Abstract
Growth hormone is one of few pharmacologic agents known to augment milk production in humans. We hypothesized that recombinant human GH (rhGH) increases the expression of cell proliferation and milk protein synthesis genes. Sequential milk and blood samples collected over four days were obtained from five normal lactating women. Following 24 h of baseline milk and blood sampling, rhGH (0.1 mg/kg/day) was administered subcutaneously once daily for 3 days. Gene expression changes were determined by microarray studies utilizing milk fat globule RNA isolated from each milk sample. Following rhGH administration, DNA synthesis and cell cycle genes were induced, while no significant changes were observed in the expression of milk synthesis genes. Expression of glycolysis and citric acid cycle genes were increased by day 4 compared with day 1, while lipid synthesis genes displayed a circadian-like pattern. Cell cycle gene upregulation occurred after a lag of ∼2 days, likely explaining the failure to increase milk production after only 3 days of rhGH treatment. We conclude that rhGH induces expression of cellular proliferation and metabolism genes but does not induce milk protein gene expression, as potential mechanisms for increasing milk production and could account for the known effect of rhGH to increase milk production following 7–10 days.
Keywords: microarray, lactation, circadian
exclusive breast feeding is advocated for the first six mo of life because of its nutritional and immunologic benefits (1). However, many women struggle with establishing full lactation, despite being highly motivated and having improved breastfeeding techniques (20). Pharmacologic agents such as metoclopramide increase maternal prolactin (PRL) concentration but its use is limited due to efficacy issues and potential side effects to the mother and infant (20, 38, 39).
Despite the knowledge that multiple endocrine hormones impact mammary gland development and lactation (19, 42), their exact roles and interactions remain unknown. Growth hormone (GH) is lactogenic in many species (31, 42) and improves commercial milk production. GH is galactopoietic in several species, including rats (10), mice (21), ruminants (6, 7, 14) and primates (30). Recombinant human GH (rhGH) administration for seven days increased milk production by 8–35% in normal lactating women (28, 38), up to 36% in women with lactation insufficiency (39) and 31% in mothers of preterm infants (20).
GH exerts many of its physiological functions (46) by regulating the transcription of genes for a variety of proteins, including insulin-like growth factor 1 (IGF-1), transcription factors and metabolic enzymes, well-described in the mouse liver (46) and in the human skeletal muscle (49). However, the mechanism(s) by which rhGH acts on the mammary gland to increase milk production remains unknown.
We recently reported the isolation of RNA from the milk fat globule (MFG) (34) and showed that the MFG is a unique source to study metabolic gene expression in the mammary epithelial compartment (MEC) (33). Preliminary studies in our lab using quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) failed to show increased expression of the milk protein gene, alpha lactalbumin (LALBA), a purported key regulator of lactose synthesis and milk volume, following rhGH administration to normal lactating women. The present study was undertaken to determine changes in MEC gene expression, using microarray analysis, in response to three days of rhGH administration in normal, lactating women. We hypothesized that rhGH will increase the expression of milk protein and cell proliferation genes. Data from the first 24 h of the study (pre-rhGH administration) has been published previously but are included here for the evaluation of changes from baseline (33).
MATERIALS AND METHODS
Subjects
Following approval from the Institutional Review Board (IRB) and the Scientific Advisory Committee of the General Clinical Research Center (GCRC) at Baylor College of Medicine, written informed consent was obtained from six healthy lactating women. All participants underwent screening tests to exclude diabetes, anemia, renal or hepatic dysfunction, and current pregnancy. All were 18–35 yr old and between 6–12 wk postpartum. Each of the women had singleton uncomplicated pregnancies and delivered at term (≥37 wk). Their infants were healthy and being exclusively breastfed at the time of the study.
The first subject enrolled was a 21 yr old Hispanic woman with a body mass index (BMI) of 30 kg/m2. During the study, the subject was on an ad libitum diet. After the 2nd dose of rhGH, she developed hyperglycemia (>200 mg/dl). The last dose of rhGH was withheld, and we documented a return to normal blood sugars 36 h after the 2nd dose of rhGH. The subject had an HbA1c of 5.8% (n = 4.3–5.7%), which suggests that this woman may have had unrecognized glucose intolerance and the added insulin resistance induced by the rhGH treatment resulted in frank hyperglycemia. This incident was reported to the IRB and Office for Human Research Protections at the National Institutes of Health. The subject was given dietary advice and scheduled to visit her primary care physician after discharge. As a result, the inclusion criteria were modified to include women with a BMI ≤ 27 kg/m2 and a normal oral glucose tolerance test on screening. Thus, all our reported results were from the five women who met these criteria and completed the study.
Study Design
Following admission to the GCRC, subjects were maintained on a regular diet (35 kcal/kg/day). Water and calorie-free drinks were available ad libitum. The women and their infants were admitted to the Texas Children's Hospital GCRC on the day of the study (day 1, 0800). After a light breakfast, the mothers breastfed their infants from both breasts. An intravenous line was inserted into the ante-cubital vein under Ela-Max analgesia (Ferndale Laboratories, Ferndale, MI) and was infused with 0.9% NaCl at low rates to keep the vein open for blood sampling. Blood samples (2.5 ml) were collected every 3 h (q3h, 30 min after the start of breast pumping) until 0830 on day 3. At 1100 and q3h until 0800 on day 3, breast milk was collected (see below). On day 3, milk and blood sample collections were decreased to q6h until 0800 of day 4. Blood was centrifuged at 3,000 rpm for 10 min at 4°C, the plasma was separated and transferred to a new tube then stored at −80°C. rhGH (Norditropin; Novo Nordisk, Princeton, NJ) at 0.1 mg/kg/day was administered by subcutaneous injection once daily at ∼0800 for 3 days. Premeal blood glucose determination was performed by YSI 2300 Stat Plus glucose analyzer (YSI, Yellow Springs, OH) after the administration of rhGH commenced to monitor for hyperglycemia. Plasma IGF-1 was determined using the Quantikine IGF-1 Immunoassay via standard solid-phase ELISA methods (R&D Systems, Minneapolis, MN). Plasma PRL, insulin, and C-peptide were measured using an electrochemiluminescence assay (Elecsys 1010; Roche Diagnostics, Indianapolis, IN).
Milk Collection
Ten milliliters of milk was collected simultaneously from both breasts using a standard breast pump (Playtex Embrace, Dover, DE). The infants then breastfed (∼10–12 min each side), and milk collection was resumed into the same bottles until the breasts were emptied. The milk bottles, weighed before and after milk collection on a Mettler AE50 balance (Mettler-Toledo, Greifensee, Switzerland), were kept on ice until processed. Total milk volume was calculated as the sum of the volume of milk pumped and the change in the infant's weight (g) over a feeding, based on the density of mature human milk (density = 1.031 g/ml). Approximately 10 ml milk was transferred into sterile, RNase-free tubes, and centrifuged (Sorvall Legend T, Germany) at 3,000 rpm for 10 min at 4°C. The supernatant fat layer was transferred using a sterile spatula to a new tube. One ml of TRIzol (Invitrogen Life Technology, Carlsbad, CA) was added prior to storing at −80°C.
Milk Samples
RNA isolation.
Total RNA was isolated from TRIzol-treated milk fat following the manufacturer's suggested procedures and as previously described (10). RNA concentration was measured using NanoDrop spectrophotometer (NanoDrop Technologies) and quality was assessed using the Experion RNA StdSens Analysis Kit manufactured (Bio-Rad Laboratories, Hercules, CA).
