Transcription factor KLF13 regulates the elevated numbers of iNKT cells in the BALB/c versus C57BL/6 thymus that results in production of sufficient levels of IL-4 to generate memory-like CD8+ T cells.
Abstract
“Memory-like T cells” are a subset of thymic cells that acquire effector function through the maturation process rather than interaction with specific antigen. Disruption of genes encoding T cell signaling proteins or transcription factors have provided insights into the differentiation of such cells. In this study, we show that in BALB/c, but not C57BL/6, mice, a large portion of thymic CD4-CD8+ T cells exhibit a memory-like phenotype. In BALB/c mice, IL-4 secreted by invariant natural killer T (iNKT) cells is both essential and sufficient for the generation of memory-like T cells. In C57BL/6 mice, iNKT cells are less abundant, producing IL-4 that is insufficient to induce thymic memory-like CD8+ T cells. BALB/c mice deficient in the transcription factor Kruppel-like factor (KLF) 13 have comparable numbers of iNKT cells to C57BL/6 mice and extremely low levels of thymic memory-like CD8+ T cells. This work documents the impact of a small number of KLF13-dependent iNKT cells on the generation of memory-like CD8+ T cells.
C57BL/6 and BALB/c mice are widely used in biomedical research. It has been observed that the two strains respond very differently to pathogens and allergens. For example, BALB/c mice are resistant to mouse adenovirus type 1 (Guida et al., 1995) and measles virus (Niewiesk et al., 1993), whereas C57BL/6 mice are susceptible to these viruses. In contrast, BALB/c mice are susceptible to herpes simplex virus-1 (Brenner et al., 1994) and most strains of Leishmania (Monroy-Ostria et al., 1994; Laskay et al., 1995; Scott et al., 1996), whereas C57BL/6 mice are resistant. The difference in resistance to Leishmania between BALB/c and C57BL/6 mice has been attributed to the predisposition to Th2 responses in BALB/c mice and to Th1 responses in C57BL/6 mice (Heinzel et al., 1989), as efficient Th1 responses were believed to be essential for clearing the pathogen. However, it remains unclear why BALB/c mice are more prone to Th2 responses and C57BL/6 mice are more prone to Th1 responses.
Multipotent progenitor T cells derived from bone marrow migrate to the thymus and develop into mature αβ or γδ T cells. For conventional αβ T cells, CD4+CD8+ double-positive (DP) thymocytes receive positive and negative selection signals through their TCRs from thymic stromal cells and progress to a mature state in which either CD4 or CD8 surface molecules are retained. The mature CD4+ and CD8+ single-positive (SP) cells leave the thymus and migrate to peripheral lymphoid organs where they encounter pathogens and allergens and develop into effecter T cells. In the periphery, naive CD8+ T cells that are activated by specific antigens and co-stimulatory molecules develop into effecter T cells that kill target cells through release of cytotoxic granules and secretion of cytokines, including IFN-γ and TNF. A small portion of activated CD8+ T cells develops into memory T cells that provide long-term protection against reinfection. The inflammatory environment derived from the innate immune system influences development into effector or memory CD8+ T cells. For example, the inflammatory cytokine IL-12 inhibits the differentiation of memory cells by repressing the expression of transcription factor Eomesodermin (Eomes) and by maintaining a high level of the transcription factor T-bet (Takemoto et al., 2006), which represses the expression of IL-7Rα (Joshi et al., 2007), a receptor essential for the survival of memory T cells (Williams and Bevan, 2007). In addition to IL-7, IL-15 is also required for the maintenance of memory CD8+ T cells (Williams and Bevan, 2007). Based on residence preference, memory CD8+ T cells can be divided into central memory T cells and effecter memory T cells (Sallusto et al., 2004). The former express high a level of CD62L and reside mainly in peripheral lymphoid organs, whereas the latter express a low level of CD62L and reside mainly in nonlymphoid tissues.
In addition to conventional αβ T cells, there are also several subsets of thymic T cells that acquire functions as a result of their maturation process rather than though antigen activation. A subset of these cells, referred to as “memory-like” or “innate-like” T cells, has been described in a few gene knockout animal models and shares characteristics with conventional memory CD8+ T cells, including high levels of expression of surface markers such as CD44 and CD122 and rapid effector responses after TCR stimulation (Prince et al., 2009; Readinger et al., 2009). In contrast to the conventional T cells that are positively and negatively selected by thymic stromal cells, these cells are believed to be selected by hematopoietic cells and are dependent on signals transduced through signaling lymphocyte activation molecule (SLAM) family members (Prince et al., 2009; Readinger et al., 2009). Another subset of the “unconventional” T cells is promyelocytic leukemia zinc finger (PLZF)–expressing NKT cells, including CD1d-restricted iNKT cells (Kovalovsky et al., 2008; Savage et al., 2008) and Vγ1.1+Vδ6.3+ γδ NKT cells (Felices et al., 2009). These cells express activation markers (CD44, CD69) and generate cytokines rapidly after TCR engagement. The numbers of these PLZF-expressing cells vary greatly among individuals (Azuara et al., 2001; Grajewski et al., 2008) and may contribute to the difference in immune responses among different mouse stains.
KLF13 is a member of the Kruppel-like family of transcription factors. We first identified KLF13 as an essential transcription factor controlling the late expression of CCL5 (RANTES) in human T lymphocytes (Song et al., 1999). Indeed, mice deficient in KLF13 showed impaired CCL5 generation in T cells after TCR stimulation (Zhou et al., 2007), and a very recent study documents that microRNA-125a regulates CCL5 expression by targeting KLF13 in activated T cells in humans and impacts RANTES expression in patients with systemic lupus erythematosus (Zhao et al., 2010). In this study, we found that KLF13 is essential for the maintenance of the large population of CD24−CD4−CD8+ memory-like T cells in the thymi of BALB/c mice. In thymocytes from C57BL/6 mice, this population is much smaller but can be expanded by provision of IL-4. In BALB/c mice, IL-4 is provided by thymic iNKT cells whose numbers are also elevated compared with C57BL/6 animals. In the absence of KLF13 in BALB/c mice, there are fewer iNKT cells and less IL-4 is produced. Disruption of either the Il4 gene or the Cd1d gene in BALB/c mice also leads to a reduction in thymic CD8+ SP T cells, demonstrating the requirement for IL-4 produced by iNKT cells. Thus, the elevated number of iNKT cells and the resulting high production of IL-4 in BALB/c mice leads to the generation of memory-like CD8+ T cells and also offers a possible explanation for the Th2 predisposition in BALB/c mice.
RESULTS
KLF13 regulates CD8+ SP cells in BALB/c mice in a cell nonautonomous manner
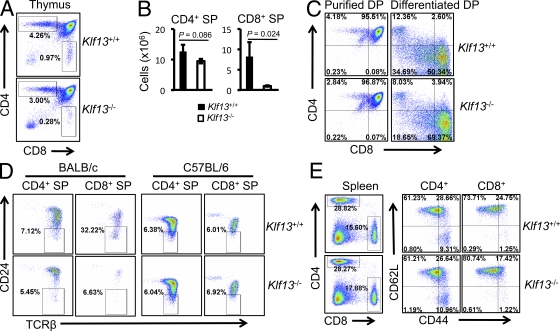
Although development of CD4+CD8− and CD4−CD8+ SP T cells in thymus is normal in Klf13−/− mice on a C57BL/6 background (Zhou et al., 2007), on a BALB/c background, Klf13−/− mice have significantly fewer CD4−CD8+ SP, but not CD4+CD8− SP cells, compared with wild-type mice (Fig. 1, A and B). To test if the effect of KLF13 on CD8+ SP cells is a cell autonomous or cell nonautonomous event, DP cells were purified to eliminate thymic stromal cells important for positive selection and lineage choice, and signals naturally provided by thymic stromal cells were added in vitro. The positive selection signal normally provided through the TCR was substituted with ionomycin and PMA, and an IL-7 signal essential for CD8+ SP lineage development was provided by recombinant IL-7. After in vitro culture for 4 d, the majority of DP cells from wild-type BALB/c mice developed into CD8+ SP cells (Fig. 1 C, top). In the absence of KLF13, DP cells developed into CD8+ SP cells as efficiently as in the presence of KLF13 (Fig. 1 C, bottom). We conclude that the developmental defect in CD8+ SP cells is nonintrinsic and is influenced by the environment (see also Fig. 2 E).
Figure 1.
