T-bet acts as a functional repressor in association with Bcl-6 to antagonize SOCS1, SOCS3, TCF-1, and late-stage IFN-γ to regulate Th1 development.
Abstract
The T-box transcription factor T-bet is important for the differentiation of naive CD4+ T helper cells (Th cells) into the Th1 phenotype. Much is known about T-bet’s role as a transcriptional activator, but less is known about the mechanisms by which T-bet functionally represses alternative Th cell genetic programs. In this study, we first identify Socs1, Socs3, and Tcf7 (TCF-1) as gene targets that are negatively regulated by T-bet. Significantly, T-bet’s role in the repression of these genes is through a direct interaction with their promoters. Consistent with this, we identified two T-bet DNA-binding elements in the Socs1 promoter that are functionally used to down-regulate transcription in primary Th1 cells. Importantly, T-bet’s novel role in transcriptional repression is because of its ability to physically associate with, and functionally recruit, the transcriptional repressor Bcl-6 to a subset of promoters. Furthermore, T-bet functionally recruits Bcl-6 to the Ifng locus in late stages of Th1 differentiation to repress its activity, possibly to prevent the overproduction of IFN-γ, which could result in autoimmunity. Collectively, these data establish a novel mechanism for T-bet–mediated gene repression in which two lineage-defining transcription factors, one a classical activator and one a repressor, collaborate to promote and properly regulate Th1 development.
CD4+ T cells are central to the adaptive immune response. Naive CD4+ T cells can differentiate into several distinct effector cell lineages (Murphy and Reiner, 2002). These include, but are not limited to, the original Th cell subsets, Th1 and Th2, and the more recently defined Th17, regulatory T cell (T reg cell), and T follicular helper cell (Tfh cell) populations. Not surprisingly, each of these cell types harbors a somewhat unique gene expression profile. These distinct profiles are in part regulated by lineage-defining transcription factors, sometimes deemed master regulators. These include T-bet for Th1 cells, GATA-3 for Th2 cells, Foxp3 for T reg cells, ROR-γt for Th17 cells, and more recently, the transcriptional repressor Bcl-6 for Tfh cells (Zheng and Flavell, 1997; Szabo et al., 2000; Hori et al., 2003; Ivanov et al., 2006; Johnston et al., 2009; Nurieva et al., 2009; Yu et al., 2009a). The diversity of expression profiles allows each of these cell types to play roles in a variety of immune responses ranging from immune tolerance to promoting antibody generation (Zhu et al., 2010).
The prevailing concept has been that each Th cell type is separate from the other lineages and strictly defined by the expression of lineage-defining master regulator transcription factors. However, recent data suggest that determining whether these cell types represent a plastic subset versus an endpoint lineage may not be that straightforward because of the fact that many of the lineage-defining factors are expressed in multiple subsets of Th cells as well as more divergent cell types (Zhu and Paul, 2010a,b). For example, the Th1 cell lineage–defining factor T-bet is expressed at varying levels in Th1, T reg, Th17, and Tfh cells (Szabo et al., 2000; Nurieva et al., 2009; Oldenhove et al., 2009; Wei et al., 2009). These studies suggest that instead of a single master regulator for each lineage, the expression levels of, and potential interactions between, lineage-defining transcription factors may drive the overall development and function of a given naive CD4+ T cell. This leads to the intriguing possibility that the simultaneous expression of two or more lineage-defining factors may promote plasticity between cell subsets dependent on the levels of each factor present in a given condition (Zhou et al., 2009; O’Shea and Paul, 2010). Therefore, more comprehensive studies addressing the molecular mechanisms by which these factors regulate gene expression both cooperatively and independently from one another are important for understanding the true capability of the cell.
The interplay between the expression, localization, and activity of specific transcription factors as well as their ability to bind to and influence the local chromatin structure determines the overall mechanisms of gene control during Th cell development. A subset of these transcription factors, including the Th1 cell lineage–defining factor T-bet, are able to regulate both the activation and repression of genetic loci to promote the development of lineage-specific gene expression patterns (Finotto et al., 2002; Szabo et al., 2002; Djuretic et al., 2007). Although much is known about how T-bet directly activates a few prototypic Th1 target genes, surprisingly little is known about the identity of the genes silenced by T-bet and the mechanisms by which it down-regulates gene expression. In addition, it is still unclear whether T-bet is able to directly repress target genes or rather T-bet’s ability to functionally repress gene expression is strictly indirect (Hwang et al., 2005). Therefore, identifying genes that are repressed by T-bet is long overdue and will aid in understanding the mechanisms involved in establishing Th cell genetic programs.
To begin to address these unanswered questions, we identified three biologically important genes, Socs1, Socs3, and Tcf7, that were expressed at higher levels in T-bet–deficient CD4+ Th1 cells as compared with their WT counterparts. We show that the promoters of these genes are directly bound and functionally regulated by T-bet. The mechanisms by which T-bet functionally represses these genes are related to its physical association with the Tfh cell lineage–defining transcriptional repressor, Bcl-6. Importantly, T-bet’s ability to associate with Bcl-6 allows T-bet, which normally serves as a transcriptional activator, to instead effectively direct a specifically targeted gene repression program. Indeed, we demonstrate in primary Th1 cells that a Bcl-6 repressive complex is targeted by T-bet to the promoters of Socs1, Socs3, and Tcf7, which leads to their T-bet–dependent transcriptional repression during Th1 cell development. Finally, we show that Bcl-6 also is recruited to the Ifng locus in a T-bet–dependent manner during late time points of Th1 differentiation correlating with the loss of Ifng expression. Collectively, our study provides evidence for a mechanism of transcriptional repression in which two lineage-defining factors collaborate to promote the development and proper functioning of a single Th cell lineage.