Microarray Study Design
cRNA amplification and expression microarray.
The methods utilized for cRNA amplification and microarray expression analyses were identical to that previously published (33, 34) using human Ref-8 BeadChips and the BeadStation system from Illumina (San Diego, CA). Microarrays were prepared according to the manufacturer's guidelines. Total RNA was converted to cDNA by reverse transcription using ArrayScript reverse transcriptase and T7-(dT)24 primers, followed by second-strand synthesis to generate double-stranded cDNA (Ambion). After purification, the cDNA was converted to biotin-labeled cRNA, hybridized to the BeadChip (Illumina), and stained with streptavidin-Cy3 for visualization. Over the 96 h study, 139 microarrays were analyzed [28 time points × 5 subjects (biologic replicates) − 1 missing sample]. Quality standards for hybridization, labeling, staining, background signal, and basal level of housekeeping gene expression for each chip were verified. After scanning the probe array, we analyzed the resulting image with BeadStudio software (Illumina).
Microarray data analysis.
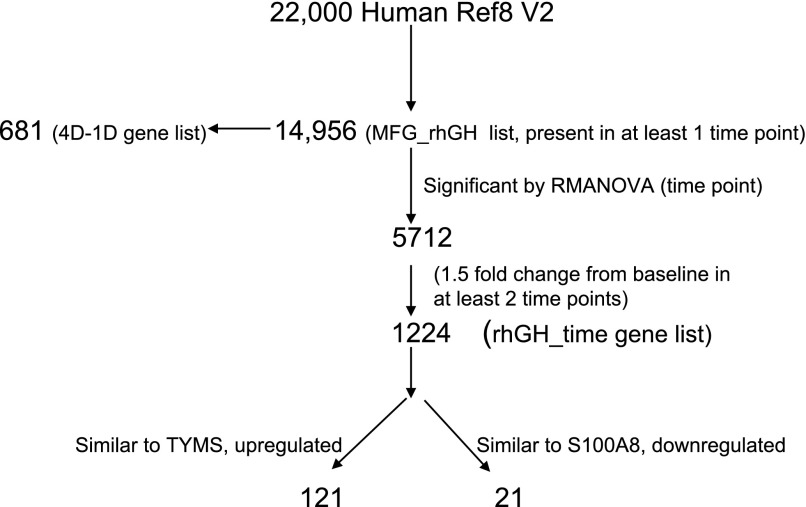
Raw microarray data were exported to GeneSpring GX 9 (Agilent Technologies, Santa Clara, CA). Signal data was q-spline normalized and filtered based on criteria that flags for each gene being present in all subjects in at least one time point. Expression data for the five subjects at each time point were averaged. An algorithm for data analysis is presented in Fig. 1. Of the ∼22,000 gene probes in the Human Ref 8 bead chip, 14,956 were present in the MFG (MFG_rhGH gene list). We used the following software for data analyses: GeneSpring GX 9, Ingenuity Pathways Analysis (IPA, Ingenuity Systems, http://www.ingenuity.com), and EDGE (50). GeneSpring GX9 was used to examine gene ontology, fold changes, and for repeated-measures ANOVA (RMANOVA). IPA was utilized to determine the networks, canonical pathways and functions associated with each gene list. The EDGE software was used for time-course analysis and for creating heat maps based on hierarchical clustering [correlation, distance = (1 − p)].
Fig. 1.
Bioinformatics flowchart. From the ∼22,000 genes and transcripts on the Illumina chip, 14,956 were expressed in the milk fat globule (MFG) RNA during the 96-h study. 4D-1D is the list of genes expressed on day 4 but not on day 1. Recombinant human growth hormone (rhGH)-time is the list of genes that were significantly changed according to repeated-measures ANOVA (RMANOVA) and at least 1.5-fold time from baseline in at least 2 time points. This list was analyzed according to fold change from baseline and time (hours) from rhGH administration. TYMS, thymidylate synthase and S100A8 (S100 calcium binding protein A8).
We analyzed this data set by creating the following gene lists: 1) genes not expressed in the first 24 h but expressed after rhGH treatment and 2) genes that changed by time (hours) from rhGH treatment. Each gene list was analyzed according to ontology, cellular functions, pathways, and networks. Expression data from the MFG_rhGH gene list were overlaid on the GH, IGF-1, and PRL signaling canonical pathways as well as on the Cell Cycle G2/M pathway to illustrate changes in gene expression across time. In addition, we evaluated 3) core circadian clock gene expression over the 4-day treatment period and 4) the expression of genes involved in metabolic functions associated with milk synthesis as identified by Rudolph et al. (47) and used in our previous publication (33) over the 4-day treatment period.
Statistical Analysis
For the plasma assays, analyses were performed by one-way ANOVA using SPSS for Windows version 16 (SPSS, Chicago, IL). RMANOVA on GeneSpring GX9 with Benjamini-Hochberg correction for multiple testing was used to determine significantly changed genes with a P value < 0.05. EDGE software uses an Optimal Discovery procedure and significant genes had a P < 0.05. IPA generated networks and pathways based on the gene lists uploaded. The significance of the pathways represented was determined by calculating the number of genes from the dataset that met the expression value cutoff that map to the pathway divided by the total number of genes that exist in the canonical pathway displayed. To maximize the sensitivity of our analyses, a Fisher's exact test was then used to calculate a P value determining the probability that the representation of the genes in the dataset compared with the canonical pathway is greater than that explained by chance alone. For our analyses, a P value <0.05 was considered significant.
RESULTS
Subject Characteristics
Five women, 25 ± 3 yr (mean ± SE), with a height of 155 ± 2 cm, weight of 59.5 ± 1.3 kg, BMI of 24.0 ± 0.5 kg/m2 were included.
Milk Volume
Milk volume did not change during the 4-day study, 865 ± 116 (day 1) and 808 ± 84, 911 ± 79 and 903 ± 70 ml on days 2, 3, and 4, respectively.
Plasma Assays
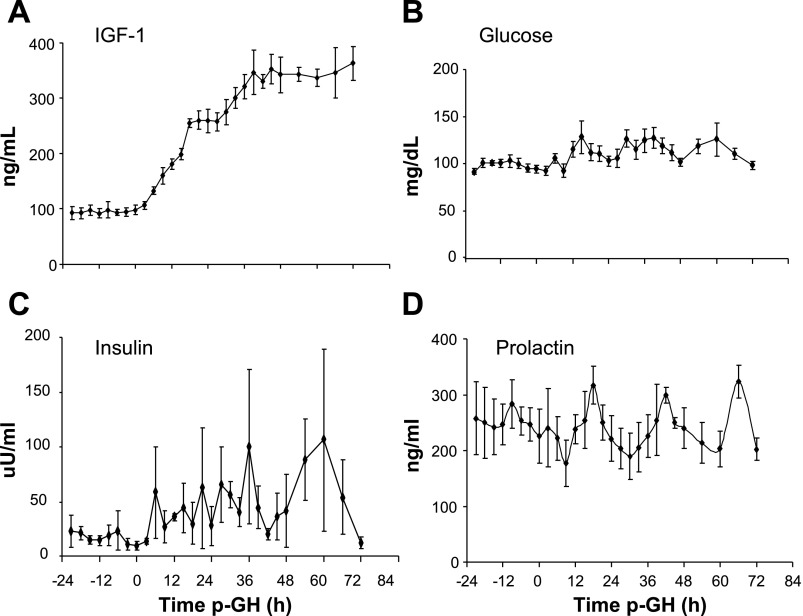
Plasma IGF-1 increased from 95 ± 1 to 347 ± 6 ng/ml (P < 0.0005) (Fig. 2A). Plasma glucose increased (P < 0.05) slightly after rhGH administration, from 5.3 ± 0.1 on day 1 to 6.1 ± 0.3 mmol/l on day 3, respectively (Fig. 2B). Plasma insulin increased (P < 0.05) over threefold from 17 ± 3 to 65 ± 21 uU/ml, (P < 0.05) (Fig. 2C). In contrast, the pattern of daily suckling-induced plasma PRL concentrations did not change during the study, 251 ± 6 and 236 ± 34 ng/ml (Fig. 2D) on day 1 and 4, respectively.
