Abstract
Gene expression signatures in blood correlate with specific diseases. Such signatures may serve as valuable diagnostic and prognostic tools in disease management. Blood gene expression signatures associated with heart failure may be applied to predict prognosis, monitor disease progression, and optimize treatment. Blood gene expression profiles were generated for 71 subjects with heart failure and 15 controls without heart failure, using the Affymetrix GeneChip U133Plus2.0. Survival analysis identified 197 “mortality genes” that were significantly associated with patient outcome. Functional categorization showed that genes associated with T cell receptor signaling were most significantly overpresented. Cluster analysis of these T cell receptor signaling genes significantly categorized heart failure patients into three risk groups (P = 0.031) that were distinct from the three risk groups categorized by New York Heart Association (NYHA) Classification (P = 0.0002). By combining the analysis of clinical assessment (NYHA class) with T cell receptor signaling gene expression, we proposed a model that demonstrated an even greater differentiation of patients at risk (P = 0.0001). In this discovery study, we identified blood expression signatures associated with heart failure patient outcomes. Characterization of these mortality genes helped identify a set of T cell receptor signaling genes that may be of utility in predicting survival of heart failure patients. These data raise the possibility of prospectively risk stratifying patients with heart failure by integrating blood gene expression signatures with current clinical assessment.
Keywords: gene expression profiling, microarray, survival analysis
heart failure remains a major public health concern, with the incidence of this disorder continuing to increase. While advances in the management of heart failure have improved patient outcomes, heart failure remains the leading hospital admission diagnosis in elderly patients and carries a 5 yr mortality rate as high as 50% (17). The morbidity and mortality of this disease thus remain unacceptably high, and better diagnostic and prognostic strategies are needed to improve the management and treatment of patients with heart failure.
Heart failure has been long known to affect circulating levels of numerous neurohormones, cytokines, and inflammatory markers (3, 12). These factors directly and adversely affect intracellular signaling and ultimately gene expression in the myocardium. Genome-wide expression profiling has provided great insights into the pathophysiology of heart failure (13). Specifically, gene expression profiling studies in patients with heart failure have demonstrated a significant elevation in the expression of genes involved in cell growth, signal transduction, and cell defense in myocardial tissue (5, 7). Thus myocardial gene expression likely reflects direct tissue changes associated with the cardiomyopathic process as well as consequent alterations in gene expression secondary to the humoral response of the disease state.
Given the systemic nature of heart failure and the protean nature of neurohormonal signaling, other tissues are also affected by the heart failure state. In this study, we have taken advantage of this fact to discover and characterize differential expression of blood genes in patients with heart failure. Gene expression profiling of blood cells has shown promise in the identification of differentially expressed genes in coronary artery disease (9) and expression patterns correlated with blood lipid levels (8). In this study, we identified and characterized expression signatures of peripheral blood cells associated with outcomes in patients with heart failure. We hypothesize that blood gene expression profiling can contribute toward the determination of heart failure patient prognosis and thus could be used to optimize patient management.
METHODS
Study Population
Heart failure patients.
Potential participants with heart failure were identified in our ambulatory clinics or at the time of hospital admission with a primary diagnosis of heart failure. Adult patients (age 21–89 yr) with history of heart failure were eligible for enrolment. All patients had left ventricular functional assessment as part of routine cardiac care prior to enrolment. Heart failure severity was clinically characterized in accordance with the New York Heart Association (NYHA) classification. Patients with NYHA class I–III heart failure were identified in our outpatient clinic at the time of routine follow up. NYHA class IV patients were identified at the time of hospital admission for decompensated heart failure. Exclusion criteria were acute myocardial infarction, unstable angina, dialysis-dependent renal failure, disseminated cancer, and sepsis. The goal of recruitment was approximately equal representation in each heart failure class.
Control group.
Control subjects were selected from patients referred for evaluation of atypical or noncardiac chest pain and who had no prior diagnosis of cardiac disease. Normal ventricular function and absence of significant coronary disease in the control patient group were confirmed by stress echocardiogram or nuclear perfusion imaging.
This study was approved by the Committees in Human Research at the University of Vermont. All participants provided written, informed consent.
Blood Collection, RNA Extraction, and Gene Expression Profiling
Approximately 20 ml blood was taken from an antecubital vein after overnight fasting. Blood samples were collected with a Vacutainer tube (Becton Dickinson, Franklin Lakes, NJ) and stored on ice until RNA extraction (performed within 6 h after blood collection). Red blood cells were ruptured with hypotonic hemolysis buffer (1.6 mM EDTA, 10 mM KHCO3, 153 mM NH4Cl, pH 7.4), and white blood cells were separated by centrifugation. Total RNA of white blood cells was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA) in accordance with the manufacturer's instructions. The quality of RNA samples was assessed on an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA) using RNA 6000 Nano Chips (Agilent Technologies). The quantity of RNA was determined by UV spectrophotometry (Beckman-Coulter DU640, Beckman-Coulter, Fullerton, CA). Five micrograms of total RNA from each sample was used for hybridization on a GeneChip U133Plus2 microarray (Affymetrix, Santa Clara, CA) in accordance with the manufacturer's instructions.