RESULTS
T-bet binds to the Socs1, Socs3, and Tcf7 promoter regions and represses their transcription
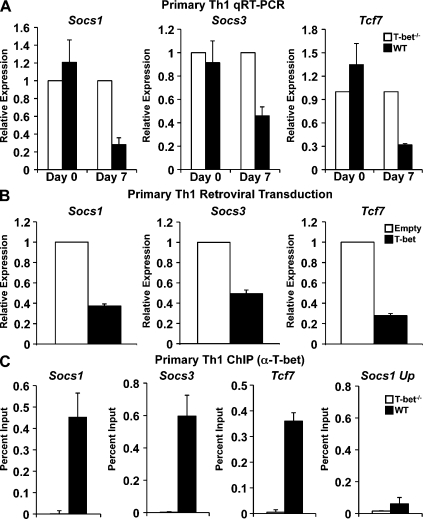
We first wanted to identify gene targets repressed by T-bet in Th1 cells for the purpose of subjecting them to comprehensive transcriptional experiments. This will provide a model system to start to illuminate the detailed transcriptional regulation mechanisms that T-bet utilizes to repress alternative Th cell genetic programs and ultimately promote Th1 cell lineage differentiation. First, similar to a previous study (Jenner et al., 2009), we performed a microarray analysis comparing WT with T-bet−/− CD4+ T cells skewed for 7 d in Th1 conditions to identify candidate targets for the in depth molecular experiments. We identified several genes that had a higher expression level in T-bet−/− as compared with WT Th1 cells, and we selected three for further analysis. Interestingly, the suppressor of cytokine signaling family members Socs1 and Socs3 were functionally repressed by T-bet expression in Th1 cells (Fig. 1 A). As their names suggest, Socs proteins are involved in the inhibition of cytokine signaling pathways, and both Socs1 and Socs3 have been linked to the negative regulation of the canonical Th1 cytokine, IFN-γ (Alexander et al., 1999; Eyles et al., 2002; Seki et al., 2003; Yamamoto et al., 2003; Harada et al., 2007; Palmer and Restifo, 2009). In addition to Socs1 and Socs3, the gene Tcf7 was more highly expressed in T-bet−/− Th1 cells (Fig. 1 A). Significantly, the protein product of theTcf7 gene, TCF-1, is a transcription factor that was recently shown to promote the development of the Th2 lineage (Yu et al., 2009b). Thus, the biological roles for Socs1, Socs3, and Tcf7 make them logical targets to be repressed by T-bet to promote Th1 differentiation, and they provide a good model for determining the mechanisms by which T-bet mediates the repression of alternative Th cell fates.
Figure 1.
T-bet binds to and represses the transcription of Socs1, Socs3, and Tcf7. (A) RNA was isolated from WT and T-bet–deficient primary CD4+ T cells either directly ex vivo or after polarization in Th1 conditions for 7 d. Socs1, Socs3, and Tcf7 transcript levels were analyzed by quantitative RT-PCR. Results were normalized to the values obtained for Actb and are expressed as a ratio relative to the T-bet−/− sample. (B) RNA was isolated from T-bet−/− CD4+ T cells transduced with an empty expression vector or one containing T-bet. Transcript levels were determined by quantitative RT-PCR, and the data were normalized and represented as in A. (C) T-bet association with the promoters of Socs1, Socs3, and Tcf7 was assessed by ChIP. Chromatin was isolated from either WT or T-bet−/− CD4+ T cells stimulated under Th1 conditions for 7 d. Chromatin samples were immunoprecipitated with antibodies to T-bet or a nonspecific antibody control. After purification, immunoprecipitated DNA was quantitated by qPCR. qPCR signals were normalized to the nonspecific antibody control as well as a standardized aliquot of the input chromatin. (A–C) Data represent the mean of three (A and B) or five (C) independent experiments (error bars indicate SEM).
To confirm that Socs1, Socs3, and Tcf7 were functionally repressed by T-bet, we performed a quantitative RT-PCR analysis comparing RNA isolated from WT or T-bet−/− CD4+ T cells skewed under Th1 conditions for 7 d (Fig. 1 A). Consistently higher transcript levels for Socs1, Socs3, and Tcf7 were detected in the T-bet–deficient cells as compared with their WT counterparts at day 7 of Th1 differentiation. As a control, we also examined gene expression directly ex vivo (day 0) in CD4+ T cells that did not yet express high levels of T-bet. Importantly, in these freshly isolated naive CD4+ T cells, we observed no difference in Socs1, Socs3, or Tcf7 transcript levels between WT and T-bet−/− T cells. These results suggest that the expression of Socs1, Socs3, and Tcf7 is functionally repressed by T-bet during the course of Th1 differentiation.