KLF13 is critical for the maintenance of CD4−CD8+CD24− T cells in BALB/c thymus. (A) CD4 and CD8 expression in thymocytes from wild-type and Klf13−/− BALB/c mice. (B) Absolute numbers of CD4+CD8− single positive (CD4+ SP) and CD4−CD8+ (CD8+ SP) cells in the thymi of wild-type and Klf13−/− BALB/c mice. P < 0.05 is considered significant. Error bars are SDs. (C) Purified CD4+CD8+ double-positive (DP) cells were differentiated with PMA, ionomycin, and IL-7 to CD8+ SP cells in vitro. (D) Expression of CD24 and TCRβ in the indicated thymic populations from BALB/c and C57BL/6 mice. Gates were based on an isotype control antibody. (E) Expression of CD44 and CD62L in CD4+ SP and CD8+ splenocytes from BALB/c mice. Data are representative of three (B and C) and four (A, C, and E) independent experiments with three mice in each genotype group.
Figure 2.
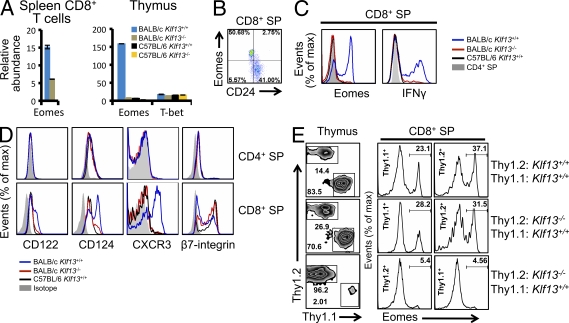
CD8+ memory-like T cells exist in the thymi of BALB/c, but not C57BL/6 mice. (A) Abundance of Eomes and T-bet mRNA relative to HPRT1 in splenic CD8+ T cells and total thymocytes from wild-type and Klf13−/− mice on BALB/c or C57BL/6 backgrounds measured by quantitative real time PCR. Error bars are generated from real-time PCR triplicates from the same RNA sample. Error bars are SDs from triplicate PCR. (B) Expression of CD24 and Eomes in CD8+ SP thymocytes from wild-type BALB/c mice. (C) Expression of Eomes and IFN-γ in CD8+ SP thymocytes from BALB/c and C57BL/6 mice. CD4+ SP thymocytes from BALB/c mice are used as negative control. (D) Expression of cell surface markers on CD4+ SP and CD8+ SP thymocytes from indicated strains of mice. (E) Expression of congenic surface markers Thy1.1 and Thy1.2 in thymocytes and Eomes in CD8+ SP cells from lethally irradiated mice adoptively transferred with unequally mixed bone marrow cells, as indicated. Data are representative of two (A and B) or three (C, D, and E) independent experiments with three mice in each genotype group (B–E).
CD24 is a cell surface marker reflective of the maturation state of T cells. In the thymus, as T cells develop from CD4−CD8− DN to CD4+CD8+ DP and finally to CD4+ SP or CD8+ SP cells, the expression of CD24 decreases from high to medium and then to low or negative. In BALB/c mice, CD24−CD4−CD8+ cells represent ∼30% of all CD8+ SP cells in KLF13+/+ thymus, but only ∼7% in Klf13−/− thymus (Fig. 1 D, left). This is in sharp contrast to C57BL/6 mice, in which the percentage of CD24−CD4−CD8+ SP cells is largely unchanged in the absence of KLF13 (Fig. 1 D, right). The percentages of CD24−CD4−CD8+ SP cells are very similar among Klf13+/+ C57BL/6, Klf13−/− C57BL/6, and Klf13−/− BALB/c mice (Fig. 1 D), suggesting that a unique population of CD24−CD4−CD8+ cells exists in the thymus of BALB/c mice, but not in C57BL/6 mice, and that the ablation of Klf13 removes this population from the BALB/c thymus. In contrast, CD4+ SP cells are only minimally affected by the absence of KLF13 in both BALB/c and C57BL/6 mice (Fig. 1 D).
The thymus is the central immune organ for the generation of mature naive T cells. Because there are fewer thymic CD8+ SP cells in Klf13−/− BALB/c mice, we predicted that there would be fewer CD8+ mature T cells in the periphery. However, the percentage, as well as absolute number of mature CD8+ T cells, is similar in the spleens of Klf13−/− and wild-type BALB/c mice (Fig. 1 E, left; and not depicted). Hyper-proliferation has been observed in peripheral T cells to maintain lymphocyte homeostasis in lymphopenic mice (Jameson, 2002), and these cells usually exhibit a CD44hi memory phenotype. To determine if the normal number of CD8+ T cells observed in the periphery of Klf13−/− BALB/c mice was caused by homeostatic proliferation, we stained splenocytes with CD44 and CD62L antibodies. Peripheral CD8+ T cells in Klf13−/− BALB/c mice did not adopt a memory-like phenotype (Fig. 1 E, right). We also considered the possibility that the Klf13−/− cells in the periphery survive longer or proliferate faster than their wild-type counterparts. T cells from central and peripheral lymphoid organs were stained with Annexin V, which detects apoptotic cells. No difference in survival was observed in Klf13−/− cells compared with wild-type cells (Fig. S1, top). We also injected mice i.p. with BrdU and analyzed BrdU incorporation, which reflects cell proliferation. Cells from Klf13−/− mice did not show any signs of hyperproliferation 16 h after BrdU injection (Fig. S1, bottom). These data indicate that both peripheral T cell renewal and survival are normal in Klf13−/− lymphocytes on a BALB/c background.
A population of KLF13-dependent memory-like CD8+ T cells exists in the thymi of BALB/c mice
To test if some unknown genes regulated by KLF13 in peripheral CD8+ T cells are responsible for their normal number in Klf13−/− mice, we performed cDNA microarray analysis on FACS-sorted splenic CD8+ T cells from Klf13+/+ and Klf13−/− BALB/c mice. In addition to Klf13, two other genes were differentially expressed more than twofold (unpublished data). One was Rcn3 (BC025602), and the other was Eomesodermin (Eomes), a T-box transcription factor proposed to promote memory formation in CD8+ T cells (Intlekofer et al., 2005; Takemoto et al., 2006). The reduced expression of Eomes in peripheral Klf13−/− BALB/c CD8+ T cells was confirmed by real-time RT-PCR (Fig. 2 A). We also checked Eomes expression in thymus. Eomes, but not T-bet, a related T-box transcription factor, showed high mRNA abundance in thymocytes from wild-type BALB/c mice but not from Klf13−/− BALB/c, wild-type C57BL/6, or Klf13−/− C57BL/6 mice (Fig. 2 A). Expression of high levels of Eomes mRNA and increased numbers of CD24−CD4−CD8+ cells in the thymi of wild-type BALB/c, but not in the thymi from Klf13−/− BALB/c or C57BL/6 (either wild-type or Klf13−/−), suggested that the CD24−CD4−CD8+ population might be CD8+ memory-like T cells. To confirm this, we measured Eomes levels after intracellular staining. Nearly all CD24−CD8+ SP cells from BALB/c thymus expressed Eomes (Fig. 2 B). Although Eomes is highly expressed in CD8+ SP cells in wild-type BALB/c thymocytes, there was very little Eomes in thymocytes from Klf13−/− BALB/c or C57BL/6 mice (Fig. 2 C, left). The CD8+ SP cells in the thymi from wild-type BALB/c mice, but not from Klf13−/− mice or C57BL/6 mice, generated large amounts of IFN-γ upon stimulation with ionomycin and PMA (Fig. 2 C, right), further suggesting that these are memory-like CD8+ T cells.
The maintenance of memory T cells is dependent on γc family cytokines, such as IL-7 and IL-15 (Rochman et al., 2009). The receptors for these cytokines are usually up-regulated in memory CD8+ T cells. Both CD122 (IL-2Rβ), a shared receptor chain for both IL-2 and IL-15, and CD124 (IL-4R) were up-regulated in BALB/c CD8+ SP cells, but not in CD4+ SP cells (Fig. 2 D). The staining pattern of β7-integrin, an adhesion molecule that is essential for trafficking of activated CD8+ T cells to gut-associated lymphoid tissues (Wagner et al., 1996; Lefrançois et al., 1999), and CXCR3, a chemokine receptor associated with recruitment of activated T cells to sites of inflammation (Xie et al., 2003), also reflected the memory phenotype of CD8+ SP cells in BALB/c mice (Fig. 2 D). Collectively, these findings demonstrate the existence of a population of memory-like CD8+ T cells in the thymus of BALB/c, but not C57BL/6, mice and this population was dependent on KLF13 in BALB/c mice.
KLF13 controls memory-like CD8+ SP thymocytes extrinsically
Our findings suggest that KLF13 regulates the development of CD8+ SP thymocytes in a cell-nonautonomous manner (Fig. 1 C). To elucidate whether KLF13 intrinsically or extrinsically regulates the generation of memory-like CD8+ SP thymocytes, we set up unequal mixed bone marrow chimeras, as described previously (Weinreich et al., 2009). Bone marrow cells from Klf13+/+ or Klf13−/− mice were mixed at a 1:9 or a 9:1 ratio and injected into lethally irradiated BALB/c hosts. The congenic markers Thy1.1 and Thy1.2 were used to track donor cells. The minority CD8+ SP thymocytes adopted the phenotype of their majority counterparts, regardless of their own genotype (Fig. 2 E and not depicted), indicating that KLF13 regulates CD8+ SP cells indirectly through the environment.