Fig. 2.
Plasma hormone concentrations (mean ± SE). Plasma IGF-1 increased 3-fold (A); plasma glucose (B) and plasma insulin (C) increased significantly after rhGH administration. D: plasma prolactin did not increase and maintained a diurnal peak. Significant values had P < 0.05, RMANOVA of the average values per day of rhGH treatment.
Microarray Data Analysis
Baseline vs. 4 days (4D-1D gene list).
Of the ∼22,000 gene probes in the Human Ref 8 BeadChip, 14,956 were expressed in MFG, (MFG_rhGH gene list). These data have been deposited in National Center for Biotechnology Information's Gene Expression Omnibus (GEO) (13) and are accessible through GEO series accession number GSE15208 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE15208). The MFG_rhGH gene list includes the 14,070 genes reported in our previous publication at baseline (prior to rhGH treatment) plus an additional 681 unique gene probes whose expressions were altered subsequent to rhGH administration (4D-1D, Supplement 1).1 The most enriched networks in this list of 681 genes were for cell cycle, DNA replication, recombination, and repair, and cancer.
Analysis according to time (hours) from rhGH treatment (rhGH_time gene list).
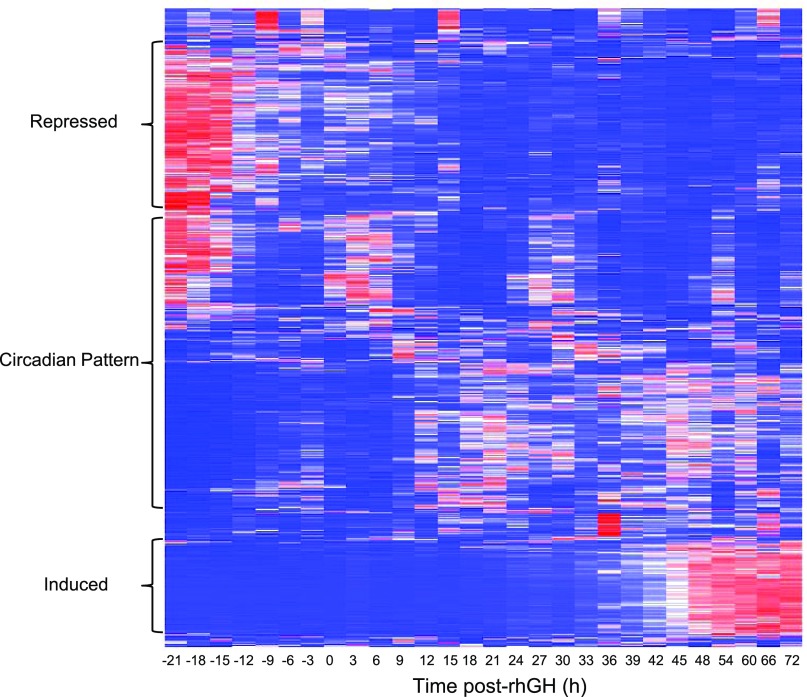
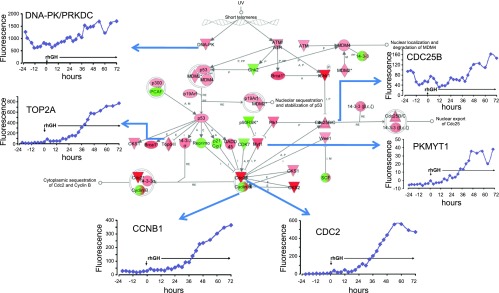
We found 1,224 gene probes (rhGH_time gene list, Supplement 2) to be changed ≥1.5-fold during the 96 h study. The heat map (Fig. 3) depicts the main clusters that illustrate changes associated with rhGH treatment. Each row represents a gene and each column its expression relative to baseline across time. The top portion of the heat map shows genes initially highly expressed (in red) and then decreased, while the bottom shows genes with initial low expression (in blue) that were induced (essentially in the last 24–30 h of the study). Interestingly, the middle of the heat map reflects genes that exhibit a circadian pattern. The top cellular functions for this rhGH_time gene list were: 1) cell cycle (P = 6.6 E-19), 2) cellular growth and proliferation (P = 7.4 E-15), 3) cell death (P = 7.0 E-14), 4) DNA replication, recombination and repair (P = 4.7 E-10), and 5) cell signaling (P = 1.9 E-7). To illustrate, the cell cycle pathway at G2/M phase (pathways from IPA) is shown on Fig. 4. Colored nodes indicate genes that were expressed and detected in the MEC. Many genes in this pathway were induced by rhGH treatment (shown in inset).
Fig. 3.
Heat map of the 1,224 genes that were significantly changed by rhGH administration (P < 0.05), ≥ 1.5-fold compared with baseline in at least 2 time points. Each column denotes the time from rhGH treatment and each row represents a gene. In blue are low-expressing genes, and in red are the highly expressed genes compared with the baseline sample. Data are the average fluorescence of 5 subjects. See Supplement 2 for a list of these genes.
Fig. 4.
Genes of the cell cycle expressed in the MFG before and after rhGH treatment. Ingenuity analysis of the Cell Cycle at G2/M phase. Colored nodes indicate genes that were expressed and detected in the microarray. Green indicates lower expression, while red indicates higher expression at the end of the study compared with the baseline, respectively. Inset graphs [Fluorescence(y-axis) vs. time p-rhGH (x-axis)] are genes whose expressions were altered after rhGH administration.
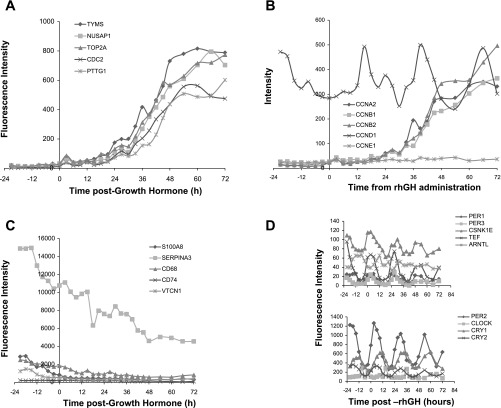
Of the genes induced by rhGH treatment, TYMS (thymidilate synthase) had the largest (∼11-fold) increase on day 4 compared with baseline expression (Fig. 5A). A total of 121 genes were similarly expressed with a Pearson correlation of 0.95 to TYMS. This induced list includes genes that function in 1) the cell cycle, 2) DNA replication, recombination, and repair, 3) cellular movement, 4) cellular assembly and organization, and 5) cellular growth and proliferation. On the other hand, those whose gene expression was repressed following rhGH administration, S100 calcium binding protein A8 (s100A8), experienced the greatest change (3.4-fold reduction in gene expression) on day 4 compared with day 1 (Fig. 5C). The 21 genes that were repressed in a similar manner to s100A8 function in cellular movement, cell cycle, free-radical scavenging, and cell death. Figure 5A shows five (TYMS, NUSAP1, TOP2A, CDC2, and PPTG1) of the 121 genes induced by rhGH treatment. Figure 5B shows the expression of induced cyclin genes (CCNA2, CCNB1, CCNB2, CCNE1). Figure 5C shows five (S100A8, SERPINA3, CD68, CD74, and VTCN1) of 21 genes repressed by rhGH treatment.