To further confirm that the T-bet–dependent transcriptional repression of these genes is cell intrinsic and not caused by another secondary defect in the T-bet−/− cells, we performed retroviral transduction experiments to express WT T-bet in T-bet−/− CD4+ T cells. Consistent with a role for T-bet in repressing Socs1, Socs3, and Tcf7 gene transcription, retroviral transduction of T-bet into T-bet−/− CD4+ T cells skewed in Th1 conditions caused a decrease in their expression as compared with an empty vector control (Fig. 1 B). Collectively, these data suggest that T-bet has the ability to functionally repress Socs1, Socs3, and Tcf7 transcription.
We next performed chromatin immunoprecipitation (IP [ChIP]) assays to examine whether T-bet associates with the Socs1, Socs3, and Tcf7 promoters to determine whether the T-bet–mediated functional repression may be the result of a direct role for T-bet. Importantly, T-bet associated with the promoter regions of Socs1, Socs3, and Tcf7 in primary CD4+ Th1 cells (Fig. 1 C). As negative controls, there were no detectable signals in the T-bet–precipitated chromatin at these promoters in T-bet−/− Th1 cells, and there was no enrichment at a region upstream of the Socs1 promoter in the T-bet–precipitated sample from the WT Th1 cells (Fig. 1 C). These data suggest that the repression of Socs1, Socs3, and Tcf7 has the potential to be, at least in part, caused by direct T-bet binding at each locus.
T-bet recruits permissive chromatin-remodeling complexes to target promoters
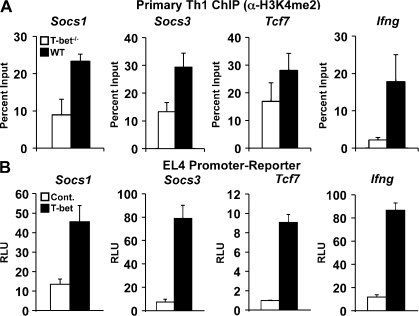
Previous studies have demonstrated that T-bet binding to positively regulated target promoters leads to altered histone modifications that are consistent with a permissive chromatin environment. Specifically, T-bet physically recruits chromatin-modifying complexes to induce the permissive histone 3-lysine 4 dimethyl (H3K4me2) mark and an SWI/SNF-dependent general chromatin-remodeling event (Lewis et al., 2007; Miller et al., 2008, 2010). It is possible that T-bet may recruit the same permissive chromatin-remodeling complexes to all target genes, with a downstream event accounting for target gene–specific activation versus repression. Alternatively, T-bet may functionally recruit a unique, repressive chromatin-remodeling complex to the repressed target genes to effectively close the chromatin structure. To explore these possibilities, we performed ChIP assays in the presence and absence of T-bet in primary Th1 cells to determine the relative levels of the permissive H3K4me2 modification. Importantly, at the negatively regulated Socs1, Socs3, and Tcf7 loci, as well as the positively regulated Ifng locus, there were higher levels of the permissive H3K4me2 modification in WT Th1 cells in comparison with the T-bet−/− Th1 cells (Fig. 2 A). Additionally, we used a restriction enzyme accessibility assay to measure the T-bet and SWI/SNF-dependent general chromatin remodeling at the promoters (Miller et al., 2010). In both primary Th1 cells that express T-bet and EL4 T cells transfected with a T-bet expression vector, we observed increased accessibility at the promoters of both the positively (Ifng) and negatively (Socs1, Socs3, and Tcf7) regulated target genes when compared with their T-bet–deficient counterpart cells (Fig. S1). Collectively, these data suggest that T-bet functionally induces a more accessible chromatin environment independent from the final outcome on the target gene expression.
Figure 2.
The T-bet–dependent repression of Socs1, Socs3, and Tcf7 is independent of the chromatin environment and requires a corepressor. (A) H3K4me2 levels at the promoters of Socs1, Socs3, Tcf7, and Ifng were assessed by ChIP. Chromatin was isolated from either WT or T-bet−/− CD4+ T cells skewed in Th1 conditions. Chromatin samples were immunoprecipitated with antibodies specific to the H3K4me2 mark or a nonspecific antibody control and quantitated as described in Fig. 1. (B) T-bet–dependent promoter-luciferase reporter activity was examined for Socs1, Socs3, Tcf7, and Ifng. EL4 T cells were transfected with the indicated pGL3 promoter-reporter construct and either an empty vector control (Cont.) or a T-bet expression plasmid. After P/I stimulation, luciferase values were measured and normalized to the activity obtained for a cotransfected renilla control. (A and B) Data represent the mean of four (A) or six (B) independent experiments (error bars indicate SEM). RLU, relative light units.
Overexpression of T-bet alone does not functionally repress Socs1, Socs3, and Tcf7 promoter activity
Another mechanism for T-bet–mediated gene repression could involve competition for an overlapping binding site. In this case, T-bet binding would displace a transcriptional activator by partially or completely blocking its binding site in the promoter. In this scenario, T-bet expression alone would be sufficient to repress the negatively regulated target genes. To test this mechanism, we cloned the Socs1, Socs3, and Tcf7 promoters upstream of a luciferase reporter vector (pGL3) and performed promoter-reporter assays. For comparison, we also examined an Ifng promoter-reporter construct, which is activated by T-bet (Shnyreva et al., 2004). The reporter constructs were cotransfected into EL4 T cells with either a T-bet expression plasmid or a control empty expression vector. Interestingly, T-bet overexpression resulted in enhanced promoter-reporter activity for all promoters tested (Fig. 2 B).