Analysis of Blood Gene Expression Profiles
After scanning, probe-level expression data (CEL files) generated from the raw microarray hybridization images were processed by GC-robust multichip average (RMA) (20) using GeneSpring v7.3 software (Agilent Technologies). Genes with unreliable measurements, assessed by cross-gene error model (http://www.chem.agilent.com/cag/bsp/products/gsgx/Downloads/pdf/error_model.pdf), were removed from further analysis. Survival analysis was conducted using MedCalc (MedCalc Software, http://www.medcalc.be). The expression data for each gene and patient survival data were correlated, using a Cox proportional hazards regression model. Significance of associations between gene expression and survival time was assessed using a log-rank test. Multitest correction was performed using the q value package in Comprehensive R Archive Network (18). A q value of 0.2 was chosen as the significance cutoff for the selection of mortality genes. Differentially expressed mortality genes between controls and NYHA I–II and between controls and NYHA III–IV were identified by a Welch t-test. Significance was identified when P was <0.05.
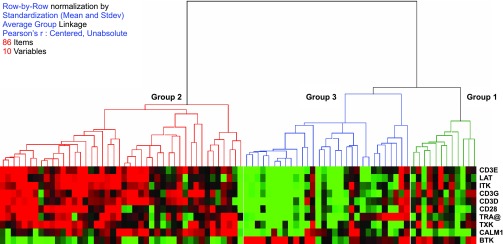
Functional categorization of the mortality genes was conducted using Ingenuity Pathway Analysis software (Ingenuity Systems, Redwood City, CA). Genes in significantly overpresented functional group(s) were subjected to cluster analysis using Pearson correlation and average group linkage by Hierarchical Clustering Explorer (Human Computer Interaction Lab, University of Maryland). The 86 samples were then reclassified into three groups based on the cluster analysis of the mortality genes. Survival analysis was applied to the reclassified groups by cluster analysis. In addition survival analysis was also applied to the NYHA classified groups (control, NYHA I–IV). A Kaplan-Meier plot was drawn using MedCalc. Significance was assessed using a log-rank test.
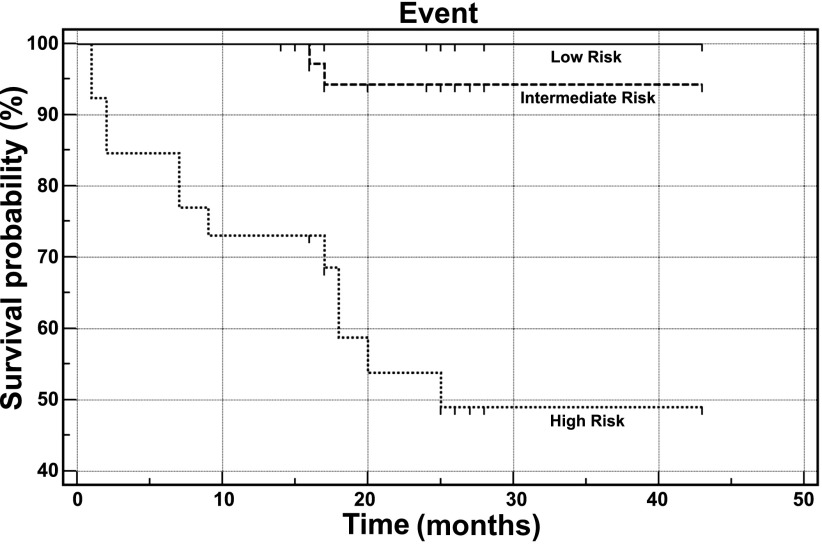
A risk stratification model was proposed that integrated the clinical severity parameter (NYHA) and the gene expression signatures associated with patient outcomes. In this model, a clinical score was assigned according to each subject based on NYHA classification as follows: 0 (control), 1–4 (NYHA = I–IV); an expression score was assigned according to the cluster analysis of the 10 T cell receptor signaling genes (Fig. 2) as either 1 (group 1), 2 (group 2), or 3 (group 3); an integrated risk index (IRI) was created as defined as the sum of the clinical score and the expression score of each subject. IRI ranged from 1 to 7, and subjects were categorized into three risk groups by IRI as low risk (IRI = 1–3), intermediate risk (IRI = 4–5), and high risk (IRI = 6–7) (see Table 3). A Kaplan-Meier plot was applied to the three risk groups using the Survival package, and the significance was assessed by a log-rank test.
Fig. 2.
Cluster analysis of the 10 mortality genes involved in T cell receptor signaling segregated the patients into 3 groups.
Table 3.