Memory-like CD8+ SP thymocytes require IL-4
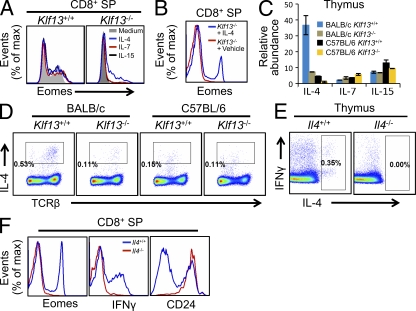
Because CD122 and CD124 are highly expressed in memory-like CD8+ T cells, we hypothesized that these cells were induced by γc family cytokines. Therefore, we treated thymocytes from wild-type and Klf13−/− BALB/c mice with IL-4, IL-7, or IL-15, the three most important γc cytokines for the homeostasis of memory T cells (Jameson, 2002; Rochman et al., 2009). IL-4, but not IL-7 or IL-15, induced increased Eomes expression in Klf13−/− CD8+ SP cells (Fig. 3 A, right). However, in wild-type CD8+ SP cells, in which Eomes levels are already high, none of the three cytokines affected Eomes expression (Fig. 3 A, left). These data indicate that IL-4 alone up-regulates Eomes in vitro. To test the effect of IL-4 in vivo, we injected Klf13−/− mice i.p. with 1 µg of IL-4 complexed with anti–IL-4 antibody every other day for 10 d. Analysis of thymocytes after the last injection showed that IL-4 efficiently induced the expression of Eomes in CD8+ SP cells (Fig. 3 B), proving that IL-4 alone is sufficient to induce Eomes in vivo. In vivo supplied IL-4 was also able to induce Eomes expression in C57BL/6 CD8+ SP cells (unpublished data). To examine IL-4 expression in vivo, we performed real-time RT-PCR on thymocytes from mice of different genetic backgrounds and genotypes. We found that IL-4 mRNA was high in wild-type BALB/c thymus but low in the thymi of Klf13−/− BALB/c mice and C57BL/6 mice, regardless of KLF13 expression (Fig. 3 C). Expression of IL-7 and IL-15 was similar among these stains (Fig. 3 C). At the protein level, IL-4–producing cells were much more abundant in thymocytes from wild-type BALB/c mice than from Klf13−/− BALB/c mice or mice on a C57BL/6 background (Fig. 3 D), suggesting that IL-4 was indeed the inducer of thymic memory-like CD8+ T cells. To conclusively demonstrate that thymic memory-like CD8+ T cells are induced by IL-4, we compared thymocytes from wild-type BALB/c mice and from Il4−/− mice on a BALB/c background. IL-4 was indeed absent in the thymi of Il4−/− mice (Fig. 3 E), and the memory-like phenotype of CD8+ SP cells was totally reversed to “normal,” manifested by low levels of intracellular Eomes, basal interferon-γ production, and loss of the CD24− population (Fig. 3 F). We therefore conclude that thymic IL-4 is both essential and sufficient for the generation of thymic memory-like CD8+ SP cells.
Figure 3.
IL-4 is both essential and sufficient to sustain memory-like CD8+ T cells in BALB/c thymus. (A) Expression of Eomes after culture of thymocytes from wild-type and Klf13−/− BALB/c mice with IL-4, IL-7, IL-15, or medium. (B) Eomes expression in CD8+ SP cells from thymi of Klf13−/− BALB/c mice after injection with 1 µg IL-4 immune complex or vehicle every other day for 10 d. (C) Relative abundance of IL-4, IL-7, or IL-15 mRNA relative to HPRT1 in thymi from the indicated stains of mice as measured by real-time RT-PCR. Error bars are SDs from triplicate PCR. (D) IL-4 and TCRβ expression in thymi from the indicated strains of mice after treatment with ionomycin and PMA in the presence of brefeldin A for 4 h. (E) FACS analysis of the expression of interferon-γ and IL-4 in wild-type and Il4−/− thymus from BALB/c mice. Thymocytes were treated with ionomycin and PMA in the presence of brefeldin A for 4 h before analysis. (F) Expression of Eomes, IFN-γ, and CD24 in CD8+ SP thymocytes from wild-type and Il4−/− BALB/c mice. Data are representative of two (A and C) or three (B–F) independent experiments with two (B) or three (D, E, and F) mice in each genotype group.
iNKT cells are the source of IL-4 and are required for the generation of memory-like CD8+ SP thymocytes
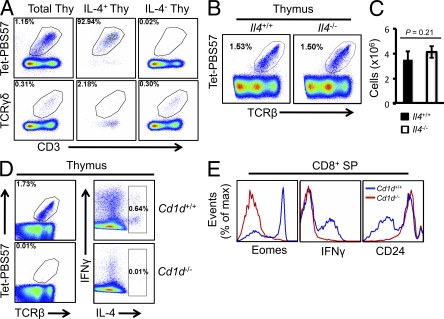
Although IL-4 has been reported to be expressed by CD4+ SP cells in Klf2−/− thymi (Weinreich et al., 2009) and by γδ T cells in Itk−/− mice (Felices et al., 2009), we found that in wild-type BALB/c mice the source of IL-4 was CD1d-restricted iNKT cells (Fig. 4 A, top) and not γδ T cells (Fig. 4 A, bottom). The percentage and the absolute number of iNKT cells was identical in thymi from Il4−/− and wild-type mice (Fig. 4, B and C), suggesting that the loss of memory-like CD8+ SP cells in Il4−/− mice was caused by the absence of IL-4 and not by some unidentified defects in iNKT cells. BALB/c mice lacking CD1d, which are devoid of CD1d-restricted NKT cells including iNKT cells (Fig. 4 D, left), did not generate IL-4 in thymocytes (Fig. 4 D, right), further proving that the source of IL-4 in BALB/c mice was iNKT cells. The loss of memory-like CD8+ SP cells in Klf13−/− (Fig. 2, C and D) and Il4−/− (Fig. 3 F) mice was also observed in Cd1d−/− mice (Fig. 4 E), proving that IL-4 generated by iNKT cells causes the expansion of memory-like T cells in BALB/c mice and that KLF13 is a major regulatory molecule in iNKT cells in BALB/c mice.
Figure 4.
iNKT cells are the source of IL-4 in the thymus. (A) Representation of iNKT cells and γδ T cells in gates of total thymocytes, IL-4+ thymocytes, and IL-4− thymocytes from wild-type BALB/c mice after stimulation with ionomycin and PMA in the presence of brefeldin A for 4 h. Tet, CD1d tetramer. (B) iNKT cells from the thymi of wild-type or IL4−/− BALB/c mice as measured by Tetramer-PB557 binding. (C) Absolute number of iNKT cells from the thymi of wild-type or IL4−/− BALB/c mice. Error bars are SDs. (D) Thymocytes from wild-type and Cd1d−/− BALB/c mice stained with Tetramer-PB557, anti–IFN-γ, anti–IL-4, and anti-TCRβ after stimulation as in A. (E) Expression of Eomes, IFN-γ, and CD24 on CD8+ SP cells from wild-type and Cd1d−/− BALB/c mice. Data are representative of three independent experiments with three mice in each genotype group (A–E).
KLF13 is intrinsically required to sustain iNKT cells in BALB/c mice
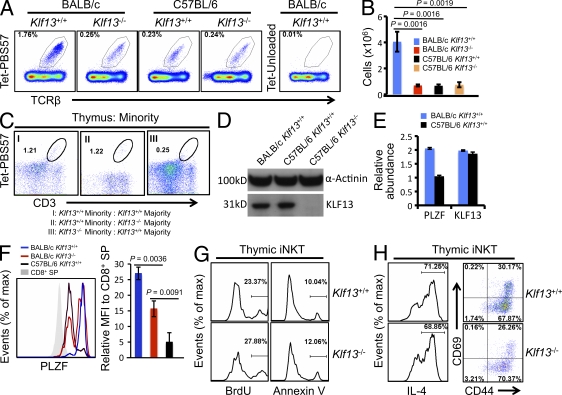
We next examined iNKT cells in thymi from mice with different genetic backgrounds and genotypes. In terms of percentage (Fig. 5 A) and absolute number (Fig. 5 B), iNKT cells were approximately sevenfold more abundant in the thymi of mice on a BALB/c background compared with mice on a C57BL/6 background. KLF13 ablation reduced the percentage and absolute number of iNKT cells in the thymi of BALB/c mice to levels similar to those in thymi of wild-type C57BL/6 mice (Fig. 5, A and B). KLF13 ablation did not further reduce the number of iNKT cells in C57BL/6 thymi (Fig. 5, A and B), presumably because of the already low number of iNKT cells in C57BL/6 mice. The percentage of iNKT cells was also slightly lower in spleens and livers of Klf13−/− BALB/c mice compared with their wild-type counterparts, but no difference was observed in lymph nodes (Fig. S2).