It is important to note that the overexpression of T-bet causes the up-regulation of several endogenous T-bet target genes in EL4 cells including other transcription factors (Beima et al., 2006). Therefore, we hypothesized that the activation of the promoter-reporter constructs for the negatively regulated genes was most likely caused by the up-regulation of another secondary endogenous regulatory factor rather than the direct transactivation capabilities of T-bet at their promoters. To test this possibility, we examined promoter-reporter activity in response to a T-bet mutant that cannot activate endogenous target genes. Importantly, this T-bet mutant is deficient in its ability to interact with chromatin-remodeling complexes but is still capable of binding to DNA and performing general chromatin-independent transactivation events (Miller et al., 2008). The T-bet mutant construct did not activate the Socs1, Socs3, or Tcf7 promoters but was still able to induce activity of the Ifng promoter-reporter as a control (Fig. S2). These data suggest that the T-bet–dependent up-regulation of a secondary endogenous factor is responsible for the activation observed for the Socs1, Socs3, and Tcf7 promoter-reporter constructs. Significantly, the T-bet mutant did not further repress the negatively regulated promoter-reporters, suggesting that T-bet alone does not inherently repress these promoters by blocking the binding of an essential activator (Fig. S2).
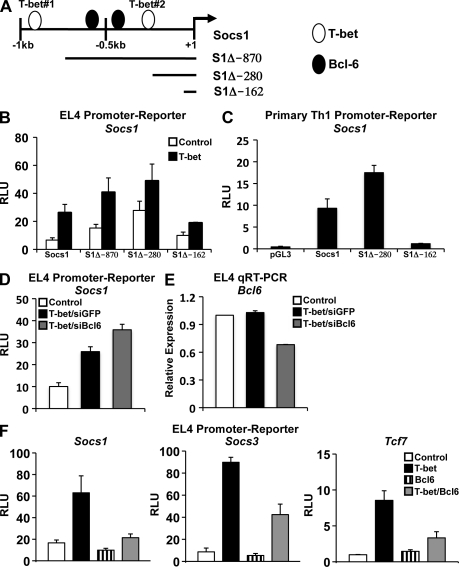
The data presented thus far strongly suggest that T-bet does not independently repress the transcription of these negatively regulated gene targets. Rather, the data support a model in which T-bet may act in conjunction with a transcriptional corepressor to inhibit gene expression. We next wanted to identify putative candidates that might contribute to the T-bet–dependent repression of these target genes. To address this question, we examined the composition of potential DNA-binding motifs for T-bet and transcriptional repressors in the Socs1 promoter using a computer software program (Genomatix). Interestingly, there are two putative T-bet binding sites as well as two potential binding elements for the transcriptional repressor Bcl-6 in the Socs1 promoter (Fig. 3 A and Fig. S3). Significantly, both the Socs3 and Tcf7 promoters also contain T-bet and Bcl-6 binding sites (Fig. S3 A).
Figure 3.
Bcl-6 represses the promoter activity of Socs1, Socs3, and Tcf7. (A) The schematic indicates the location of predicted T-bet binding sites in the Socs1 promoter as well as the Socs1 truncation mutant constructs (see Fig. S3 for precise location). (B and C) EL4 (B) or primary WT Th1 (C) cells were transfected with the indicated Socs1 truncation or full-length promoter-reporter construct. In addition, EL4 cells were cotransfected with either an empty expression vector (control) or one expressing T-bet. Luciferase reporter activity was normalized to the activity obtained for the cotransfected renilla control. (D) EL4 cells were transfected with the Socs1 promoter-reporter in conjunction with a control or T-bet expression vector and a control siRNA (siGFP) or one specific to Bcl-6 (siBcl6). After P/I stimulation, luciferase promoter-reporter values were normalized to the renilla control. (E) EL4 cells were transfected and treated as in D. RNA was isolated, and Bcl6 transcript levels were analyzed by quantitative RT-PCR. Results were normalized to the values obtained for Actb and are expressed as a ratio relative to the control sample. (F) EL4 T cells were cotransfected with the indicated pGL3 promoter-reporter construct and an empty vector control, T-bet, Bcl-6, or T-bet and Bcl-6 in combination. After P/I stimulation, luciferase promoter-reporter values were normalized to the renilla control. (B–F) Data represent the mean of three (B–E) or four (F) independent experiments (error bars indicate SEM). RLU, relative light units.
To ascertain the role, if any, for these sites in Socs1 regulation, we engineered truncation mutant Socs1 promoter-reporter constructs progressively deleting both the T-bet and Bcl-6 binding sites (Fig. 3 A). Intriguingly, progressive truncation of these sites enhanced promoter-reporter activity in comparison with the full-length Socs1 construct in both the EL4 cell line and primary Th1 cells (Fig. 3, B and C). These data indicate that a potential negative regulatory element may be located within this region (−1 kb to −280 bp) of the Socs1 promoter.