Risk analysis with NYHA class and T cell receptor gene clustering
| NYHA | T Cell Receptor Gene Clustering | IRI | n | |
|---|---|---|---|---|
| Low risk (n = 24) | control | group 1 | 1 | 1 |
| 13 M/11 F | control | group 2 | 2 | 12 |
| 100% survival | control | group 3 | 3 | 2 |
| I | group 1 | 2 | 1 | |
| I | group 2 | 3 | 6 | |
| II | group 1 | 3 | 2 | |
| Intermediate risk (n = 36) | I | group 3 | 4 | 4 |
| 22 M/14 F | II | group 2 | 4 | 13 |
| 94% survival | II | group 3 | 5 | 4 |
| III | group 1 | 4 | 7 | |
| III | group 2 | 5 | 6 | |
| IV | group 1 | 5 | 2 | |
| High risk (n = 26) | III | group 3 | 6 | 9 |
| 20 M/6 F | IV | group 2 | 6 | 6 |
| 49% survival | IV | group 3 | 7 | 11 |
IRI, integrated risk index; M, male; F, female. See text for discussion.
RESULTS
Subject Recruitment
A total of 87 subjects participated in this study: 72 heart failure patients and 15 nonheart failure controls. One heart failure patient was excluded after dying from a nonheart failure-related operation. All were Caucasian with the exception of one patient who was of Middle Eastern decent. Heart failure subjects were clinically categorized into the four NYHA classes. The clinical characteristics and medications for all subjects are summarized in Table 1. The underlying etiology of heart failure was ischemic heart disease in 39 patients and nonischemic in 32 patients. Valvular heart disease was a contributing factor in four patients (two with mitral regurgitation and two with aortic stenosis). The incidence of ischemic cardiomyopathy, diabetes, and atrial fibrillation was all higher in the NYHA III–IV group, reflecting either the effect of these confounding factors in advanced heart failure or the effect of heart failure on these comorbidities. Not surprisingly, loop diuretic use was also higher in more advanced heart failure. In the follow-up period (median 26 mo) 14 heart failure patients died. Thirteen of the 14 patients died of cardiovascular causes. In the 14th patient, heart failure was the secondary cause of death. In eight of the 14 deaths the cause of heart failure was ischemic heart disease.
Table 1.
Demographic characteristics and medications of heart failure and control subjects
| Control | NYHA I | NYHA II | NYHA III | NYHA IV | |
|---|---|---|---|---|---|
| n | 15 | 11 | 20 | 21 | 19 |
| Age: mean, range | 58.0 (33–81) | 51.0 (28–66) | 64.8 (29–87) | 68.5 (50–85) | 64.0 (46–81) |
| Sex: male/female | 7/8 | 8/3 | 12/8 | 13/8 | 15/4 |
| LVEF %: mean, range | 63 (55–77) | 33 (22–40) | 25 (10–64) | 23 (10–60) | 20 (10–40) |
| Ischemic cardiomyopathy, % | 0 | 45.5 | 40.0 | 66.7 | 63.2 |
| Diabetes mellitus, % | 20.0 | 27.3 | 20.0 | 52.4 | 52.6 |
| Hypertension, % | 60.0 | 72.7 | 75.0 | 66.7 | 89.5 |
| Renal insufficiency, % | 0.0 | 8.3 | 15.0 | 19.1 | 57.9 |
| Atrial fibrillation, % | 0.0 | 27.3 | 25.0 | 42.9 | 63.2 |
| Loop diuretic, % | 20.0 | 18.2 | 45.0 | 81.0 | 79.0 |
| β-Blocker, % | 40.0 | 63.6 | 90.0 | 85.7 | 79.0 |
| ACE-I/ARB, % | 26.7 | 81.8 | 90.0 | 90.5 | 63.2 |
| Statin, % | 26.7 | 27.2 | 60.0 | 42.9 | 63.2 |
NYHA, New York Heart Association class; LVEF %, left ventricular ejection fraction percentage; renal insufficiency, serum creatinine ≥1.5; ACE-I, angiotensin converting enzyme inhibitor; ARB, angiotensin II receptor blocker; statin, HMG CoA reductase inhibitor.
Analysis of Blood Gene Expression Profiles
Affymetrix GeneChip U133Plus2.0 is a whole genome microarray containing more than 56,000 probe sets. A cross-gene error model was applied to the 87 blood expression profiles processed with GC-RMA. After we removed probe sets with unreliable measurements, ∼27,000 probe sets remained for further analysis.
Of these probe sets, survival analysis and subsequent multitest correction identified a total of 197 genes significantly (q < 0.2) associated with patient survival, and these were defined as “mortality genes.” Table 2 shows a representative sample of the mortality genes, listing those associated with known signaling pathways (a complete list of the mortality genes is part of an online data supplement). Ten mortality genes were found to be differentially expressed (P < 0.05) in the compensated heart failure group, and 113 mortality genes were differentially expressed in the decompensated heart failure group. Of these 123 genes that were differentially expressed in the heart failure groups, the majority, 89 genes, were downregulated in heart failure patients. Interestingly, 83 of the mortality genes while associated with patient survival were not differentially expressed in heart failure groups as defined by NYHA class.