Fig. 5.
A: microarray expression (fluorescence intensity) of 5 (of 121) genes that increased in response to rhGH treatment. Data are the average from 5 subjects. TYMS, NUSAP1, TOP2A, CDC2, PTTG1 were among the most highly induced genes. B: microarray expression data on cyclin genes CCNA2, CCNB1, CCNB2, CCND1, and CCNE1. Of these cyclins, CCNA2, CCNB1, CCNB2, and CCNE1 were induced by rhGH treatment, while CCND1 expression was not increased further. C: microarray expression (fluorescence intensity) of 5 (of 21) genes downregulated in response to rhGH treatment - S100A8, SERPINA3, CD68, CD74, VTCN1. D: core circadian clock gene expression in the mammary epithelial compartment before and after rhGH treatment. Circadian expression was not changed by rhGH administration. Values are the means at each time point for the 5 subjects. Note different scales on the y-axis.
Circadian clock genes.
The expressions of the core circadian clock genes were examined in the MFG (Fig. 5D). PER1, PER3, CSNK1ε, TEF, ARNTL, PER2, CLOCK, CRY1, and CRY2 were all detected and their expression patterns were not altered by 3 days of rhGH treatment.
Milk synthesis and metabolic genes.
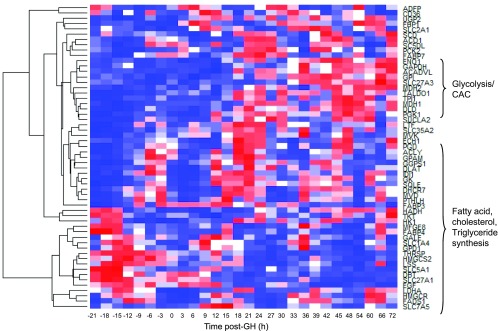
rhGH treatment did not change the expression of any genes previously identified as being involved in milk synthesis >1.5-fold. However, by EDGE time-course analysis, 57 of the 98 milk synthesis genes displayed a time-dependent gene expression pattern (P < 0.05, Fig. 6). Several genes involved in glycolysis and the citric acid cycle were more highly expressed towards the end of the study (e.g., ENO1, GAPDH, GPI). Genes involved in fatty acid, cholesterol and triglyceride synthesis (e.g., FADS1, HMGCR, GPAM) exhibited a circadian pattern of expression. Most milk protein genes were not changed by rhGH treatment. Figure 7 depicts the canonical signaling pathways for GH, IGF-1, and PRL overlayed with the expression data after rhGH treatment. Note that GHR expression is absent; IGF1-R is present but not upregulated by rhGH treatment. Cell growth and cell cycle progression nodes were induced (Fig. 7, B and C), whereas the b-casein gene, a milk-protein gene, was not induced (Fig. 7C).
Fig. 6.
Milk synthesis genes significantly (P < 0.05) regulated as analyzed by EDGE time-course analysis. Hierarchical clustering, correlation (distance = 1 − p) generated on the EDGE software. Genes involved in glycolysis and citric acid cycle (CAC) were more highly expressed toward the end of the study. Genes involved in the synthesis of fatty acid, cholesterol, and triglycerides exhibited a circadian pattern.
Fig. 7.
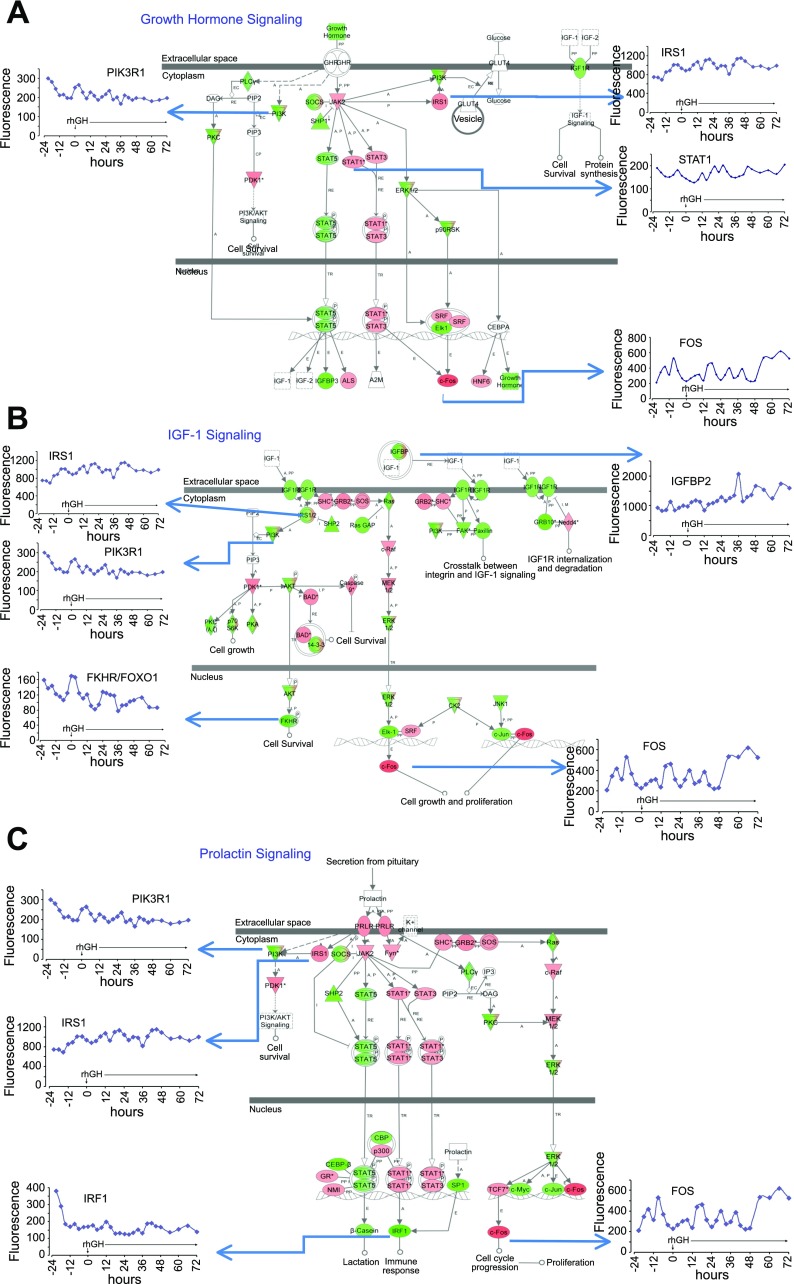
A: growth hormone signaling in the mammary epithelial compartment (MEC). Figure is taken from Ingenuity Pathways. Note that GLUT4 is known not to be expressed in the mammary epithelium and is not expressed in our MEC samples. Colored nodes indicate genes that were expressed and detected in the microarray. Green indicates lower expression, and red indicates higher expression at the end of the study compared with baseline, respectively. Inset graphs [Fluorescence (y-axis) vs. time p-rhGH (x-axis)] are genes whose expressions were altered after rhGH administration. B: insulin-like growth factor (IGF-1) Signaling in the MEC. Figure is taken from Ingenuity Pathways. Colored nodes indicate genes that were expressed and detected in the microarray. Green indicates lower expression and Red indicates higher expression at the end of the study compared with baseline, respectively. Inset graphs [Fluorescence (y-axis) vs. time p-rhGH (x-axis)] are genes whose expressions were altered after rhGH administration. C: prolactin signaling in the MEC. Figure is taken from Ingenuity Pathways. Colored nodes indicate genes that were expressed and detected in the microarray. Green indicates lower expression and Red indicates higher expression at the end of the study compared with baseline, respectively. Inset graphs [Fluorescence (y-axis) vs. time p-rhGH (x-axis)] are genes whose expressions were altered after rhGH administration.