Bcl-6 represses promoter activity of Socs1, Socs3, and Tcf7
Given the failure of T-bet overexpression alone to repress promoter activity, we examined the functional consequence that Bcl-6 has on Socs1 promoter-reporter activity in the presence of T-bet (Fig. 3, D–F). We first examined Socs1 promoter-reporter activity in response to Bcl-6 knockdown. Importantly, when a small interfering RNA (siRNA) specific to Bcl-6 was transfected in combination with T-bet, there was a modest enhancement of the Socs1 promoter-reporter activity when compared with cotransfection with a control siRNA (Fig. 3 D). Although the observed effect was modest, it was proportional to the knockdown in Bcl-6 levels in these cells (Fig. 3 E). Importantly, an increase in Tcf7 promoter-reporter activity was also observed upon Bcl-6 siRNA knockdown (Fig. S4 A). These data are consistent with a role for Bcl-6 in the repression of these genes.
To complement the knockdown results, we also performed overexpression experiments. Strikingly, simultaneous expression of Bcl-6 and T-bet substantially inhibited Socs1 promoter activity in comparison with T-bet transfection alone (Fig. 3 F). Furthermore, Bcl-6 and T-bet overexpression also inhibited the activity of the Socs3 and Tcf7 promoter-reporter constructs. Importantly, expression of Bcl-6 alone only minimally inhibited overall promoter activity in the absence of T-bet. Collectively, these data suggest that a combination of T-bet and Bcl-6 represses the promoter activity of Socs1, Socs3, and Tcf7.
T-bet–dependent repression is independent of Bcl-6 DNA-binding activity
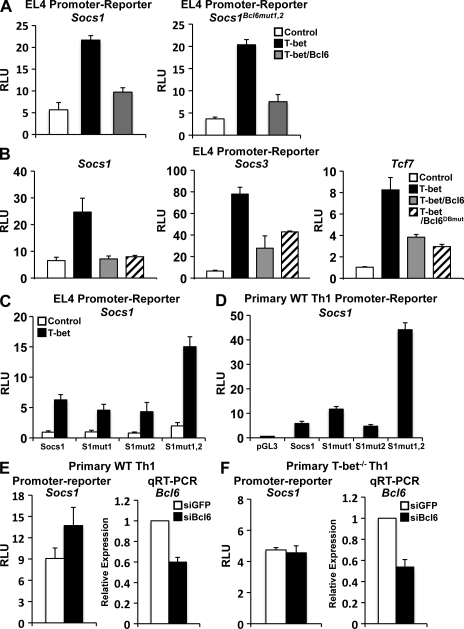
To ascertain whether the Bcl-6–mediated repression is caused by direct binding of Bcl-6 to the Socs1 promoter, we made a Socs1 promoter-reporter construct with mutations in the Bcl-6 DNA-binding elements (Fig. 4 A and Fig. S3 B). Surprisingly, this mutant Socs1 promoter was still repressed by the simultaneous overexpression of T-bet and Bcl-6 (Fig. 4 A). These data suggest that Bcl-6 may be functionally recruited to the Socs1 promoter by another factor. To confirm this unexpected result and to rule out the possibility that a cryptic Bcl-6 DNA-binding element is found in the Socs1 promoter, we next created a Bcl-6 construct with a point mutation in its DNA-binding domain. This mutation disrupts the DNA-binding activity of Bcl-6 but keeps its transcriptional repression capacity intact (Mascle et al., 2003). Significantly, in the presence of T-bet, both WT Bcl-6 and the Bcl-6 DNA-binding mutant (Bcl6DBmut) repressed the T-bet–dependent activity of the Socs1 promoter to similar levels (Fig. 4 B and Fig. S5 B). In addition, both the Socs3 and Tcf7 promoter-reporters were repressed to an equivalent degree by the WT Bcl-6 and Bcl-6 DNA binding–deficient proteins (Fig. 4 B). To further confirm these results, we also examined a second independent Bcl-6 DNA-binding point mutant construct and observed similar results (Figs. S5 and S6). Importantly, as a control, WT Bcl-6 but not the two Bcl6DBmut proteins repressed the promoter activity of Gzmb, a known direct Bcl-6 gene target (Fig. S6; Yoshida et al., 2006). These data indicate that in contrast to Gzmb, the repressive capability of Bcl-6 at the Socs1, Socs3, and Tcf7 promoters does not require its direct DNA-binding activity despite the presence of Bcl-6 DNA-binding elements. Rather, these data highly suggest that Bcl-6 is recruited to these promoters by another factor. Because of the data presented thus far demonstrating a functional role for T-bet in the repression of Socs1, Socs3, and Tcf7 in primary Th1 cells, we hypothesized that T-bet is required for the recruitment of Bcl-6 to these loci.
Figure 4.
Bcl-6–dependent repression of Socs1, Socs3, and Tcf7 is independent of Bcl-6 DNA binding. (A) The Socs1Bcl6mut1,2 or WT promoter-reporter constructs were cotransfected with an empty expression vector (control), T-bet, or T-bet and Bcl-6. (See Fig. S3 for precise location of the Bcl-6 binding site mutations in the Socs1Bcl6mut1,2 construct.) (B) EL4 T cells were transfected with the indicated pGL3 promoter construct and an empty vector control, T-bet, T-bet and Bcl-6, or T-bet and the Bcl-6 DNA-binding mutant (Bcl6DBmut). (C) The indicated Socs1 point mutants (in T-bet binding sites) or WT promoter-reporter constructs were cotransfected with either an empty expression vector (control) or one expressing T-bet. (A–C) The samples were stimulated with P/I, and the reporter activity was measured and normalized to the activity of a renilla control. (D) The indicated Socs1 point mutant (in T-bet binding sites) or WT promoter-reporter constructs were transfected into primary WT Th1 cells. After overnight incubation, reporter activity was measured and normalized to a renilla control. (E and F) Primary WT (E) or T-bet−/− (F) Th1 cells were transfected with the Socs1 promoter-reporter and either a control siRNA (siGFP) or one specific to Bcl-6 (siBcl6). Luciferase promoter-reporter values were normalized to the renilla control. RNA was isolated from the primary Th1 cells, and Bcl6 transcript levels were analyzed by quantitative RT-PCR. Results were normalized to the values obtained for Actb and are expressed as a ratio relative to the control sample. (A–F) Data represent the mean of three (A and D) or four (B, C, E, and F) independent experiments (error bars indicate SEM). RLU, relative light units.