Table 2.
Selected genes associated with survival time in heart failure patients
| Survival Analysis |
Ctrl vs. NYHA I–II |
Ctrlvs. NYHA III–IV |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene Title | Gene Symbol | Entrez Gene | P, Log-rank Test | q Value | HR (95% CI) | P | Fold Change | P | Fold Change | Pathways |
| Bruton agammaglobulinemia tyrosine kinase | BTK | 695 | 1.40E-04 | 5.45E-02 | 14.5 (3.7, 57.3) | 8.3E-02 | 1.11 | 1.1E-03 | 1.29 | 1, 2, 3, 7 |
| Calmodulin 1 | CALM1 | 801 | 2.42E-03 | 1.98E-01 | 0.1 (0.0, 0.4) | 2.3E-02 | 0.88 | 1.1E-03 | 0.83 | 1, 7, 9, 11 |
| CD22 molecule | CD22 | 933 | 2.25E-03 | 1.93E-01 | 21.6 (2.3, 207.3) | 3.5E-01 | 0.96 | 8.8E-01 | 0.99 | 7 |
| CD28 molecule | CD28 | 940 | 8.63E-05 | 4.23E-02 | 0.5 (0.3, 0.7) | 8.5E-01 | 1.01 | 1.5E-02 | 0.58 | 1 |
| CD3d molecule, delta | CD3D | 915 | 1.66E-03 | 1.63E-01 | 0.3 (0.2, 0.7) | 5.9E-01 | 0.95 | 7.1E-03 | 0.75 | 1, 8 |
| CD3e molecule, epsilon | CD3E | 916 | 7.88E-04 | 1.23E-01 | 0.3 (0.2, 0.6) | 7.9E-01 | 0.96 | 1.5E-03 | 0.68 | 1, 8 |
| CD3 g molecule, gamma | CD3G | 917 | 2.00E-03 | 1.81E-01 | 0.4 (0.2, 0.7) | 4.8E-01 | 0.90 | 3.4E-03 | 0.68 | 1, 8 |
| Cyclin-dependent kinase 2 | CDK2 | 1017 | 1.55E-04 | 5.64E-02 | 0.0 (0.0, 0.1) | 3.1E-01 | 0.95 | 1.7E-01 | 0.94 | 6 |
| Cyclin-dependent kinase inhibitor 2A | CDKN2A | 1029 | 1.55E-04 | 5.64E-02 | 8.8 (2.5, 30.6) | 1.4E-01 | 1.04 | 2.0E-02 | 1.12 | 6 |
| Heat shock 70-kDa protein 4 | HSPA4 | 3308 | 4.64E-04 | 9.67E-02 | 2.9 (1.5, 5.7) | 9.1E-01 | 1.00 | 7.1E-02 | 1.29 | 8 |
| IL-2-inducible T cell kinase | ITK | 3702 | 5.47E-04 | 1.04E-01 | 0.3 (0.2, 0.6) | 9.7E-01 | 1.00 | 2.9E-03 | 0.71 | 1, 3 |
| Inositol 1,4,5-triphosphate receptor, type 2 | ITPR2 | 3709 | 1.52E-03 | 1.56E-01 | 38.6 (3.9, 379.0) | 3.0E-02 | 1.12 | 1.8E-02 | 1.12 | 9 |
| Integrin, alpha 4 | ITGA4 | 3676 | 9.70E-04 | 1.32E-01 | 0.2 (0.1, 0.5) | 2.3E-01 | 1.10 | 7.5E-01 | 0.98 | 3 |
| Linker for activation of T cells | LAT | 27040 | 8.25E-04 | 1.26E-01 | 0.2 (0.1, 0.5) | 4.9E-01 | 0.96 | 1.7E-03 | 0.77 | 1, 2, 4 |
| Nuclear receptor coactivator 3 | NCOA3 | 8202 | 8.34E-04 | 1.26E-01 | 5.0 (1.7, 14.4) | 4.4E-01 | 0.98 | 5.3E-01 | 1.07 | 8 |
| Phospholipase C, gamma 2 | PLCG2 | 5336 | 8.92E-04 | 1.31E-01 | 27.7 (4.0, 191.5) | 1.5E-01 | 0.94 | 3.0E-01 | 1.06 | 2, 3, 4, 5, 7, 10, 11 |
| Protein kinase C, alpha | PRKCA | 5578 | 1.52E-03 | 1.56E-01 | 0.4 (0.2, 0.7) | 9.1E-01 | 0.98 | 6.2E-02 | 0.74 | 2, 3, 4, 5, 9, 10, 11 |
| Protein kinase C, beta 1 | PRKCB1 | 5579 | 1.45E-05 | 2.26E-02 | 29.1 (7.3, 117.0) | 4.2E-01 | 1.04 | 3.6E-01 | 1.08 | 2, 3, 4, 5, 7, 9, 10, 11 |
| Protein kinase C, eta | PRKCH | 5583 | 6.39E-04 | 1.11E-01 | 0.2 (0.1, 0.5) | 6.2E-02 | 0.91 | 7.3E-04 | 0.82 | 2, 3, 4, 5, 9 |
| SMAD family member 3 | SMAD3 | 4088 | 2.24E-03 | 1.92E-01 | 0.1 (0.0, 0.4) | 5.4E-01 | 0.94 | 4.0E-01 | 0.95 | 6 |