DISCUSSION
We previously reported gene expression in the MFG during the initial 24 h of this study, which demonstrated that the transcription products in the MFG reflect those of the MEC (33, 34). As with the mouse mammary gland (14), the MFG expressed milk protein transcripts at very high levels and most of the genes required for glycolysis, citric acid cycle, and fatty acid synthesis. In the present report, we describe the changes in gene expression following 3 days of high-dose rhGH.
Multiple hormones, including GH, PRL, insulin, and steroid and thyroid hormones, are necessary to ensure successful lactation (42). Because this study was performed in normal, lactating women, with presumed normal hormonal secretion, there is an inherent difficulty in attributing and separating the observed effects (in milk production and gene expression changes) to a unique action of a single hormone. There is a complex interplay in the metabolic and proliferative actions of GH and IGF-1, with GH increasing plasma IGF-1, which subsequently mediates many of GH's systemic effects. For a more in-depth discussion, we direct you to a review by Mauras and Haymond (36). In addition, the receptors for both GH and PRL belong to the receptor cytokine superfamily and may share similar STAT signaling pathways (Fig. 7, A and C) (18). Human GH activates both the GHR and the PRLR, while PRL mediates its effects through the PRLR (53). Although PRL is essential for lactation, plasma PRL concentrations do not correlate with the rate of milk secretion (42).
In this study, we opted to use a supraphysiological dose of rhGH (0.1 mg/kg/day), a dose used in a previous study by our group (23). This higher dose was given for a shorter duration, compared with other studies looking at the effects on milk production (20, 28, 38, 39) to elicit maximal response in gene expression in a shorter amount of time that could be clearly associated with the rhGH administration. In the five women enrolled in this study, milk production was unchanged after 3 days of GH treatment but each woman reported subjective feeling of increased breast fullness within 24–48 h after rhGH treatment as we previously reported as well (24). Three days of rhGH treatment did not change PRL concentrations or pattern of secretion (Fig. 2D) but significantly raised the plasma insulin and IGF-1 concentrations as expected (Fig. 2, A and C). Although we did not measure milk IGF-1 concentration to determine local changes in hormone concentration, we would expect to see much lower concentrations with smaller changes compared with plasma IGF-1 (9). Serum GH concentrations were not measured in this study, but our group has previously shown that GH treatment in nonlactating subjects increased only slightly (from 4.7 ± 1.5 to 6.9 ± 0.6 ng/ml) (23), compared with serum PRL at 200–300 ng/ml in lactating women, therefore we expect minimal binding of GH to the PRLR.
The Ingenuity diagrams for GH, IGF-1, and PRL signaling pathways were utilized and overlaid with the expression values of the MFG_rhGH gene list (Fig. 7, A–C). The nodes are colored according to their expression at the end of the study relative to the beginning of the study. Red nodes indicate higher expression, while green nodes indicate lower expression compared with the first sample obtained. The GHR mRNA is absent, but molecules downstream (e.g., FOS and IRS1) are expressed and regulated (Fig. 7A). In contrast, the IGF1R and the PRLR are present (Fig. 7, B and C) and do not change significantly during the study period. We have shown that the mRNA for IGF-1R, but not GHR nor IGF-1, is expressed in the MFG, suggesting the absence of the growth hormone receptor (GHR) protein in the MEC (34). Although the GHR has been reported to be expressed in both the stroma and MEC in mouse mammary gland (25) and in in vitro human mammary epithelial cell lines (43), we did not detect GHR in the MFG samples (34) from normal, lactating women. Similarly in vitro, INS-1 cells downregulate GHR expression with the addition of bGH and rPRL (4). In addition, Ilkbahar et al. (25) showed that GHR expression in the mouse is lower in the mammary epithelium compared with the stroma and that GHR expression was at its lowest during lactation. This latter observation could be attributable to a reduction in the relative stromal fraction due to the increased parenchymal content during lactation. Thus, because of the absence of the GHR in the MEC, we believe that the colored nodes in Fig. 7A downstream to the GHR are likely due to stimulation of other cytokines, likely PRL, which shares similar JAK/STAT signaling pathways with GH. Because serum PRL levels are already maximal during lactation and did not increase further during this 4-day study, the increase in the expression of cell cycle genes (e.g., FOS, Fig. 7, B and C) are likely to be attributable to increased IGF-1 signaling.
GH increases IGF-1 production from the liver and other tissues. We propose that the human mammary stroma produces IGF-1, which acts by a paracrine effect on the MEC mediating the effects of rhGH (42). Kleinberg et al. (29) similarly proposed that GH in the presence of estrogen induces IGF-1 from the stroma, which then acts through specific IGF-1 epithelial cell receptors to drive ductal development in the rat. A similar mechanism may also be at work in the mammary gland during lactation since overexpression of IGF-I in the mammary tissue of transgenic mice during prolonged lactation increased milk production (22) as did the administration of either recombinant LR3-IGF-I or recombinant murine GH (21). Although these data make the above scenario plausible, they do not prove whether the effect of rhGH are indirectly mediated by IGF-1. In addition, IGF-1 signaling from the stroma may be usurped in postpubertal stages in the mouse (45), thus high circulating IGF-1 may be responsible for the IGF-1 signaling in this study.
In rats, GH synergizes with PRL to maintain lactation and regulate milk composition (10, 16). Likewise in mice, treatment with recombinant murine GH increased milk production and mammary gland weight during prolonged lactation (21). Mouse GH also increases milk protein secretion in mouse explants and MEC cultures (17, 35). However, GH treatment of PRLR+/− mice during late pregnancy failed to increase mammary tissue β-casein content or acetyl-CoA carboxylase-α activity but instead increased mammary development and DNA content, suggesting that during late pregnancy and early lactation, GH supports proliferation rather than differentiation (3). These results are also consistent with studies conducted in HC11 cells (41) derived from midpregnant mice and with our observation of increased cell cycle gene expression without increased milk synthesis gene expression or milk production after 3 days of rhGH treatment. It is possible that the effects of GH on mammary epithelial proliferation versus milk synthesis and secretion depend on the stage of lactation and duration of treatment.
Many of the 681 unique genes not expressed in the baseline 24 h sampling but induced by rhGH were involved in the cell cycle signaling pathway. In response to rhGH administration, ∼121 genes were induced (Fig. 5A), and those with the largest changes are involved with DNA synthesis and the cell cycle. For example, TYMS catalyzes the methylation of deoxyuridylate to deoxythymidylate critical for DNA replication and repair. NUSAP1 (nucleolar and spindle associated protein 1) is a microtubule-associated protein. It associates with chromosomes and promotes the organization of mitotic spindle microtubules. TOP2A (DNA topoisomerase II) encodes an enzyme that controls and alters the topological states of DNA during transcription.