To start to test this hypothesis, we first examined whether the putative T-bet DNA-binding elements are required for the negative regulation of the Socs1 promoter. Significantly, as the data from Fig. 3 indicate, there is an increase in Socs1 promoter activity in the truncation constructs that do not contain the T-bet binding elements. We next made a series of Socs1 reporter constructs mutating the most distal T-bet site (S1mut1), the most proximal T-bet site (S1mut2), or both in combination (S1mut1,2; Fig. S3 C). We transfected these T-bet point mutant constructs into EL4 cells in either the presence or absence of T-bet. Interestingly, the elimination of both T-bet binding sites in the S1mut1,2 construct resulted in a striking increase in Socs1 promoter activity, whereas the mutation of either T-bet site individually (S1mut1 or S1mut2) had little to no effect on promoter activity (Fig. 4 C). Importantly, we also examined the same Socs1 point mutant constructs in the more physiological setting of primary Th1 cells. Once again, the simultaneous mutation of both the proximal and distal T-bet sites led to a significant increase in promoter activity in comparison with the WT Socs1 promoter-reporter construct (Fig. 4 D). Consistent with the conclusion that the repression mediated through these T-box DNA-binding elements is T-bet dependent, we did not observe any enhancement of the Socs1 T-bet binding site mutant construct in comparison with the WT control in T-bet−/− primary Th1 cells (Fig. 4 D vs. Fig. S7). These results suggest that the two T-bet binding elements are critical for the repression of the Socs1 promoter.
We next wanted to determine whether the repressive capability of Bcl-6 requires a T-bet–dependent activity. To test this possibility, we transfected WT versus T-bet−/− primary Th1 cells with either a control siRNA or one specific to Bcl-6. In WT Th1 cells, the knockdown of Bcl-6 enhanced Socs1 promoter-reporter activity (Fig. 4 E). Significantly, Bcl-6 knockdown had no effect on Socs1 activity in the absence of T-bet in the T-bet−/− Th1 cells (Fig. 4 F). Importantly, the Bcl-6 knockdown levels were equivalent in WT and T-bet−/− Th1 cells, with the degree of Bcl-6 reduction proportional to the enhancement of Socs1 promoter-reporter activity in the WT cells (Fig. 4, E and F). Collectively, these data suggest that the Bcl-6–mediated repression of the Socs1 promoter is dependent on T-bet.
T-bet recruits Bcl-6 to the Socs1, Socs3, and Tcf7 promoters in Th1 cells
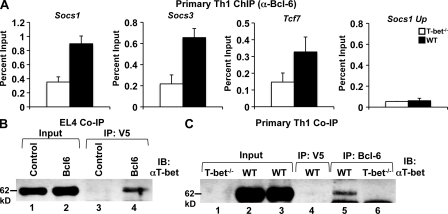
The data we have presented thus far strongly imply that both a T-bet DNA binding–dependent and a Bcl-6 DNA binding–independent activity are necessary for the repression of the Socs1, Socs3, and Tcf7 genes. To test whether T-bet is required for the recruitment of Bcl-6 to these promoters in Th1 cells, we performed ChIP experiments to examine Bcl-6 association with the Socs1, Socs3, and Tcf7 promoters in WT versus T-bet−/− primary Th1 cells. Significantly, there was a higher level of Bcl-6 association with all three promoters in the WT versus T-bet−/− Th1 cells (Fig. 5 A). As a negative control, we did not detect Bcl-6 at a location upstream of the Socs1 promoter (which also is not bound by T-bet; Fig. 1 C), indicating that the targeting of Bcl-6 is specific to the T-bet–bound area in the promoter region. As an additional control, the protein and RNA expression levels of Bcl-6 were similar in WT and T-bet−/− Th1 cells (Fig. S8), indicating that alterations in the amount of Bcl-6 present in each cell type cannot explain the observed differences. Collectively, these data strongly suggest that the recruitment of Bcl-6 to the Socs1, Socs3, and Tcf7 promoters is dependent on the presence of T-bet.
Figure 5.