| Spleen tyrosine kinase | SYK | 6850 | 3.98E-05 | 3.24E-02 | 75.5 (10.2, 556.2) | 6.5E-01 | 1.02 | 1.9E-02 | 1.13 | 2, 4, 7 |
| T cell receptor alpha | TRA© | 6955 | 8.33E-04 | 1.26E-01 | 0.3 (0.1, 0.6) | 5.0E-01 | 0.98 | 1.1E-04 | 0.61 | 1 |
| TXK tyrosine kinase | TXK | 7294 | 1.54E-03 | 1.56E-01 | 0.6 (0.4, 0.8) | 3.7E-01 | 0.92 | 1.3E-03 | 0.61 | 1, 3 |
| v-myc myelocytomatosis viral oncogene homolog | MYC | 4609 | 8.89E-04 | 1.31E-01 | 0.3 (0.2, 0.6) | 8.3E-01 | 1.03 | 3.6E-01 | 0.90 | 5, 6, 10 |
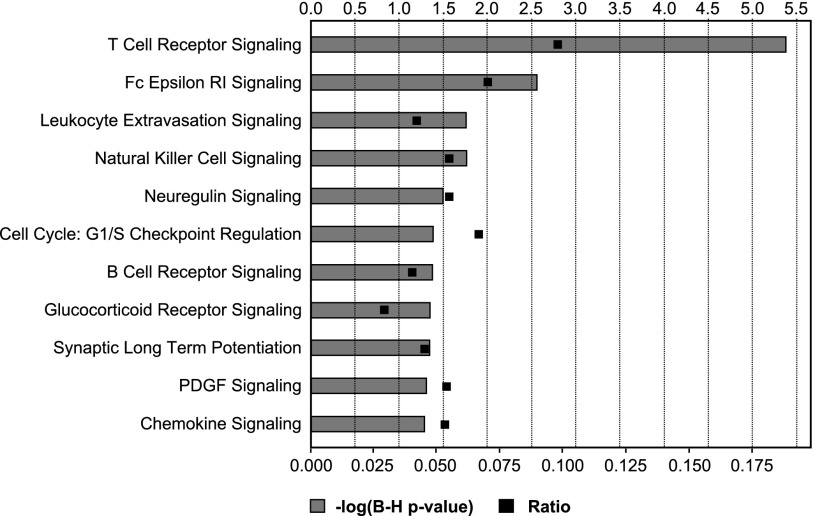
HR, hazard ratio; CI, confidence interval. Pathways: 1, T cell receptor signaling; 2, Fc Epsilon RI signaling; 3, Leukocyte extravasation signaling; 4, Natural killer cell signaling; 5, Neuregulin signaling; 6, Cell cycle: G1/S; 7, B cell receptor signaling; 8, Glucocorticoid receptor signaling; 9, Synaptic long term potentiation; 10, PDGF signaling; 11, Chemokin signaling.
A complete list of genes associated with survival is part of the online data supplement.
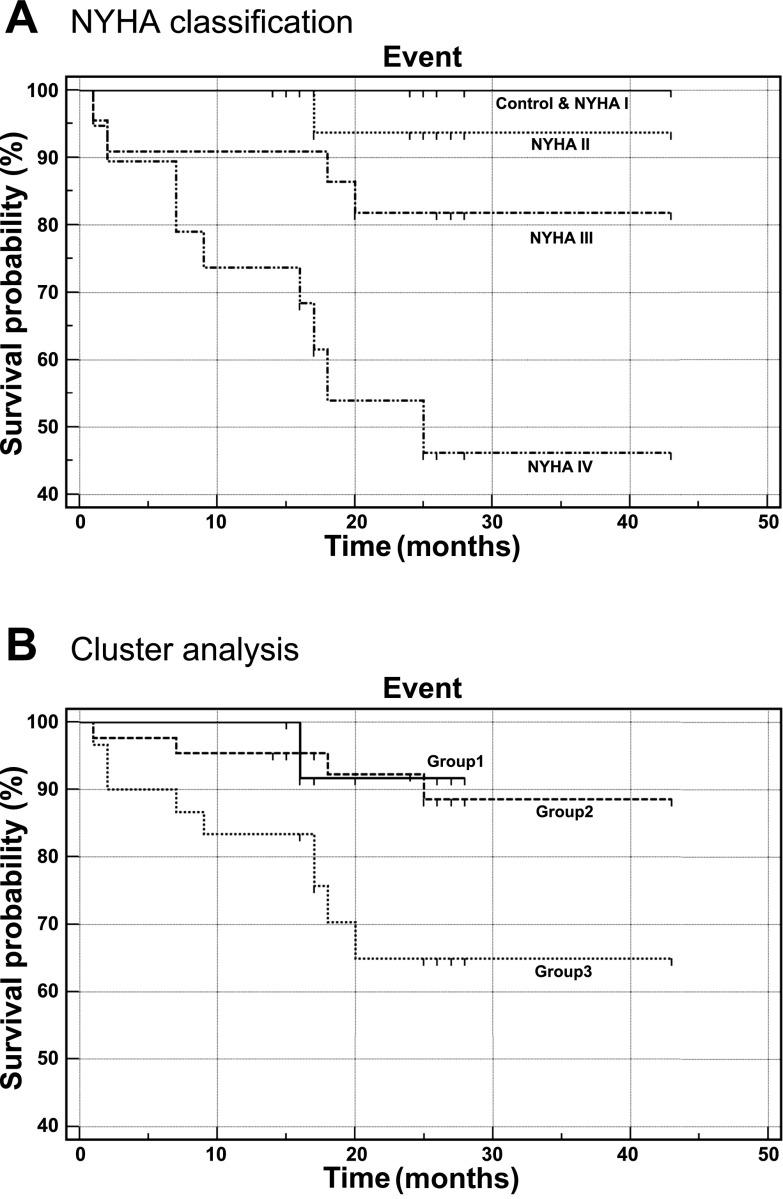
Functional categorization of the mortality genes showed that 11 canonical pathways were significantly overpresented (P < 0.05, Fisher's exact test, Benjamini-Hochberg multiple testing correction). The most significantly overpresented canonical pathway involved T cell receptor signaling (Fig. 1). Ten mortality genes were identified in this functional group. Cluster analysis of these 10 genes divided the 86 subjects into three groups (Fig. 2). Survival analysis demonstrated that the survival of study subjects was associated with disease severity as described by NYHA classification as well as with the three groups defined by the 10 T cell receptor signaling-related mortality genes (Fig. 3B). In contrast, in this patient cohort left ventricular ejection fraction was not predictive of survival (data not shown). There was no sex difference in survival (P = 0.2). Survival analysis for NYHA-defined groups (P = 0.0002) was enhanced by mortality gene clustering (P = 0.031) such that the risk stratification model using IRI provided a marked improvement in differentiating risk groups with the P value of a log-rank test being 0.0001, an increase from using either NYHA or the cluster analysis of 10 T cell receptor signaling-related genes (Fig. 4, Table 3). This approach provided good segregation of heart failure patients with 49% survival in the high risk group, 94% in the intermediate group, and 100% in the low risk group. Based on their gene expression clustering and confirmed by their survival to the end of the study period, nine heart failure patients were allocated to the low risk group.
Fig. 1.