Figure 5.
KLF13 regulates iNKT cells in BALB/c mice. (A) iNKT cells (Tet-PBS57+) in thymi of wild-type and Klf13−/− on BALB/c or C57BL/6 backgrounds. (B) Absolute number of thymic iNKT cells in indicated mice. (C) Minority iNKT cells (Tet-PBS57+) from lethally irradiated mice adoptively transferred with unequally mixed bone marrow cells, as indicated. (D) Expression of indicated genes in DP thymocytes from indicated mice measured by Western blot. (E) Abundance of indicated genes in FACS-sorted iNKT cells from indicated mice measured by quantitative real-time PCR. Error bars are generated from real-time PCR triplicates from the same RNA sample. (F) PLZF expression in thymic iNKT cells from the indicated mice (left). The mean fluorescence intensity (MFI) of PLZF in iNKT cells relative to CD8+ SP cells was determined using three mice for each point (right). (G) BrdU uptake and Annexin V binding to thymic iNKT cells from wild-type and Klf13−/− BALB/c mice. (H) Expression of IL-4, CD69, and CD44 by thymic iNKT cells from wild-type and KLF13−/− BALB/c mice. IL-4 was analyzed 4 h after stimulation with Ionomycin and PMA in the presence of brefeldin A. Data are representative of three independent experiments (A–H) with three mice in each genotype group (A–C and F–H). Error bars are SDs (B and F) or SDs from triplicate (E).
Mice adoptively transferred with unequally mixed bone marrow cells (Fig. 2 E) were also analyzed for iNKT cells. The percentage of minority iNKT cells was significantly reduced in the absence of KLF13, regardless of the genotype of co-injected majority cells (Fig. 5 C), suggesting that KLF13 intrinsically regulates iNKT cells in BALB/c mice.
KLF13 regulates iNKT cells in BALB/c mice through PLZF
The observations that BALB/c mice have more iNKT cells than C57BL/6 mice and Klf13−/− BALB/c mice have a similar number of iNKT cells to C57BL/6 mice suggest that BALB/c mice might harbor a higher level of KLF13. However, KLF13 abundance is very similar between the two strains, both in DP thymocytes (Fig. 5 D) and in thymic iNKT cells (Fig. 5 E). However, PLZF, a transcription factor that is essential for the development of iNKT cells (Kovalovsky et al., 2008; Savage et al., 2008), is expressed at significantly lower levels in C57BL/6 mice than in BALB/c mice (Fig. 5 E). Lower expression of PLZF in C57BL/6 thymic iNKT cells was confirmed by intracellular staining (Fig. 5 F). Further, KLF13 ablation reduces the PLZF level in thymic iNKT cells on a BALB/c background (Fig. 5 F), but does not further decrease PLZF in thymic iNKT cells on a C57BL/6 background (not depicted).
The level of PLZF decreases when iNKT cells develop through stages I (CD44loNK1.1−), II (CD44hiNK1.1−) and III (CD44hiNK1.1+; Kovalovsky et al., 2008; Savage et al., 2008). It is possible that the higher level of PLZF reflects a more immature state of thymic iNKT cells in BALB/c mice. Because the NK1.1 surface molecule does not exist in BALB/c mice, we tested two other pan-NK cell markers that are present in both C57BL/6 and BALB/c mice to determine whether they can be used to identify iNKT cell developmental stages. One marker, DX5, co-stains ∼5% of iNKT cells with NK1.1 in C57BL/6 mice (Fig. S3 A, left), whereas the other marker, NKG2D, co-stains ∼60% of iNKT cells with NK1.1 in C57BL/6 mice (Fig. S3 B, left). The staining patterns of DX5 in iNKT cells are very similar among mice with different backgrounds and genotypes (Fig. S3 A, right), suggesting that DX5 is not a useful marker for determining iNKT developmental stages I through III as defined by NK1.1 and CD44. Therefore, we used NKG2D to assess the differences in developmental stages of iNKT cells in C57BL/6 and BALB/c mice. We found that the percentage of mature CD44hiNKG2D+ iNKT cells in BALB/c mice is considerably lower than that in C57BL/6 mice (Fig. S3 B, right), suggesting a correlation between an immature state and higher PLZF expression in BALB/c thymic iNKT cells. In mature peripheral splenic iNKT cells, PLZF levels are similar between C57BL/6 and BALB/c mice (Fig. S4), further proving that the PLZF expression difference in BALB/c and C57BL/6 mice is a result of a more immature state of thymic iNKT cells in BALB/c mice.
In Klf13−/− BALB/c mice, however, the percentage of the CD44hiNKG2D+ mature thymic iNKT cells is comparable to that in wild-type BALB/c mice (Fig. S3 B, right). In each specific stage as defined by CD44 and NKG2D, PLZF expression is always lower in Klf13−/− BALB/c mice than in wild-type BALB/c mice (Fig. S3 C), suggesting that KLF13 ablation does not affect the maturation state of iNKT cells and that lower expression of PLZF is a direct result of KLF13 ablation. Lower levels of PLZF may account for the decreased number of iNKT cells in Klf13−/− BALB/c and C57BL/6 mice.
To determine if KLF13-dependent PLZF regulates the number of iNKT cells in BALB/c mice by affecting cell proliferation and survival, we assessed the BrdU incorporation and cellular apoptosis that were revealed by Annexin V binding in Klf13−/− iNKT cells. Both cell proliferation and survival of Klf13−/− iNKT cells were normal (Fig. 5 G). When compared with wild-type counterparts, the Klf13−/− BALB/c iNKT cells generated a similar amount of IL-4 and expressed normal cell surface activation markers (Fig. 5 H), suggesting that KLF13-dependent PLZF does not regulate IL-4 directly in iNKT cells and that iNKT cells function normally in the absence of KLF13. These data suggest that the intracellular level of PLZF only affects the generation of iNKT cells. A higher level of PLZF may expedite the transition into iNKT cells, leading to accumulation of early stage immature iNKT cells. Once iNKT cells are committed, they function normally, regardless of intracellular PLZF levels or maturation stages. Thus, the decreased IL-4 production in Klf13−/− BALB/c thymi is a direct result of fewer iNKT cells. This is further supported by the strong correlation between IL-4 generation and iNKT cell number in mice from different genetic backgrounds and genotypes (compare Fig. 3 D and Fig. 5 A).
DISCUSSION
In this study, we identify a thymic memory-like Eomes-expressing CD8+ T cell population in BALB/c but not C57BL/6 mice. This population is regulated by IL-4 secreted by iNKT cells. KLF13 regulates the abundance of iNKT cells in BALB/c mice through PLZF, resulting in IL-4–dependent induction of memory-like CD8+ T cells. The strain-specific differential importance of KLF13 in regulating iNKT cells suggests a new potential mechanism for genetic control of human diseases.
