swensen J, Shattuck-Eidens D, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266(5182):66-71 [DOI] [PubMed] [Google Scholar]
- 2. Chen A, Kleiman FE, Manley JL, Ouchi T, Pan ZQ. Autoubiquitination of the BRCA1/BARD1 RING ubiquitin ligase. J Biol Chem. 2002;277(24):5287-92 [DOI] [PubMed] [Google Scholar]
- 3. Wu-Baer F, Lagrazon K, Yuan W, Baer R. The BRCA1/BARD1 heterodimer assembles polyubiquitin chains through an unconventional linkage involving lysine residue K6 of ubiquitin. J Biol Chem. 2003;278(37):34743-6 [DOI] [PubMed] [Google Scholar]
- 4. Zhong Q, Chen CF, Li S, et al. Association of BRCA1 with the hRad50-hMre11-p95 complex and the DNA damage response. Science. 1999;285(5476):747-50 [DOI] [PubMed] [Google Scholar]
- 5. Zheng L, Pan H, Li S, et al. Sequence-specific transcriptional corepressor function for BRCA1 through a novel zinc finger protein. Mol Cell. 2000;6(4):757-68 [DOI] [PubMed] [Google Scholar]
- 6. Ouchi T, Monteiro AN, August A, Aaronson SA, Hanafusa H. BRCA1 regulates p53-dependent gene expression. Proc Natl Acad Sci U S A. 1998;95(5):2302-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ouchi T, Lee SW, Ouchi M, Aaronson SA, Horvath CM. Collaboration of signal transducer and activator of transcription 1 (STAT1) and BRCA1 in differential regulation of IFN-gamma target genes. Proc Natl Acad Sci U S A. 2000;97(10):5208-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Monteiro AN, August A, Hanafusa H. Evidence for a transcriptional activation function of BRCA1 C-terminal region. Proc Natl Acad Sci U S A. 1996;93(24):13595-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ouchi T. BRCA1 phosphorylation: biological consequences. Cancer Bio Ther. 2006;5(5):470-5 [DOI] [PubMed] [Google Scholar]
- 10. Murray MM, Mullan PB, Harkin DP. Role played by BRCA1 in transcriptional regulation in response to therapy. Biochem Soc Trans. 2007;35(Pt 5):1342-6 [DOI] [PubMed] [Google Scholar]
- 11. Manke IL, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302(5645):636-9 [DOI] [PubMed] [Google Scholar]
- 12. Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302(5645):639-42 [DOI] [PubMed] [Google Scholar]
- 13. Rodriguez MC, Songyang Z. BRCT domains: phosphopeptide binding and signaling modules. Front Biosci. 2008;13:5905-15 [DOI] [PubMed] [Google Scholar]
- 14. Aglipay JA, Martin SA, Tawara H, Lee SW, Ouchi T. ATM activation by ionizing radiation requires BRCA1-associated BAAT1. J Biol Chem. 2006;281(14):9710-8 [DOI] [PubMed] [Google Scholar]
- 15. Lugovskoy AA, Zhou P, Chou JJ, McCarty JS, Li P, Wagner G. Solution structure of the CIDE-N domain of CIDE-B and a model for CIDE-N/CIDE-N interactions in the DNA fragmentation pathway of apoptosis. Cell. 1999;99(7):747-55 [DOI] [PubMed] [Google Scholar]
- 16. So EY, Ouchi T. Functional interaction of BRCA1/ATM-associated BAAT1 with DNA-PKcs. Exp Ther Med. In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y. GammaH2AX and cancer. Nat Rev Cancer. 2008;9(12):957-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Srivastava N, Gochhait S, de Boer P, Bamezai RN. Role of H2AX in DNA damage response and human cancers. Mutat Res. 2009;681(2-3):180-8 [DOI] [PubMed] [Google Scholar]
- 19. Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimmer dissociation. Nature. 2003;421(6922):499-506 [DOI] [PubMed] [Google Scholar]
- 20. Pellegrini M, Celeste A, Difilippantonio S, Guo R, Wang W, Feigenbaum L, Nussenzweig A. Autophosphorylation at serine 1987 is dispensable for murine Atm activation in vivo. Nature. 2006;443(7108):222-5 [DOI] [PubMed] [Google Scholar]
- 21. Daniel JA, Pellegrini M, Lee JH, Paull TT, Feigenbaum L, Nussenzweig A. Multiple autophosphorylation sites are dispensable for murine ATM activation in vivo. J Cell Biol. 2008;183(5):777-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Collis SJ, DeWeese TL, Jeggo PA, Parker AR. The life and death of DNA-PK. Oncogene. 2005;24(6):949-61 [DOI] [PubMed] [Google Scholar]
- 23. Douglas P, Zhong Y, Ye R, Moorhead GB, Xu X, Lees-Miller SP. Protein phosphatase 6 interacts with the DNA-dependent protein kinase catalytic subunit and dephosphorylates gamma-H2AX. Mol Cell Biol. 2010;30(6):1368-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Strunnikov AV, Jessberger R. Structural maintenance of chromosomes (SMC) proteins: conserved molecular properties for multiple biological functions. Eur J Biochem. 1999;263(1):6-13 [DOI] [PubMed] [Google Scholar]
- 25. Losada A, Hirano T. Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev. 2005;19(11):1269-87 [DOI] [PubMed] [Google Scholar]
- 26. Watrin E, Peters JM. Cohesin and DNA damage repair. Exp Cell Res. 2006;312(14):2687-93 [DOI] [PubMed] [Google Scholar]
- 27. Wong RW. An update on cohesin function as a ‘molecular glue’ on chromosomes and spindles. Cell Cycle. 2010;9(9):1754-8 [DOI] [PubMed] [Google Scholar]
- 28. Kitagawa R, Bakkenist CJ, McKinnon PJ, Kastan MB. Phosphorylation of SMC1 is a critical downstream event in the ATM-NBS1-BRCA1 pathway. Genes Dev. 2004;18(12):1423-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yazdi PT, Wang Y, Zhao S, Patel N, Lee EY, Qin J. SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes Dev. 2002;16(5):571-82 [DOI] [PMC free article] [PubMed] [Google Scholar]