Functional categorization of mortality genes. The 197 mortality genes identified in survival analysis were processed with the Ingenuity Pathway Analysis software. A total of 11 signaling pathways were found significantly overpresented in the mortality genes. Significantly overpresented pathways were defined as the ones with P values (Fisher's exact test) <0.05 after Benjamini-Hochberg multiple testing correction. Ratio was calculated between the number of mortality genes belonged to a specific pathway and the total number of genes belonged to that pathway.
Fig. 3.
Kaplan Meier survival plots of the study subjects. A: study subjects were classified by NYHA classification. B: study subjects were classified by the cluster analysis of the 10 mortality genes involved in T cell receptor signaling: the solid plot indicates group 1; the dashed plot indicates group 2; the dotted plot indicates group 3.
Fig. 4.
Kaplan Meier survival plots by combined risk analysis. Subject classified by low (solid), intermediate (dashed), and high risk (dotted) using integrated risk indexing as defined in Table 3, see text for discussion.
DISCUSSION
Neurohormonal activation in heart failure is well established and involves activation of the renin-angiotensin-aldosterone network and the sympathetic nervous system. Associated with this process is the enhanced production of cytokines and other inflammatory factors that have been directly implicated in the pathophysiological progression of heart failure (2, 6, 11, 15). These factors have both direct and indirect effects on cardiac remodeling and can also affect gene expression in other tissues (19). Due to the systemic nature of the disease, we tested the hypothesis that noncardiac tissue can exhibit specific gene expression signatures that associate with cardiovascular events.
By performing blood gene expression profiling analysis, we analyzed >27,000 transcripts in blood and demonstrated for the first time that blood gene expression varies as a function of patient outcomes (see online data supplement).1 A total of 197 mortality genes were discovered. Intriguingly, a large number (eighty-three) of these genes were not differentially expressed between NYHA groups. As heart failure outcomes are determined by multiple factors, some of these factors are likely independent of NYHA class as is made evident by the correlation of expression the above 83 genes with patient survival in this study. Of the mortality genes that are differential expressed across NYHA groups, the differences in gene expression lay almost exclusively between the control group and the decompensated group (NYHA class III–IV). Aronow et al. (1) showed little concordance of altered gene expression in four separate cardiac hypertrophy models in the compensated state, whereas a greater overlap of differentially expressed genes was observed between groups exhibiting more severe forms of cardiomyopathy. Similarly, blood gene expression among patients with heart failure likely exhibits a convergence of transcriptional regulation as the disease progresses.