The existence of memory-like or innate-like CD8+ SP T cells has been described in several gene-manipulated models, including Klf2 (Weinreich et al., 2009), Tec kinases Itk and Rlk (Atherly et al., 2006; Broussard et al., 2006; Dubois et al., 2006; Horai et al., 2007), and histone acetyltransferase Cbp (Fukuyama et al., 2009) knockout mice and in CIITA transgenic mice (Choi et al., 2005; Li et al., 2005). Itk and Rlk were believed to be the predominant factors that drove thymocyte development into conventional lineages versus innate lineages, including iNKT cells, CD8αα+ T cells, mucosal invariant T cells, and the aforementioned innate-like CD8+ SP T cells, which were thought to result from thymocyte/thymocyte selection caused by lack of ITK in Itk−/− mice (Horai et al., 2007). However, our findings suggest that it is also possible that the innate-like CD8+ SP cells in Itk−/− mice, as well as in the aforementioned models, were converted from conventional CD8+ SP cells by NKT-generated IL-4, similar to the mechanism we report here for BALB/c mice. Itk−/− mice exhibited elevated levels of IL-4–producing γδ NKT cells (Felices et al., 2009), and the removal of SLAM-associated protein, an adaptor molecule essential for thymocyte-selected iNKT and γδ NKT cells (Pasquier et al., 2005; Kreslavsky et al., 2009), eliminated the innate-like phenotype in Itk−/− CD8+ SP cells (Horai et al., 2007), strongly suggesting that the innate-like phenotype was an indirect bystander effect exerted by γδ NKT cells. In CIITA transgenic mice, IL-4 and STAT6 are essential to the memory-like phenotype of CD8+ SP cells (Li et al., 2005), implying that these cells were also induced by IL-4, whose source is presumably NKT cells. Because NKT cells are derived from the same hematopoietic precursor as conventional CD8+ T cells, mixed bone marrow chimera experiments cannot reveal the bystander effects in the aforementioned animal models. The exception is the Klf2 knockout model, in which an unequal mixed bone marrow chimera experiment revealed that development of the thymic memory-like phenotype was indeed cell extrinsic (Weinreich et al., 2009). With the same unequal mixed bone marrow chimera approach, two recent papers confirmed that the memory-like phenotype in CD8+ SP thymocytes from Itk−/−, CPB−/− (Weinreich et al., 2010), Sh2d1a−/− (SLAM-associated protein), and Id3−/− (Verykokakis et al., 2010) mice were indeed the result of bystander effects. The thymic memory-like CD8+ cells were also described in BALB/c mice, and the authors ascribed the memory-like phenotype to IL-4 generated by expanded PLZF+ T cells on the basis that PLZF+ T cells were able to produce IL-4, and the ablation of either IL-4R or PLZF (CD1d in the case of wild-type BALB/c mice) removed the memory-like thymic cells in Klf2−/− and Itk−/− C57BL/6 mice, as well as in wild-type BALB/c mice (Weinreich et al., 2010). Our findings in Il4−/− BALB/c mice extend their observations and rule out the possibility of IL-13–mediated effects that cannot be eliminated in the IL-4R knockout mice model (Weinreich et al., 2010) because IL-4R also responds to IL-13. Our data from in vitro cell culture and in vivo IL-4 provision further prove that NKT cell–derived IL-4 alone is sufficient to induce thymic memory-like CD8+ cells. This eliminates the possibility that in the PLZF and CD1d knockout model (Weinreich et al., 2010) other defects in NKT cells, rather than overproduction of IL-4, cause the expansion of the memory-like subset. It is interesting that IL-4 does not induce a memory-like phenotype in thymic CD4+ T cells (Fig. 2, C and D). This phenomenon was shown to be dependent on a repressive effect of GATA3, which is abundant in CD4+ T cells, but expressed at much lower levels in CD8+ T cells. When GATA3 is removed, CD4+ T cells do develop a memory-like phenotype in response to IL-4 (Yagi et al., 2010).
Although KLF2 and KLF13 are both members of the same transcription factor family, they mediate opposing functions in T cells. To date, 17 members of the mammalian KLF family have been identified (Cao et al., 2010). Each contains a highly conserved DNA-binding domain comprised of three C2H2 zinc fingers at the C terminus and bind to the CACCC element and GC box consensus sequences. In addition, there is a highly conserved seven-residue sequence between the zinc fingers (TGEKP[Y/F]X), but the non–DNA-binding regions are quite divergent (Cao et al., 2010). KLF2 is expressed in lung, endothelial cells, and lymphocytes, and KLF2 deficiency is lethal. KLF2 has been reported to regulate aspects of T cell quiescence, survival, activation, and migration (Cao et al., 2010). KLF2 is expressed in quiescent T cells and is down-regulated soon after activation (Buckley et al., 2001; Kuo et al., 1997). In contrast, KLF13 is not expressed in resting T cells but is produced within 24–48 h of activation through the T cell receptor (Song et al., 2002). KLF13 deficiency is not lethal (Zhou et al., 2007). KLF13 regulates the late expression of CCL5 in T cells by recruiting a large trans-activating complex, including p300/CBP, PCAF, MAPK Nemo-like kinase, and the ATPase BRG-1 (Ahn et al., 2007). The reciprocal effects of KLF2 and KLF13 on thymic NKT cell number and on the activation state of T cells suggest that these closely related transcription factors exert a Yin/Yang effect on the immune system.
BALB/c and C57BL/6 mice are differentially susceptible to many diseases. Many of the differences can be ascribed to the predisposition to Th1 or Th2 immune responses in C57BL/6 and BALB/c mice, respectively. Th1 cells are induced by IL-12 and generate IFN-γ, which activates macrophages and other cells to control intracellular pathogens (Medzhitov, 2007). Th2 cells are induced by IL-4 and secrete IL-4, IL-5, and IL-13. These cytokines are important for the activation of eosinophils, basophils, and the mucosal epithelia, which are involved in protection against multicellular parasites (Medzhitov, 2007). IL-4 stimulates a class switch in B cells to produce IgE, which mediates the functions of eosinophils and basophils. However, excessive Th2 responses are also coupled with allergy development mediated by IgE-activated eosinophils and basophils. In the presence of robust Th2 responses, Th1 cells are usually not sufficient to control intracellular pathogens, as in the case of Leishmania infection of BALB/c mice. IL-4 is key in the balance between Th1 and Th2 immune responses. The presence of more IL-4–generating thymic iNKT cells may explain in part why BALB/c mice are more prone to a Th2 response. iNKT cells can stimulate IgE production by secreting IL-4 (Yoshimoto et al., 1995), and the basal serum IgE level is much higher in BALB/c mice (∼500 ng/ml) than in C57BL/6 mice (<100 ng/ml; Ota et al., 2009). The development of allergic airway hyperresponse, an animal model of asthma, is more severe in BALB/c mice than in C57BL/6 mice in response to various allergens (Pinelli et al., 2001; Atochina et al., 2003; Shinagawa and Kojima, 2003; Kierstein et al., 2006; Fukunaga et al., 2007; Page et al., 2007; Conrad et al., 2009; Gueders et al., 2009; Van Hove et al., 2009; Sahu et al., 2010), and iNKT cells are absolutely required for allergic airway hyperresponse in BALB/c mice (Umetsu and Dekruyff, 2010). Elevated numbers of iNKT cells were observed in the lungs of moderate-to-severe persistent asthma patients (Akbari et al., 2006). Furthermore, when iNKT cells are activated with α-galactosylceramide during Leishmania donovani infection in C57BL/6 mice, experimental visceral leishmaniasis is exacerbated (Stanley et al., 2008), implying that the decreased number of iNKT cells contributes to resistance to Leishmania in C57BL/6 mice. Our data suggest the differential expression of PLZF in iNKT cells from C57BL/6 and BALB/c mice might account for the different iNKT cell numbers in the two strains. However, PLZF transgenic mice on a C57BL/6 background did not show more abundant iNKT cells (Savage et al., 2008; Kovalovsky et al., 2010), presumably because of an insufficient effect of ectopic PLZF expression in iNKT cells where endogenous PLZF is already high. In summary, our observations suggest that thymic iNKT cells might play an important role in susceptibility to diseases among different individuals. Our finding that KLF13 regulates iNKT cell numbers may have potential diagnostic and therapeutic implications.
MATERIALS AND METHODS
Mice.
Klf13−/− mice on C57BL/6 background were previously described (Zhou et al., 2007). The mice were backcrossed for >10 generations to a BALB/c background. C57BL/6 mice with EGFP replacing the first intron of IL-4 (Hu-Li et al., 2001), here referred to as Il4−/− mice, were backcrossed to a BALB/c background (Line 185) and maintained by Taconic–National Institute of Allergy and Infectious Disease exchange program. Cd1d−/− mice on a BALB/c background were purchased from The Jackson Laboratory. BALB/c Thy1.1+ mice (provided by G. Punkosdy, National Institutes of Health, Bethesda, MD) were previously described (Huter et al., 2008). The mice were housed in a conventional facility and littermates were used when 6–8 wk old, unless otherwise indicated in the text. The National Cancer Institute Animal Care and Use Committee approved all animal experimental protocols, and all mice were cared for in accordance with the National Institutes of Health guidelines.
In vivo treatment of Klf13−/− mice with IL-4.
5-wk-old Klf13−/− mice were injected i.p. with IL-4C (1 µg IL-4 in 100 µl vehicle) every other day for 10 d. IL-4C, which has longer half-life in vivo than IL-4, was prepared as previously described (Morris et al., 2009) with slight modifications. In brief, 20 µg of recombinant IL-4 (PeproTech) powder was reconstituted with 80 µl water and mixed with 120 µl anti–IL-4 antibody (1 mg/ml; clone 11B11; eBioscience). The mixture was incubated for 10 min at room temperature, and then diluted 10-fold with PBS containing 1% serum from BALB/c mice (Innovative Research) and kept at 4°C until used.
Flow cytometry.