Repeated sampling in this study allowed us to explore how the GH/IGF-1 axis influences cyclin gene expression. Cyclin genes (CCNA2, CCNB1, CCNB2, and CCNE1) were induced in the last 36 h of the study (Fig. 5B); CCND1, on the other hand, demonstrated a circadian pattern of expression. The molecular events that control the cell cycle are ordered and directional and the cyclins determine a cell's progress through the cycle. It is known that GH induces cyclins D1 and D3, which push the cell from G1 to S phase via activation of the NF-κB pathway (5, 26). IGF-1's role in cell proliferation and survival is mediated primarily via the PI3-kinase-Akt pathway (5). IGF-1 induces cyclin D1, E, A2, and B1 in ventricular myocytes and skeletal muscle satellite cells (11, 44). In the mouse whole mammary gland, IGF-1 induced type D cyclins, as well as cyclins E, A2, and B1 (51). Although IGF-1 may play a role in cell-cycle progression at several phases (11, 48, 51), it's reportedly more important for G2/M progression in the mouse uterus and in the mammary epithelium in the intact mammary gland cultured in vitro (2, 51). Figure 4 shows Ingenuity canonical cell cycle pathway at G2/M phase and the expression data in response to rhGH treatment. This figure clearly shows increased expression of target genes (cdc2 and cyclin B). Therefore, in our study, we suggest that an increase in systemic and local GH and IGF1 concentration would then promote cellular proliferation and thus increase the epithelial compartment capable of increased milk production.
GH administration repressed 21 genes (Fig. 5C) S100A8 (S100 calcium binding protein A8) involved in chemotaxis, and apoptosis was decreased the most. SERPINA3 (Serpin peptidase inhibitor or alpha-1-antichymotrypsin) codes for a plasma protease inhibitor and is required for the regulation of leukocyte proteases and in inflammatory responses (24). CD68 (CD68 molecule) encodes a protein member of the lysosomal/endosomal-associated membrane glycoprotein family and is a marker of tissue macrophages. It is known that breast milk contains a mixture of bioactive components such as cytokines and inflammatory cells that may influence the infant's immune development (15). The decrease in CD68 gene expression and others genes involved in inflammation may be due to MEC proliferation resulting in a decrease or dilution of macrophage mRNA in breast milk. Macrophages may also function to clear apoptotic cells, but since rhGH treatment increased mammary epithelial survival and/or proliferation, macrophage recruitment into the mammary gland may have decreased. We previously checked for the expression of CD68 and CD11c by qRT-PCR to determine macrophage contamination in single samples of 10 and 100 ng of MFG RNA. The expressions were negligible. Recent work has suggested that immediately postweaning, the mammary epithelial cell itself may differentiate into a phagocytic epithelial phenotype, involved in clearing apoptotic mammary epithelial cells and residual milk products (40). These cells express markers that are traditionally thought of as macrophage or inflammatory markers. We could on the basis of these data conjecture that rhGH increases cell cycling and proliferation and either absolutely or relatively decreases apoptosis resulting in a decrease in phagocytic cells, explaining the downregulation of traditional inflammatory markers.
Frequent sampling also enabled the analysis of circadian genes (Fig. 5D). Many biological processes in mammals and in mammalian cells are subject to daily oscillations, and some of these are controlled by endogenous oscillation mechanisms governed by the circadian clock. The master clock resides in the suprachiasmatic nuclei, which synchronizes circadian oscillators in peripheral tissues. Circadian clock gene expression has been demonstrated in almost every cell type (8), and we reported its expression in human lactating mammary epithelium during the first 24 h of this study (33) Although Sjogren et al. (49) reported changes in CLOCK and PERIOD expression in the human skeletal muscle after rhGH treatment, we did not observe significant alterations in circadian gene expression following rhGH administration in MFG RNA (Fig. 5D). In our study, serum PRL maintained its pulsatile expression with breastfeeding and pumping performed every 3 h (Fig. 2D). It is possible that entrainment to the timing of milk demand and PRL signaling and/or the physical act of suckling were stronger stimuli than the increased plasma concentrations of IGF-1. Regardless, rhGH administration did not alter circadian clock gene expression.
GH and IGF-1 influence several metabolic pathways. In the muscle of hypopituitary men, rhGH administered for 2 wk, increased the expression of IGF-1 while reducing the expression of enzymes regulating metabolic genes involved in lipid and energy production (49). There are no previous studies of the in vivo effects of rhGH on the temporal pattern of gene expression of metabolic genes in the human MEC. In the present study, utilizing the MFG, 57 of 98 metabolic genes (providing energy for milk production) changed with time, but none more than 1.5-fold (Fig. 6). Genes that function in glycolysis and the citric acid cycle were more highly expressed toward the end of the study. Interestingly, many genes that function in fatty acid, triglyceride, and cholesterol synthesis maintained a circadian rhythm, and their overall expression was altered little by rhGH treatment. Mechanisms for fat metabolism may be set at this stage of lactation due to the continuous synthesis of large amounts of lipid for milk production. This same pattern has been shown in the mouse liver (32, 52). Although GH is known to induce lipolysis in peripheral adipose tissue (27), there was no change in the concentration of breast milk fat (data not shown). Expression of milk protein genes did not increase, supporting our preliminary data on LALBA expression by qRT-PCR (data not shown). This is illustrated in Fig. 7C, where the β-casein gene, annotated in the PRL pathway, was noticeably not upregulated by rhGH administration (green node, Fig. 7C).
Finally, insulin and IGF-1 concentrations were significantly increased about three- to fourfold by rhGH administration. These ligands are capable of cross-reacting with their respective receptors and may activate a highly similar set of downstream intracellular events. It is not possible to delineate the effects of each hormone utilizing our study design in normal lactating women. However, in cultured fibroblasts, these hormones are able to trigger distinct cellular responses with IGF 1 being a more potent mitogen than is insulin (12). In bovine mammary explants, microarray studies showed that insulin regulated genes primarily involved in cellular metabolic processes (37), not those for cellular proliferation . The change of gene expression of cell cycle, growth, and proliferation indicate that the effects of the GH/IGF-1axis most likely predominated as well.
Our data are unique in that the frequent samples collected over 96 h allowed us to discern clearly patterns of gene expression across time, and in response to acute, short-term hormonal treatment. The present studies have elucidated the intricacies of the biological response of in vivo human tissue in response to a commonly used biological product. We have demonstrated that rhGH regulates genes for cellular proliferation but does not increase milk protein gene expression. Unraveling the primary and secondary genomic effects of rhGH or other hormones and drugs as well as determining their effects on women with lactation difficulties will be the primary challenge over the next decade.
GRANTS
This project was supported by National Institute of Diabetes and Digestive and Kidney Diseases (NIHDDK) Grant 5 RO1 DK-055478. This work is a publication of the USDA/Agricultural Research Service, CNRC, Department of Pediatrics, Baylor College of Medicine, Houston, TX, USA. The contents of this publication do not necessarily reflect the views of policies of the USDA, nor does mention of trade names, commercial products, or organizations imply endorsement from the US Government.
DISCLOSURES
The authors declare that there is no conflict of interest that would prejudice the impartiality of scientific work. P. D. Maningat, P. Sen, M. Rijnkels, D. L. Hadsell, and M. S. Bray have nothing to declare. M. W. Haymond received an NIDDK RO1 for this study and is on the Novo Nordisk International Advisory Committee. Novo Nordisk did not make any financial contributions in this study.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank the following for their invaluable help: Karen Jones, William Jeong, Michael Kueht, Brent Manning, Susan Sharma, Amy Pontius, Cindy Bryant, Linda Peasant, and the nursing staff of the GCRC.