Bcl-6 is recruited to the promoters of Socs1, Socs3, and Tcf7 through a physical interaction with T-bet in Th1 cells. (A) Chromatin was isolated from either WT or T-bet−/− CD4+ T cells stimulated in Th1 conditions for 7 d. Chromatin samples were immunoprecipitated with either an antibody to Bcl-6 or a nonspecific antibody control. Immunoprecipitated DNA was quantitated by qPCR and normalized to the nonspecific antibody control as well as a standardized aliquot of the input chromatin. Data represent the mean of five independent experiments (error bars indicate SEM). (B) EL4 T cells were transfected with an untagged T-bet expression construct in combination with either a V5 epitope–tagged Bcl-6 expression vector or empty vector control. Lysates were prepared and immunoprecipitated with an antibody for the V5 epitope tag and then probed with a T-bet–specific antibody. (C) Lysates from either WT or T-bet−/− primary Th1 cells were immunoprecipitated with either a Bcl-6 (lanes 5 and 6) or control antibody (lane 4). After IP, a Western blot analysis was performed with a T-bet–specific antibody. (B and C) Data are representative of three (B) or five (C) independent experiments. IB, immunoblot.
T-bet and Bcl-6 physically interact to repress transcription
Collectively, the aforementioned data suggest that T-bet is required to recruit Bcl-6 to the negatively regulated promoters. We next wanted to determine whether T-bet targets Bcl-6 to these promoters by physically interacting with Bcl-6. To start to test this possibility, we performed a co-IP experiment in which we cotransfected EL4 T cells with T-bet in combination with either an empty vector construct or one expressing Bcl-6. Importantly, T-bet specifically precipitated with Bcl-6, indicating that T-bet and Bcl-6 can be found in the same complex in this overexpression system (Fig. 5 B). We then wanted to explore whether T-bet and Bcl-6 form a stable complex at their endogenous expression levels in primary CD4+ Th1 cells. In co-IP experiments, T-bet coprecipitated with Bcl-6 in WT Th1 cells (Fig. 5 C). As a negative control, the T-bet signal was not detected in either WT Th1 cells immunoprecipitated with a nonspecific antibody control or in T-bet−/− CD4+ Th1 cells (Fig. 5 C). Collectively, these experiments suggest that T-bet and Bcl-6 have the ability to form a repressive complex to inhibit the transcription of Socs1, Socs3, and Tcf7.
The C-terminal domain of Bcl-6, which contains six zinc fingers, is required for its association with T-bet
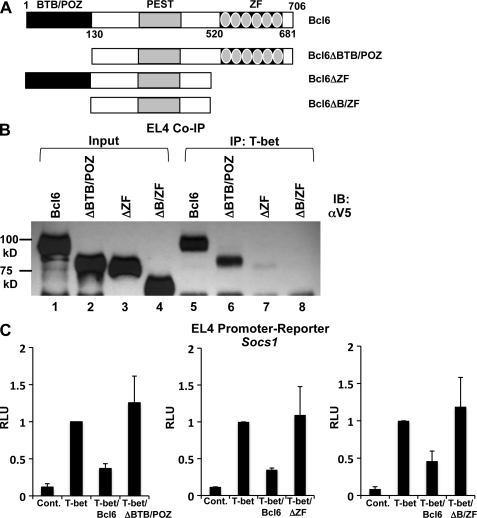
We next wanted to determine which domain or domains within Bcl-6 mediate its interaction with T-bet. To accomplish this, we created Bcl-6 truncation constructs and tested their ability to form a complex with T-bet in EL4 co-IP experiments (Fig. 6 A). A Bcl-6 construct lacking the repressive BTB/POZ domain still associated with T-bet, with only a modest reduction in its interaction as compared with WT Bcl-6 (Fig. 6 B). Interestingly, Bcl-6 mutant constructs with a C-terminal truncation deleting its six zinc fingers were no longer able to associate with T-bet (Fig. 6 B). These data suggest that the C-terminal domain of Bcl-6 is required for its interaction with T-bet. This is an intriguing result because the six zinc fingers within this domain are known to mediate both DNA binding as well as other protein–protein interactions (Mascle et al., 2003). It will be important to determine in future experiments whether the association between T-bet and Bcl-6 impedes the DNA-binding activity of Bcl-6, which could preferentially redirect its targeting to T-bet binding elements.
Figure 6.
The C-terminal zinc finger domain of Bcl-6 is required for its association with T-bet. (A) The schematic indicates the location of the primary functional domains contained within Bcl-6. These include an N-terminal BTB/POZ domain, a C-terminal domain containing six individual zinc fingers (displayed as gray ovals), and a centrally located PEST domain. Diagrams of the Bcl-6 truncation mutant constructs are shown below the WT schematic. (B) EL4 T cells were transfected with an untagged T-bet expression construct in combination with V5 epitope–tagged WT Bcl-6 (lanes 1 and 5) or the Bcl-6 truncation mutant expression vectors as indicated (lanes 2–4 and 6–8). Lysates were prepared and immunoprecipitated with an antibody to T-bet (lanes 5–8). The blot was then probed with a V5 epitope tag–specific antibody. Data are representative of three independent experiments. (C) EL4 T cells were transfected with the full-length Socs1 promoter-reporter construct and an empty vector control, T-bet, T-bet and Bcl-6, or T-bet and the indicated Bcl-6 truncation mutants. After P/I stimulation, luciferase promoter-reporter values were normalized to the renilla control. Data represent the mean of three independent experiments (error bars indicate SEM). IB, immunoblot; RLU, relative light units.