Cells in single-cell suspension were treated with BD Fc Block, and then stained with fluorophore-labeled antibodies against surface molecules with standard methods. For intracellular staining of IL-4 and IFN-γ, cells were stimulated with 1 µM ionomycin and 10 ng/ml PMA (both from Sigma-Aldrich) in the presence of GolgiPlug (BD) for 4 h before surface and intracellular antigens were stained sequentially with a Fixation and Permeabilization kit from eBioscience according to the manufacturer’s instructions. Eomes was stained with eBioscience’s Foxp3 staining buffer according to the manufacturer’s instructions. PLZF was stained as previously described (Savage et al., 2008). Data were collected on a LSR II machine (BD), and analyzed with FlowJo software (Tree Star). Antibodies against CD3 (145-2C11), CD4 (GK1.5), CD8 (53–6.7), TCRβ (H57-597), TCRγδ (eBioGL3), CD24 (M1/69), CD44 (IM7), CD62L (MEL-14), CD122 (5H4), IL-4 (11B11), IFN-γ (XMG1.2), and Eomes (Dan11mag) were purchased from eBioscience. Antibodies against β7-integrin (M293) and CD124 (mIL4R-M1) were purchased from BD. Antibody against CXCR3 (clone 220803) was purchased from R&D Systems. PBS57-loaded CD1d tetramer provided by the National Institutes of Health Tetramer Facility was used to detect iNKT cells. For detection of liver iNKT cells, liver lymphocytes were purified by spinning total liver cells in single-cell suspension over 37.5% Percoll (GE Healthcare) at 690 g for 12 min. The pelleted cells were treated with ACK buffer to remove red blood cells before surface staining. In vivo BrdU labeling and detection were performed according to supplier’s instructions (BrdU Flow kits; BD). Mice were analyzed 16 h after BrdU injection. Annexin V staining was performed following supplier’s instructions (eBioscience).
Cell sorting, microarray, and real-time PCR.
Splenic CD8+ T cells were sorted on a BD FACSAria machine based on CD8+CD4−CD19−. Thymocytes from 10 mice were pooled and stained with PE-labeled, PBS57-loaded CD1d tetramer. iNKT cells were enriched with anti-PE beads (Miltenyi Biotec) and then sorted on a FACSAria instrument based on expression of CD3 and tetramer. RNA was prepared from sorted cells with RNEasy kit (QIAGEN). Microarrays on Affymetrix GeneChip Mouse Gene 1.0 ST slides were performed by National Cancer Institute-Frederick Advanced Technology Program. The results were analyzed with GeneSpring GX software (Agilent Technologies) and deposited in Gene Expression Omnibus under accession no. GSE25502. For real-time PCR, RNA was reverse-transcribed with iScript cDNA Synthesis kit (Bio-Rad Laboratories). A PCR master mix containing SYBR Green was used to amplify the resulting cDNA on a 7900HT machine (Applied Biosystems). Data were plotted using the 2−ΔΔCt method. Primers used were as follows: Eomes, 5′-GGCCTACCAAAACACGGATA-3′ and 5′-GACCTCCAGGGACAATCTGA-3′; T-bet, 5′-TCAACCAGCACCAGACAGAG-3′ and 5′-ATCCTGTAATGGCTTGTGGG-3′; IL-4, 5′-TGAACGAGGTCACAGGAGAA-3′ and 5′-CGAGCTCACTCTCTGTGGTG-3′; IL-7, 5′-GCCACATTAAAGACAAAGAAGGT-3′ and 5′-TGGTTCATTATTCGGGCAAT-3′; IL-15, 5′-AGCACTGCCTCTTCATGGTC-3′ and 5′-CTGCCATCCATCCAGAACTC-3′; HPRT1, 5′-TCCTCCTCAGACCGCTTTT-3′ and 5′-CATAACCTGGTTCATCATCGC-3′; PLZF, 5′-AGAAGTTCAGCCTCAAGCACC-3′ and 5′-AGTAGTCTCGGGAGCGCTGG-3′; KLF13, 5′-TGCGAGAAAGTTTACGGGAA-3′ and 5′-TGCGAACTTCTTGTTGCACT-3′.
In vitro stimulation of thymocytes.
107 freshly isolated thymocytes were cultured in 4 ml complete medium (DME + 10% fetal bovine serum) with 20 ng/ml IL-4, 20 ng/ml IL-7, or 50 ng/ml IL-15 (all from PeproTech) for 20 h. The cells were then stained with Eomes and analyzed with FACS.
In vitro differentiation of CD8+ SP cells.
The protocol was based on that used by the Singer laboratory (National Institutes of Health) with slight modifications. DP cells were purified with cell panning. In brief, a 10 cm Petri dish was coated with 1.5 µg/ml peanut agglutinin (Sigma-Aldrich) in PBS overnight at 37°C, and then blocked with blocking buffer (HBSS with 5% fetal bovine serum) for 30 min at room temperature. 5 × 107 thymocytes in 10 ml blocking buffer were loaded onto the plate and incubated for 2 h at 4°C. The unbound cells were discarded and the plates were washed gently by swirling three times with blocking buffer, and the bound cells were eluted with complete culture medium (RPMI with 10% fetal bovine serum) containing 0.2 M d-galactose (Sigma-Aldrich) by incubating the plate for 5 min at room temperature. The resulting DP cells were usually >95% pure, and the main contaminants were CD4+ SP cells. CD8+ SP cell contamination was <1%. The purified DP cells were suspended in complete medium at 5 × 106 cells/ml and stimulated with 0.3 ng/ml ionomycin and 0.3 ng/ml PMA for 16 h, and then ionomycin and PMA were washed away with medium. The cells were cultured in complete medium for 16 h to recover. 10 ng/ml IL-7 was then added to the medium and the cells were cultured for another 24 h before treatment with 0.01% pronase (EMD) in HBSS for 12 min at 37°C to remove surface CD4 and CD8. The cells were cultured for another 24 h with IL-7 and freshly expressed CD4 and CD8 surface markers were analyzed with FACS.
Mixed bone marrow chimera.
These experiments were performed essentially as previously described (Verykokakis et al., 2010) except that the recipient BALB/c mice were irradiated at a dose of 500 rad twice, with a 3-h interval.
Statistics.
P < 0.05 from Student’s unpaired, two-tailed t test were considered significant.
Online supplemental material.
Fig. S1 shows T cell proliferation and survival in Klf13−/− BALB/c mice. Fig. S2 shows percentages of peripheral iNKT cells in Klf13−/− BALB/c mice. Fig. S3 shows the development stages of thymic iNKT cells from Klf13+/+, Klf13−/− BALB/c mice, and C57BL/6 mice. Fig. S4 shows PLZF levels in splenic and thymic iNKT cells from BALB/c and C57BL/6 mice. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20101527/DC1.
Acknowledgments
We thank Drs. A. Singer, P. Schwartzberg, Y. Wan, W. Ouyang, and J. Lin for helpful discussions; Dr. G. Punkosdy for providing BALB/c Thy1.1 mice; Dr. X. Tai for help with in vitro differentiation of CD8+ SP cells, and the National Institutes of Health tetramer facility for providing CD1d tetramers.