Current addresses: P. D. Maningat, Rockefeller Univ., 1230 York Ave., New York, NY 10021 (mmaningat@mail.rockefeller.edu); M. S. Bray, Univ. of Alabama at Birmingham, Dept. of Epidemiology, 1665 University Blvd., Ryals Public Health Bldg., Ste. 220E, Birmingham AL 35294 (mbray@uab.edu).
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1.CDC CfDCaP. Breastfeeding Trends and Updated National Health Objectives for Exclusive Breastfeeding — United States, Birth Years 2000–2004 In: MMWR Morb Mortal Wkly Rep 2007, p. 760–763. [PubMed] [Google Scholar]
- 2.Adesanya OO, Zhou J, Samathanam C, Powell-Braxton L, Bondy CA. Insulin-like growth factor 1 is required for G2 progression in the estradiol-induced mitotic cycle. Proc Natl Acad Sci USA 96: 3287–3291, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allan GJ, Tonner E, Barber MC, Travers MT, Shand JH, Vernon RG, Kelly PA, Binart N, Flint DJ. Growth hormone, acting in part through the insulin-like growth factor axis, rescues developmental, but not metabolic, activity in the mammary gland of mice expressing a single allele of the prolactin receptor. Endocrinology 143: 4310–4319, 2002. [DOI] [PubMed] [Google Scholar]
- 4.Asfari M, De W, Postel-Vinay MC, Czernichow P. Expression and regulation of growth hormone (GH) and prolactin (PRL) receptors in a rat insulin producing cell line (INS-1). Mol Cell Endocrinol 107: 209–214, 1995. [DOI] [PubMed] [Google Scholar]
- 5.Baixeras E, Jeay S, Kelly PA, Postel-Vinay MC. The proliferative and antiapoptotic actions of growth hormone and insulin-like growth factor-1 are mediated through distinct signaling pathways in the Pro-B Ba/F3 cell line. Endocrinology 142: 2968–2977, 2001. [DOI] [PubMed] [Google Scholar]
- 6.Bauman DE. Bovine somatotropin and lactation: from basic science to commercial application. Domest Anim Endocrinol 17: 101–116, 1999. [DOI] [PubMed] [Google Scholar]
- 7.Bauman DE, Vernon RG. Effects of exogenous bovine somatotropin on lactation. Annu Rev Nutr 13: 437–461, 1993. [DOI] [PubMed] [Google Scholar]
- 8.Bray MS, Shaw CA, Moore MW, Garcia RA, Zanquetta MM, Durgan DJ, Jeong WJ, Tsai JY, Bugger H, Zhang D, Rohrwasser A, Rennison JH, Dyck JR, Litwin SE, Hardin PE, Chow CW, Chandler MP, Abel ED, Young ME. Disruption of the circadian clock within the cardiomyocyte influences myocardial contractile function, metabolism, and gene expression. Am J Physiol Heart Circ Physiol 294: H1036–H1047, 2008. [DOI] [PubMed] [Google Scholar]
- 9.Breier BH, Milsom SR, Blum WF, Schwander J, Gallaher BW, Gluckman PD. Insulin-like growth factors and their binding proteins in plasma and milk after growth hormone-stimulated galactopoiesis in normally lactating women. Acta Endocrinol 129: 427–435, 1993. [DOI] [PubMed] [Google Scholar]
- 10.Caron RW, Jahn GA, Deis RP. Lactogenic actions of different growth hormone preparations in pregnant and lactating rats. J Endocrinol 142: 535–545, 1994. [DOI] [PubMed] [Google Scholar]
- 11.Chakravarthy MV, Abraha TW, Schwartz RJ, Fiorotto ML, Booth FW. Insulin-like growth factor-I extends in vitro replicative life span of skeletal muscle satellite cells by enhancing G1/S cell cycle progression via the activation of phosphatidylinositol 3′-kinase/Akt signaling pathway. J Biol Chem 275: 35942–35952, 2000. [DOI] [PubMed] [Google Scholar]
- 12.Dupont J, Khan J, Qu BH, Metzler P, Helman L, LeRoith D. Insulin and IGF-1 induce different patterns of gene expression in mouse fibroblast NIH-3T3 cells: identification by cDNA microarray analysis. Endocrinology 142: 4969–4975, 2001. [DOI] [PubMed] [Google Scholar]
- 13.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eppard PJ, Bauman DE, Bitman J, Wood DL, Akers RM, House WA. Effect of dose of bovine growth hormone on milk composition: alpha-lactalbumin, fatty acids, and mineral elements. J Dairy Sci 68: 3047–3054, 1985. [DOI] [PubMed] [Google Scholar]
- 15.Field CJ. The immunological components of human milk and their effect on immune development in infants. J Nutr 135: 1–4, 2005. [DOI] [PubMed] [Google Scholar]
- 16.Flint DJ, Tonner E, Beattie J, Panton D. Investigation of the mechanism of action of growth hormone in stimulating lactation in the rat. J Endocrinol 134: 377–383, 1992. [DOI] [PubMed] [Google Scholar]
- 17.Gallego MI, Binart N, Robinson GW, Okagaki R, Coschigano KT, Perry J, Kopchick JJ, Oka T, Kelly PA, Hennighausen L. Prolactin, growth hormone, and epidermal growth factor activate Stat5 in different compartments of mammary tissue and exert different and overlapping developmental effects. Dev Biol 229: 163–175, 2001. [DOI] [PubMed] [Google Scholar]
- 18.Goffin V, Kelly PA. Prolactin and growth hormone receptors. Clin Endocrinol (Oxf) 45: 247–255, 1996. [DOI] [PubMed] [Google Scholar]
- 19.Groner B. Transcription factor regulation in mammary epithelial cells. Domest Anim Endocrinol 23: 25–32, 2002. [DOI] [PubMed] [Google Scholar]
- 20.Gunn AJ, Gunn TR, Rabone DL, Breier BH, Blum WF, Gluckman PD. Growth hormone increases breast milk volumes in mothers of preterm infants. Pediatrics 98: 279–282, 1996. [PubMed] [Google Scholar]
- 21.Hadsell DL, Parlow AF, Torres D, George J, Olea W. Enhancement of maternal lactation performance during prolonged lactation in the mouse by mouse GH and long-R3-IGF-I is linked to changes in mammary signaling and gene expression. J Endocrinol 198: 61–70, 2008. [DOI] [PubMed] [Google Scholar]
- 22.Hadsell DL, Torres DT, Lawrence NA, George J, Parlow AF, Lee AV, Fiorotto ML. Overexpression of des(1–3) insulin-like growth factor 1 in the mammary glands of transgenic mice delays the loss of milk production with prolonged lactation. Biol Reprod 73: 1116–1125, 2005. [DOI] [PubMed] [Google Scholar]
- 23.Horber FF, Marsh HM, Haymond MW. Differential effects of prednisone and growth hormone on fuel metabolism and insulin antagonism in humans. Diabetes 40: 141–149, 1991. [DOI] [PubMed] [Google Scholar]
- 24.Horvath AJ, Irving JA, Rossjohn J, Law RH, Bottomley SP, Quinsey NS, Pike RN, Coughlin PB, Whisstock JC. The murine orthologue of human antichymotrypsin: a structural paradigm for clade A3 serpins. J Biol Chem 280: 43168–43178, 2005. [DOI] [PubMed] [Google Scholar]
- 25.Ilkbahar YN, Thordarson G, Camarillo IG, Talamantes F. Differential expression of the growth hormone receptor and growth hormone-binding protein in epithelia and stroma of the mouse mammary gland at various physiological stages. J Endocrinol 161: 77–87, 1999. [DOI] [PubMed] [Google Scholar]