We identified and characterized an expression signature of 10 genes involved in T cell receptor signaling. Perhaps most striking is that by integrating NYHA and the gene expression signature associated with patient outcomes, risk stratification can be further improved (P = 0.0001). The commonly used clinical parameter of left ventricular ejection fraction was, by contrast, not predictive of survival in this patient cohort. These data suggest that blood gene expression profiling may be of benefit in assessing patient prognosis. Importantly, this analytical approach is capable of segregating out the patients that are at the highest risk for cardiac mortality and thus may be of utility in guiding the medical management of patients with heart failure in the future.
Blood leukocytes were the primary source of RNA isolated in this study. Therefore, the differential expression of leukocyte genes in heart failure may or may not directly reflect the primary disease process. For example, our transcriptomic data may reflect relative lower lymphocyte concentrations, as have been found to predict survival in advanced heart failure (14). Irrespective, our data demonstrate that the expression profile can be used as a signature of heart failure severity. Previous studies investigating transcriptional gene expression of myocardial tissue in patients with heart failure have similarly demonstrated an overrepresentation of genes involved in inflammatory and cytokine signaling. In these studies, the gene expression in many of the inflammatory signaling pathways is enhanced, whereas in our study transcriptional expression of some of the genes involved cytokine signaling is reduced. Suppressed expression of genes in blood leukocytes may be a consequence of the overexpression and circulatory release of inflammatory factors by other tissues. Moreover, the high representation of inflammatory genes correlating with mortality shown in these data supports the concept that inflammation and cytokine signaling play an important role in the pathophysiology of heart failure (4). Many studies have demonstrated increased circulating mediators of inflammation in heart failure, including tumor necrosis factor-α, interleukin-2, and interleukin-6 (2, 4). In this study many of the intracellular signaling pathways involving the mortality genes, such as the Fc Epsilon R1 pathway and the T cell receptor signaling pathway, are directly involved in the production of tumor necrosis factor-α and interleukins.
Our results suggest that gene expression profiling in peripheral blood may predict prognosis in patients with heart failure. This approach, if validated in a larger cohort, could have direct implications in patient management. Several serum biomarkers have been shown to be elevated in heart failure and positively correlate with adverse patient outcomes. While these biomarkers may be predictive, they can be difficult to measure, with short half lives and wide sample variability, thus often limiting utility in standard clinical practice. While the commonly used heart failure biomarker brain natriuretic peptide has been demonstrated to a be sensitive discriminator in patients suspected of acute heart failure (10), a recent, large randomized trial showed brain natriuretic peptide-guided therapy to be of no benefit over standard clinical management (16). As such, accurate and reproducible biomarkers that can be used in the management and prognosis of patients with chronic heart failure remains an unmet need. As demonstrated in this study, gene expression profiling and in particular when combined with NYHA class may be of clinical utility in assessing the long term prognosis of patients with heart failure.
The small size of our control group and the larger number of female subjects in the control group limits our findings to some extent, and the study needs to be reproduced in a larger sample. However, no differences between males and females were found in mortality, our end point.
The death rate from cardiovascular diseases has steadily decreased over the past two decades. However, greater survival combined with the aging of the population has directly contributed to the increasing incidence of heart failure in western countries. Although treatment of heart failure has advanced, little progress has been made on the development of biological metrics that aid in predicting prognosis in patients with heart failure and optimizing their management. Blood gene expression profiling particularly when combined with the clinical assessment of NYHA class may provide a better delineation of patient's risk in heart failure, and thus be a valuable tool in optimizing medical management.
GRANTS
This work was supported in part by National Heart, Lung, and Blood Institute Grant 5R01HL-077637 (P. VanBuren).
DISCLOSURES
C.-C. Liew and J. Ma are both employees and C.-C. Liew is chief scientist of GeneNews, which partly sponsored this research.