The Intramural Research Programs of the National Institutes of Health, the National Cancer Institute, and the National Institute of Allergy and Infectious Diseases supported this research.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- DP
- double positive
- Eomes
- Eomesodermin
- iNKT
- invariant natural killer T
- KLF
- Kruppel-like factor
- PLZF
- promyelocytic leukemia zinc finger
- SLAM
- signaling lymphocyte activation molecule
- SP
- single positive
References
- Ahn Y.T., Huang B., McPherson L., Clayberger C., Krensky A.M. 2007. Dynamic interplay of transcriptional machinery and chromatin regulates “late” expression of the chemokine RANTES in T lymphocytes. Mol. Cell. Biol. 27:253–266 10.1128/MCB.01071-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari O., Faul J.L., Hoyte E.G., Berry G.J., Wahlström J., Kronenberg M., DeKruyff R.H., Umetsu D.T. 2006. CD4+ invariant T-cell-receptor+ natural killer T cells in bronchial asthma. N. Engl. J. Med. 354:1117–1129 10.1056/NEJMoa053614 [DOI] [PubMed] [Google Scholar]
- Atherly L.O., Lucas J.A., Felices M., Yin C.C., Reiner S.L., Berg L.J. 2006. The Tec family tyrosine kinases Itk and Rlk regulate the development of conventional CD8+ T cells. Immunity. 25:79–91 10.1016/j.immuni.2006.05.012 [DOI] [PubMed] [Google Scholar]
- Atochina E.N., Beers M.F., Tomer Y., Scanlon S.T., Russo S.J., Panettieri R.A., Jr, Haczku A. 2003. Attenuated allergic airway hyperresponsiveness in C57BL/6 mice is associated with enhanced surfactant protein (SP)-D production following allergic sensitization. Respir. Res. 4:15 10.1186/1465-9921-4-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V., Grigoriadou K., Lembezat M.P., Nagler-Anderson C., Pereira P. 2001. Strain-specific TCR repertoire selection of IL-4-producing Thy-1 dull gamma delta thymocytes. Eur. J. Immunol. 31:205–214 [DOI] [PubMed] [Google Scholar]
- Brenner G.J., Cohen N., Moynihan J.A. 1994. Similar immune response to nonlethal infection with herpes simplex virus-1 in sensitive (BALB/c) and resistant (C57BL/6) strains of mice. Cell. Immunol. 157:510–524 10.1006/cimm.1994.1246 [DOI] [PubMed] [Google Scholar]
- Broussard C., Fleischacker C., Horai R., Chetana M., Venegas A.M., Sharp L.L., Hedrick S.M., Fowlkes B.J., Schwartzberg P.L. 2006. Altered development of CD8+ T cell lineages in mice deficient for the Tec kinases Itk and Rlk. Immunity. 25:93–104 10.1016/j.immuni.2006.05.011 [DOI] [PubMed] [Google Scholar]
- Buckley A.F., Kuo C.T., Leiden J.M. 2001. Transcription factor LKLF is sufficient to program T cell quiescence via a c-Myc—dependent pathway. Nat. Immunol. 2:698–704 10.1038/90633 [DOI] [PubMed] [Google Scholar]
- Cao Z., Sun X., Icli B., Wara A.K., Feinberg M.W. 2010. Role of Kruppel-like factors in leukocyte development, function, and disease. Blood. 116:4404–4414 10.1182/blood-2010-05-285353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E.Y., Jung K.C., Park H.J., Chung D.H., Song J.S., Yang S.D., Simpson E., Park S.H. 2005. Thymocyte-thymocyte interaction for efficient positive selection and maturation of CD4 T cells. Immunity. 23:387–396 10.1016/j.immuni.2005.09.005 [DOI] [PubMed] [Google Scholar]
- Conrad M.L., Yildirim A.O., Sonar S.S., Kiliç A., Sudowe S., Lunow M., Teich R., Renz H., Garn H. 2009. Comparison of adjuvant and adjuvant-free murine experimental asthma models. Clin. Exp. Allergy. 39:1246–1254 10.1111/j.1365-2222.2009.03260.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois S., Waldmann T.A., Müller J.R. 2006. ITK and IL-15 support two distinct subsets of CD8+ T cells. Proc. Natl. Acad. Sci. USA. 103:12075–12080 10.1073/pnas.0605212103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felices M., Yin C.C., Kosaka Y., Kang J., Berg L.J. 2009. Tec kinase Itk in gammadeltaT cells is pivotal for controlling IgE production in vivo. Proc. Natl. Acad. Sci. USA. 106:8308–8313 10.1073/pnas.0808459106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga J., Abe M., Murai A., Akitake Y., Hosokawa M., Takahashi M. 2007. Comparative study to elucidate the mechanism underlying the difference in airway hyperresponsiveness between two mouse strains. Int. Immunopharmacol. 7:1852–1861 10.1016/j.intimp.2007.07.010 [DOI] [PubMed] [Google Scholar]
- Fukuyama T., Kasper L.H., Boussouar F., Jeevan T., van Deursen J., Brindle P.K. 2009. Histone acetyltransferase CBP is vital to demarcate conventional and innate CD8+ T-cell development. Mol. Cell. Biol. 29:3894–3904 10.1128/MCB.01598-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grajewski R.S., Hansen A.M., Agarwal R.K., Kronenberg M., Sidobre S., Su S.B., Silver P.B., Tsuji M., Franck R.W., Lawton A.P., et al. 2008. Activation of invariant NKT cells ameliorates experimental ocular autoimmunity by a mechanism involving innate IFN-gamma production and dampening of the adaptive Th1 and Th17 responses. J. Immunol. 181:4791–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueders M.M., Paulissen G., Crahay C., Quesada-Calvo F., Hacha J., Van Hove C., Tournoy K., Louis R., Foidart J.M., Noël A., Cataldo D.D. 2009. Mouse models of asthma: a comparison between C57BL/6 and BALB/c strains regarding bronchial responsiveness, inflammation, and cytokine production. Inflamm. Res. 58:845–854 10.1007/s00011-009-0054-2 [DOI] [PubMed] [Google Scholar]
- Guida J.D., Fejer G., Pirofski L.A., Brosnan C.F., Horwitz M.S. 1995. Mouse adenovirus type 1 causes a fatal hemorrhagic encephalomyelitis in adult C57BL/6 but not BALB/c mice. J. Virol. 69:7674–7681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzel F.P., Sadick M.D., Holaday B.J., Coffman R.L., Locksley R.M. 1989. Reciprocal expression of interferon γ or interleukin 4 during the resolution or progression of murine leishmaniasis. Evidence for expansion of distinct helper T cell subsets. J. Exp. Med. 169:59–72 10.1084/jem.169.1.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horai R., Mueller K.L., Handon R.A., Cannons J.L., Anderson S.M., Kirby M.R., Schwartzberg P.L. 2007. Requirements for selection of conventional and innate T lymphocyte lineages. Immunity. 27:775–785 10.1016/j.immuni.2007.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu-Li J., Pannetier C., Guo L., Löhning M., Gu H., Watson C., Assenmacher M., Radbruch A., Paul W.E. 2001. Regulation of expression of IL-4 alleles: analysis using a chimeric GFP/IL-4 gene. Immunity. 14:1–11 10.1016/S1074-7613(01)00084-X [DOI] [PubMed] [Google Scholar]
- Huter E.N., Stummvoll G.H., DiPaolo R.J., Glass D.D., Shevach E.M. 2008. Cutting edge: antigen-specific TGF beta-induced regulatory T cells suppress Th17-mediated autoimmune disease. J. Immunol. 181:8209–8213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer A.M., Takemoto N., Wherry E.J., Longworth S.A., Northrup J.T., Palanivel V.R., Mullen A.C., Gasink C.R., Kaech S.M., Miller J.D., et al. 2005. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 6:1236–1244 10.1038/ni1268 [DOI] [PubMed] [Google Scholar]
- Jameson S.C. 2002. Maintaining the norm: T-cell homeostasis. Nat. Rev. Immunol. 2:547–556 [DOI] [PubMed] [Google Scholar]
- Joshi N.S., Cui W., Chandele A., Lee H.K., Urso D.R., Hagman J., Gapin L., Kaech S.M. 2007. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 27:281–295 10.1016/j.immuni.2007.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierstein S., Poulain F.R., Cao Y., Grous M., Mathias R., Kierstein G., Beers M.F., Salmon M., Panettieri R.A., Jr, Haczku A. 2006. Susceptibility to ozone-induced airway inflammation is associated with decreased levels of surfactant protein D. Respir. Res. 7:85 10.1186/1465-9921-7-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalovsky D., Uche O.U., Eladad S., Hobbs R.M., Yi W., Alonzo E., Chua K., Eidson M., Kim H.J., Im J.S., et al. 2008. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat. Immunol. 9:1055–1064 10.1038/ni.1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalovsky D., Alonzo E.S., Uche O.U., Eidson M., Nichols K.E., Sant’Angelo D.B. 2010. PLZF induces the spontaneous acquisition of memory/effector functions in T cells independently of NKT cell-related signals. J. Immunol. 184:6746–6755 10.4049/jimmunol.1000776 [DOI] [PubMed] [Google Scholar]
- Kreslavsky T., Savage A.K., Hobbs R., Gounari F., Bronson R., Pereira P., Pandolfi P.P., Bendelac A., von Boehmer H. 2009. TCR-inducible PLZF transcription factor required for innate phenotype of a subset of gammadelta T cells with restricted TCR diversity. Proc. Natl. Acad. Sci. USA. 106:12453–12458 10.1073/pnas.0903895106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C.T., Veselits M.L., Leiden J.M. 1997. LKLF: A transcriptional regulator of single-positive T cell quiescence and survival. Science. 277:1986–1990 10.1126/science.277.5334.1986 [DOI] [PubMed] [Google Scholar]
- Laskay T., Diefenbach A., Röllinghoff M., Solbach W. 1995. Early parasite containment is decisive for resistance to Leishmania major infection. Eur. J. Immunol. 25:2220–2227 10.1002/eji.1830250816 [DOI] [PubMed] [Google Scholar]