- 26.Jeay S, Sonenshein GE, Postel-Vinay MC, Kelly PA, Baixeras E. Growth hormone can act as a cytokine controlling survival and proliferation of immune cells: new insights into signaling pathways. Mol Cell Endocrinol 188: 1–7, 2002. [DOI] [PubMed] [Google Scholar]
- 27.Jorgensen JO, Moller L, Krag M, Billestrup N, Christiansen JS. Effects of growth hormone on glucose and fat metabolism in human subjects. Endocrinol Metab Clin North Am 36: 75–87, 2007. [DOI] [PubMed] [Google Scholar]
- 28.Kaplan W, Sunehag AL, Dao H, Haymond MW. Short-term effects of recombinant human growth hormone and feeding on gluconeogenesis in humans. Metabolism 57: 725–732, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleinberg DL, Ruan W, Catanese V, Newman CB, Feldman M. Non-lactogenic effects of growth hormone on growth and insulin-like growth factor-I messenger ribonucleic acid of rat mammary gland. Endocrinology 126: 3274–3276, 1990. [DOI] [PubMed] [Google Scholar]
- 30.Kleinberg DL, Todd J. Evidence that human growth hormone is a potent lactogen in primates. J Clin Endocrinol Metab 51: 1009–1013, 1980. [DOI] [PubMed] [Google Scholar]
- 31.Knight CH, Fowler PA, Wilde CJ. Galactopoietic and mammogenic effects of long-term treatment with bovine growth hormone and thrice daily milking in goats. J Endocrinol 127: 129–138, 1990. [DOI] [PubMed] [Google Scholar]
- 32.Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet 5: 407–441, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maningat PD, Sen P, Rijnkels M, Sunehag AL, Hadsell DL, Bray M, Haymond MW. Gene expression in the human mammary epithelium during lactation: the milk fat globule transcriptome. Physiol Genomics 37: 12–22, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maningat PD, Sen P, Sunehag AL, Hadsell DL, Haymond MW. Regulation of gene expression in human mammary epithelium: effect of breast pumping. J Endocrinol 195: 503–511, 2007. [DOI] [PubMed] [Google Scholar]
- 35.Markoff E, Talamantes F. The lactogenic response of mouse mammary explants to mouse prolactin and growth hormone. Endocr Res Commun 7: 269–278, 1980. [DOI] [PubMed] [Google Scholar]
- 36.Mauras N, Haymond MW. Are the metabolic effects of GH and IGF-I separable? Growth Horm IGF Res 15: 19–27, 2005. [DOI] [PubMed] [Google Scholar]
- 37.Menzies KK, Lefevre C, Macmillan KL, Nicholas KR. Insulin regulates milk protein synthesis at multiple levels in the bovine mammary gland. Funct Integr Genomics 9: 197–217, 2009. [DOI] [PubMed] [Google Scholar]
- 38.Milsom SR, Breier BH, Gallaher BW, Cox VA, Gunn AJ, Gluckman PD. Growth hormone stimulates galactopoiesis in healthy lactating women. Acta Endocrinol 127: 337–343, 1992. [DOI] [PubMed] [Google Scholar]
- 39.Milsom SR, Rabone DL, Gunn AJ, Gluckman PD. Potential role for growth hormone in human lactation insufficiency. Horm Res 50: 147–150, 1998. [DOI] [PubMed] [Google Scholar]
- 40.Monks J, Henson PM. Differentiation of the mammary epithelial cell during involution: implications for breast cancer. J Mammary Gland Biol Neoplasia 14: 159–170, 2009. [DOI] [PubMed] [Google Scholar]
- 41.Mukhina S, Liu D, Guo K, Raccurt M, Borges-Bendris S, Mertani HC, Lobie PE. Autocrine growth hormone prevents lactogenic differentiation of mouse mammary epithelial cells. Endocrinology 147: 1819–1829, 2006. [DOI] [PubMed] [Google Scholar]
- 42.Neville MC, McFadden TB, Forsyth I. Hormonal regulation of mammary differentiation and milk secretion. J Mammary Gland Biol Neoplasia 7: 49–66, 2002. [DOI] [PubMed] [Google Scholar]
- 43.Raccurt M, Lobie PE, Moudilou E, Garcia-Caballero T, Frappart L, Morel G, Mertani HC. High stromal and epithelial human gh gene expression is associated with proliferative disorders of the mammary gland. J Endocrinol 175: 307–318, 2002. [DOI] [PubMed] [Google Scholar]
- 44.Reiss K, Cheng W, Pierzchalski P, Kodali S, Li B, Wang S, Liu Y, Anversa P. Insulin-like growth factor-1 receptor and its ligand regulate the reentry of adult ventricular myocytes into the cell cycle. Exp Cell Res 235: 198–209, 1997. [DOI] [PubMed] [Google Scholar]
- 45.Richert MM, Wood TL. The insulin-like growth factors (IGF) and IGF type I receptor during postnatal growth of the murine mammary gland: sites of messenger ribonucleic acid expression and potential functions. Endocrinology 140: 454–461, 1999. [DOI] [PubMed] [Google Scholar]
- 46.Rowland JE, Lichanska AM, Kerr LM, White M, d'Aniello EM, Maher SL, Brown R, Teasdale RD, Noakes PG, Waters MJ. In vivo analysis of growth hormone receptor signaling domains and their associated transcripts. Mol Cell Biol 25: 66–77, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rudolph MC, McManaman JL, Phang T, Russell T, Kominsky DJ, Serkova NJ, Stein T, Anderson SM, Neville MC. Metabolic regulation in the lactating mammary gland: a lipid synthesizing machine. Physiol Genomics 28: 323–336, 2007. [DOI] [PubMed] [Google Scholar]
- 48.Samani AA, Yakar S, LeRoith D, Brodt P. The role of the IGF system in cancer growth and metastasis: overview and recent insights. Endocr Rev 28: 20–47, 2007. [DOI] [PubMed] [Google Scholar]
- 49.Sjogren K, Leung KC, Kaplan W, Gardiner-Garden M, Gibney J, Ho KK. Growth hormone regulation of metabolic gene expression in muscle: a microarray study in hypopituitary men. Am J Physiol Endocrinol Metab 293: E364–E371, 2007. [DOI] [PubMed] [Google Scholar]
- 50.Storey JD, Xiao W, Leek JT, Tompkins RG, Davis RW. Significance analysis of time course microarray experiments. Proc Natl Acad Sci USA 102: 12837–12842, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stull MA, Richert MM, Loladze AV, Wood TL. Requirement for IGF-I in epidermal growth factor-mediated cell cycle progression of mammary epithelial cells. Endocrinology 143: 1872–1879, 2002. [DOI] [PubMed] [Google Scholar]
- 52.Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, Sugano S, Iino M, Shigeyoshi Y, Hashimoto S. A transcription factor response element for gene expression during circadian night. Nature 418: 534–539, 2002. [DOI] [PubMed] [Google Scholar]
- 53.Wennbo H, Tornell J. The role of prolactin and growth hormone in breast cancer. Oncogene 19: 1072–1076, 2000. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.