Supplementary Material
ACKNOWLEDGMENTS
We thank Patricia Bauman and Kelly Begin for technical assistance.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1.Aronow BJ, Toyokawa T, Canning A, Haghighi K, Delling U, Kranias E, Molkentin JD, Dorn GW. Divergent transcriptional responses to independent genetic causes of cardiac hypertrophy. Physiol Genomics 6: 19–28, 2001. [DOI] [PubMed] [Google Scholar]
- 2.Aukrust P, Ueland T, Lien E, Bendtzen K, Muller F, Andreassen AK, Nordoy I, Aass H, Espevik T, Simonsen S, Froland SS, Gullestad L. Cytokine network in congestive heart failure secondary to ischemic or idiopathic dilated cardiomyopathy. Am J Cardiol 83: 376–382, 1999. [DOI] [PubMed] [Google Scholar]
- 3.Braunwald E. Biomarkers in heart failure. N Engl J Med 358: 2148–2159, 2008. [DOI] [PubMed] [Google Scholar]
- 4.Braunwald E, Bristow MR. Congestive heart failure: fifty years of progress. Circulation 102: IV14–IV23, 2000. [DOI] [PubMed] [Google Scholar]
- 5.Cunha-Neto E, Dzau VJ, Allen PD, Stamatiou D, Benvenutti L, Higuchi ML, Koyama NS, Silva JS, Kalil J, Liew CC. Cardiac gene expression profiling provides evidence for cytokinopathy as a molecular mechanism in Chagas' disease cardiomyopathy. Am J Pathol 167: 305–313, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kapadia S, Dibbs Z, Kurrelmeyer K, Kalra D, Seta Y, Wang F, Bozkurt B, Oral H, Sivasubramanian N, Mann DL. The role of cytokines in the failing human heart. Cardiol Clin 16: 645–56, 1998. [DOI] [PubMed] [Google Scholar]
- 7.Kittleson MM, Minhas KM, Irizarry RA, Ye SQ, Edness G, Breton E, Conte JV, Tomaselli G, Garcia JG, Hare JM. Gene expression analysis of ischemic and nonischemic cardiomyopathy: shared and distinct genes in the development of heart failure. Physiol Genomics 21: 299–307, 2005. [DOI] [PubMed] [Google Scholar]
- 8.Ma J, Dempsey AA, Stamatiou D, Marshall KW, Liew CC. Identifying leukocyte gene expression patterns associated with plasma lipid levels in human subjects. Atherosclerosis 191: 63–72, 2007. [DOI] [PubMed] [Google Scholar]
- 9.Ma J, Liew CC. Gene profiling identifies secreted protein transcripts from peripheral blood cells in coronary artery disease. J Mol Cell Cardiol 35: 993–998, 2003. [DOI] [PubMed] [Google Scholar]
- 10.Maisel AS, Krishnaswamy P, Nowak RM, McCord J, Hollander JE, Duc P, Omland T, Storrow AB, Abraham WT, Wu AH, Clopton P, Steg PG, Westheim A, Knudsen CW, Perez A, Kazanegra R, Herrmann HC, McCullough PA. Rapid measurement of B-type natriuretic peptide in the emergency diagnosis of heart failure. N Engl J Med 347: 161–167, 2002. [DOI] [PubMed] [Google Scholar]
- 11.Mann DL. Targeted anticytokine therapy and the failing heart. Am J Cardiol 95: 9C–16C, 2005. [DOI] [PubMed] [Google Scholar]
- 12.Mann DL, Bristow MR. Mechanisms and models in heart failure: the biomechanical model and beyond. Circulation 111: 2837–2849, 2005. [DOI] [PubMed] [Google Scholar]
- 13.Margulies KB, Bednarik DP, Dries DL. Genomics, transcriptional profiling, and heart failure. J Am Coll Cardiol 53: 1752–1759, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ommen SR, Hodge DO, Rodeheffer RJ, McGregor CG, Thomson SP, Gibbons RJ. Predictive power of the relative lymphocyte concentration in patients with advanced heart failure. Circulation 97: 19–22, 1998. [DOI] [PubMed] [Google Scholar]
- 15.Ono K, Matsumori A, Shioi T, Furukawa Y, Sasayama S. Cytokine gene expression after myocardial infarction in rat hearts: possible implication in left ventricular remodeling. Circulation 98: 149–156, 1998. [DOI] [PubMed] [Google Scholar]
- 16.Pfisterer M, Buser P, Rickli H, Gutmann M, Erne P, Rickenbacher P, Vuillomenet A, Jeker U, Dubach P, Beer H, Yoon SI, Suter T, Osterhues HH, Schieber MM, Hilti P, Schindler R, Brunner-La Rocca HP. BNP-guided vs symptom-guided heart failure therapy: the Trial of Intensified vs Standard Medical Therapy in Elderly Patients With Congestive Heart Failure (TIME-CHF) randomized trial. JAMA 301: 383–392, 2009. [DOI] [PubMed] [Google Scholar]
- 17.Roger VL, Weston SA, Redfield MM, Hellermann-Homan JP, Killian J, Yawn BP, Jacobsen SJ. Trends in heart failure incidence and survival in a community-based population. JAMA 292: 344–350, 2004. [DOI] [PubMed] [Google Scholar]
- 18.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toth MJ, Ades PA, Tischler MD, Tracy RP, LeWinter MM. Immune activation is associated with reduced skeletal muscle mass and physical function in chronic heart failure. Int J Cardiol 109: 179–187, 2006. [DOI] [PubMed] [Google Scholar]
- 20.Wu Z, Irizarry RA, Gentleman R, Murillo RM, Spencer F. A model based background adjustment for oligonucleotide expression arrays. Johns Hopkins University, Dept. of Biostatistics Working Papers: Working Paper 1, 2004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.