- Lefrançois L., Parker C.M., Olson S., Muller W., Wagner N., Schön M.P., Puddington L. 1999. The role of β7 integrins in CD8 T cell trafficking during an antiviral immune response. J. Exp. Med. 189:1631–1638 10.1084/jem.189.10.1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Kim M.G., Gourley T.S., McCarthy B.P., Sant’Angelo D.B., Chang C.H. 2005. An alternate pathway for CD4 T cell development: thymocyte-expressed MHC class II selects a distinct T cell population. Immunity. 23:375–386 10.1016/j.immuni.2005.09.002 [DOI] [PubMed] [Google Scholar]
- Medzhitov R. 2007. Recognition of microorganisms and activation of the immune response. Nature. 449:819–826 10.1038/nature06246 [DOI] [PubMed] [Google Scholar]
- Monroy-Ostria A., Fuentes-Fraga I., García-Flores C., Favila-Castillo L. 1994. Infection of BALB/c, C57B1/6 mice and F1 hybrid CB6F1 mice with strains of Leishmania mexicana isolated from Mexican patients with localized or diffuse cutaneous leishmaniasis. Arch. Med. Res. 25:401–406 [PubMed] [Google Scholar]
- Morris S.C., Heidorn S.M., Herbert D.R., Perkins C., Hildeman D.A., Khodoun M.V., Finkelman F.D. 2009. Endogenously produced IL-4 nonredundantly stimulates CD8+ T cell proliferation. J. Immunol. 182:1429–1438 10.4049/jimmunol.0900638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiesk S., Brinckmann U., Bankamp B., Sirak S., Liebert U.G., ter Meulen V. 1993. Susceptibility to measles virus-induced encephalitis in mice correlates with impaired antigen presentation to cytotoxic T lymphocytes. J. Virol. 67:75–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota T., Aoki-Ota M., Duong B.H., Nemazee D. 2009. Suppression of IgE B cells and IgE binding to Fc(epsilon)RI by gene therapy with single-chain anti-IgE. J. Immunol. 182:8110–8117 10.4049/jimmunol.0900300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page K., Lierl K.M., Herman N., Wills-Karp M. 2007. Differences in susceptibility to German cockroach frass and its associated proteases in induced allergic inflammation in mice. Respir. Res. 8:91 10.1186/1465-9921-8-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquier B., Yin L., Fondanèche M.C., Relouzat F., Bloch-Queyrat C., Lambert N., Fischer A., de Saint-Basile G., Latour S. 2005. Defective NKT cell development in mice and humans lacking the adapter SAP, the X-linked lymphoproliferative syndrome gene product. J. Exp. Med. 201:695–701 10.1084/jem.20042432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinelli E., Dormans J., Fonville M., van der Giessen J. 2001. A comparative study of toxocariasis and allergic asthma in murine models. J. Helminthol. 75:137–140 [PubMed] [Google Scholar]
- Prince A.L., Yin C.C., Enos M.E., Felices M., Berg L.J. 2009. The Tec kinases Itk and Rlk regulate conventional versus innate T-cell development. Immunol. Rev. 228:115–131 10.1111/j.1600-065X.2008.00746.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Readinger J.A., Mueller K.L., Venegas A.M., Horai R., Schwartzberg P.L. 2009. Tec kinases regulate T-lymphocyte development and function: new insights into the roles of Itk and Rlk/Txk. Immunol. Rev. 228:93–114 10.1111/j.1600-065X.2008.00757.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochman Y., Spolski R., Leonard W.J. 2009. New insights into the regulation of T cells by gamma(c) family cytokines. Nat. Rev. Immunol. 9:480–490 10.1038/nri2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu N., Morales J.L., Fowell D., August A. 2010. Modeling susceptibility versus resistance in allergic airway disease reveals regulation by Tec kinase Itk. PLoS ONE. 5:e11348 10.1371/journal.pone.0011348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto F., Geginat J., Lanzavecchia A. 2004. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu. Rev. Immunol. 22:745–763 10.1146/annurev.immunol.22.012703.104702 [DOI] [PubMed] [Google Scholar]
- Savage A.K., Constantinides M.G., Han J., Picard D., Martin E., Li B., Lantz O., Bendelac A. 2008. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 29:391–403 10.1016/j.immuni.2008.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott P., Eaton A., Gause W.C., di Zhou X., Hondowicz B. 1996. Early IL-4 production does not predict susceptibility to Leishmania major. Exp. Parasitol. 84:178–187 10.1006/expr.1996.0103 [DOI] [PubMed] [Google Scholar]
- Shinagawa K., Kojima M. 2003. Mouse model of airway remodeling: strain differences. Am. J. Respir. Crit. Care Med. 168:959–967 10.1164/rccm.200210-1188OC [DOI] [PubMed] [Google Scholar]
- Song A., Chen Y.F., Thamatrakoln K., Storm T.A., Krensky A.M. 1999. RFLAT-1: a new zinc finger transcription factor that activates RANTES gene expression in T lymphocytes. Immunity. 10:93–103 10.1016/S1074-7613(00)80010-2 [DOI] [PubMed] [Google Scholar]
- Song A., Patel A., Thamatrakoln K., Liu C., Feng D., Clayberger C., Krensky A.M. 2002. Functional domains and DNA-binding sequences of RFLAT-1/KLF13, a Krüppel-like transcription factor of activated T lymphocytes. J. Biol. Chem. 277:30055–30065 10.1074/jbc.M204278200 [DOI] [PubMed] [Google Scholar]
- Stanley A.C., Zhou Y., Amante F.H., Randall L.M., Haque A., Pellicci D.G., Hill G.R., Smyth M.J., Godfrey D.I., Engwerda C.R. 2008. Activation of invariant NKT cells exacerbates experimental visceral leishmaniasis. PLoS Pathog. 4:e1000028 10.1371/journal.ppat.1000028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto N., Intlekofer A.M., Northrup J.T., Wherry E.J., Reiner S.L. 2006. Cutting Edge: IL-12 inversely regulates T-bet and eomesodermin expression during pathogen-induced CD8+ T cell differentiation. J. Immunol. 177:7515–7519 [DOI] [PubMed] [Google Scholar]
- Umetsu D.T., Dekruyff R.H. 2010. Natural killer T cells are important in the pathogenesis of asthma: the many pathways to asthma. J. Allergy Clin. Immunol. 125:975–979 10.1016/j.jaci.2010.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hove C.L., Maes T., Cataldo D.D., Guéders M.M., Palmans E., Joos G.F., Tournoy K.G. 2009. Comparison of acute inflammatory and chronic structural asthma-like responses between C57BL/6 and BALB/c mice. Int. Arch. Allergy Immunol. 149:195–207 10.1159/000199715 [DOI] [PubMed] [Google Scholar]
- Verykokakis M., Boos M.D., Bendelac A., Kee B.L. 2010. SAP protein-dependent natural killer T-like cells regulate the development of CD8(+) T cells with innate lymphocyte characteristics. Immunity. 33:203–215 10.1016/j.immuni.2010.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner N., Löhler J., Kunkel E.J., Ley K., Leung E., Krissansen G., Rajewsky K., Müller W. 1996. Critical role for beta7 integrins in formation of the gut-associated lymphoid tissue. Nature. 382:366–370 10.1038/382366a0 [DOI] [PubMed] [Google Scholar]
- Weinreich M.A., Takada K., Skon C., Reiner S.L., Jameson S.C., Hogquist K.A. 2009. KLF2 transcription-factor deficiency in T cells results in unrestrained cytokine production and upregulation of bystander chemokine receptors. Immunity. 31:122–130 10.1016/j.immuni.2009.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich M.A., Odumade O.A., Jameson S.C., Hogquist K.A. 2010. T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nat. Immunol. 11:709–716 10.1038/ni.1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M.A., Bevan M.J. 2007. Effector and memory CTL differentiation. Annu. Rev. Immunol. 25:171–192 10.1146/annurev.immunol.25.022106.141548 [DOI] [PubMed] [Google Scholar]
- Xie J.H., Nomura N., Lu M., Chen S.L., Koch G.E., Weng Y., Rosa R., Di Salvo J., Mudgett J., Peterson L.B., et al. 2003. Antibody-mediated blockade of the CXCR3 chemokine receptor results in diminished recruitment of T helper 1 cells into sites of inflammation. J. Leukoc. Biol. 73:771–780 10.1189/jlb.1102573 [DOI] [PubMed] [Google Scholar]
- Yagi R., Junttila I.S., Wei G., Urban J.F., Jr, Zhao K., Paul W.E., Zhu J. 2010. The transcription factor GATA3 actively represses RUNX3 protein-regulated production of interferon-gamma. Immunity. 32:507–517 10.1016/j.immuni.2010.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto T., Bendelac A., Watson C., Hu-Li J., Paul W.E. 1995. Role of NK1.1+ T cells in a TH2 response and in immunoglobulin E production. Science. 270:1845–1847 10.1126/science.270.5243.1845 [DOI] [PubMed] [Google Scholar]
- Zhao X., Tang Y., Qu B., Cui H., Wang S., Wang L., Luo X., Huang X., Li J., Chen S., Shen N. 2010. MicroRNA-125a contributes to elevated inflammatory chemokine RANTES levels via targeting KLF13 in systemic lupus erythematosus. Arthritis Rheum. 62:3425–3435 10.1002/art.27632 [DOI] [PubMed] [Google Scholar]
- Zhou M., McPherson L., Feng D., Song A., Dong C., Lyu S.C., Zhou L., Shi X., Ahn Y.T., Wang D., et al. 2007. Kruppel-like transcription factor 13 regulates T lymphocyte survival in vivo. J. Immunol. 178:5496–5504 [DOI] [PMC free article] [PubMed] [Google Scholar]