We next assessed the functional role for each domain in mediating T-bet–dependent repression. Significantly, the Bcl-6 C-terminal zinc finger truncation constructs defective in their ability to associate with T-bet no longer repressed Socs1 promoter-reporter activity (Fig. 6 C). In addition, the Bcl-6 BTB/POZ domain truncation also could not repress Socs1 promoter-reporter activity (Fig. 6 C). This is consistent with the required role for the BTB/POZ domain in the repressive capability of Bcl-6 (Basso and Dalla-Favera, 2010). We also observed similar losses of repressive capabilities for the Bcl-6 truncations at the Tcf7 promoter (Fig. S9). Collectively, these data are consistent with the hypothesis that Bcl-6 is functionally recruited by T-bet to repress the activity of these promoters.
Bcl-6 also represses Ifng, a target gene positively regulated by T-bet
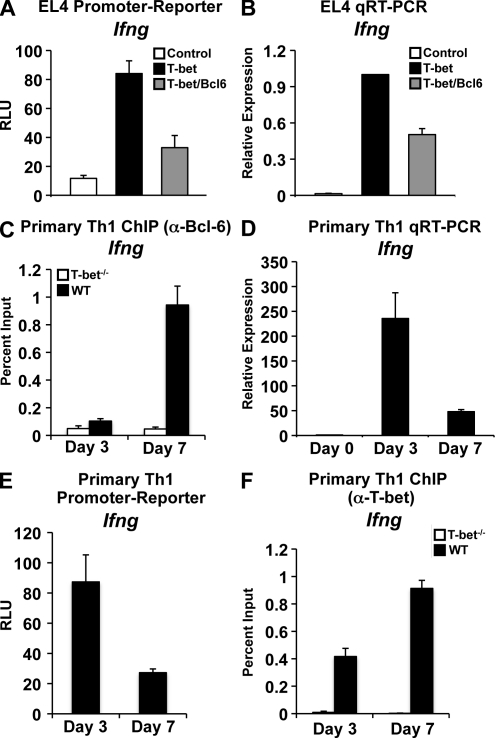
We next sought to determine whether Bcl-6 plays a role in regulating genes that are normally activated by T-bet in Th1 cells. To accomplish this goal, we first examined the effect of Bcl-6 overexpression on T-bet–dependent Ifng promoter-reporter activity. Interestingly, Bcl-6 repressed the T-bet–dependent activation of the Ifng promoter similar to its functional effect at the Socs1, Socs3, and Tcf7 promoters (Fig. 7 A). In addition, the knockdown of Bcl-6 also enhanced the T-bet–dependent activation of the Ifng promoter-reporter construct (Fig. S4). To assess whether Bcl-6 has the same effect on Ifng transcription in the context of the chromatin environment, we overexpressed T-bet either alone or in combination with Bcl-6 and assessed endogenous Ifng gene transcripts by quantitative RT-PCR. Significantly, Bcl-6 also repressed the T-bet–dependent activation of endogenous Ifng gene expression (Fig. 7 B). These results suggest that Bcl-6 plays a role in the functional repression of both the genes that are positively and negatively regulated by T-bet.
Figure 7.
T-bet–dependent recruitment of Bcl-6 to the Ifng promoter in late time points of Th1 culture correlates with a reduction in Ifng gene transcription. (A) EL4 T cells were cotransfected with the indicated pGL3 promoter-reporter construct and an empty expression vector control, T-bet, or T-bet and Bcl-6. After P/I stimulation, luciferase values were measured and normalized to a renilla control. (B) EL4 T cells were transfected and treated as in A followed by RNA isolation. A quantitative RT-PCR analysis to determine endogenous Ifng transcript levels was performed and normalized to Actb. (C) Chromatin was isolated from either WT or T-bet−/− CD4+ T cells at day 3 or 7 of Th1 differentiation. Chromatin samples were immunoprecipitated with either an antibody to Bcl-6 or a nonspecific antibody control. Immunoprecipitated DNA was quantitated by qPCR with signals for the Ifng promoter normalized to the nonspecific antibody control as well as a standardized aliquot of the input chromatin. (D) RNA samples were isolated from WT CD4+ T cells at days 3 and 7 of Th1 differentiation. A quantitative RT-PCR analysis was performed to determine Ifng transcript levels, and results were normalized to Actb. The data are represented as fold change over the Ifng transcript levels at day 0. (E) The Ifng promoter-reporter construct was transfected into primary Th1 cells at days 3 and 7 of Th1 differentiation. After overnight incubation, luciferase values were measured and normalized to both empty vector (pGL3) and renilla controls. (F) ChIP samples were treated and analyzed as in C with the exception that a T-bet antibody was used for IP. (A–F) Data represent the mean of four (A, C, and F) or three (B, D, and E) independent experiments (error bars indicate SEM). RLU, relative light units.
These intriguing observations led us to examine whether Bcl-6 may play a role in regulating Ifng expression during primary Th1 cell differentiation. We first wanted to determine whether Bcl-6 is present at the Ifng promoter in primary Th1 cells. Interestingly, in a ChIP analysis comparing WT with T-bet−/− Th1 cells (day 7), similar to the negatively regulated targets, Bcl-6 associated with the Ifng promoter in a T-bet–dependent manner (Fig. 7 C). Given these results, we hypothesized that the Bcl-6 association with the Ifng locus may temper its expression. If this is the case, Bcl-6 may be mechanistically down-regulating Ifng in the late stages of the Th1 effector cell to prevent the overproduction of IFN-γ, which could result in potential T cell–mediated autoimmune diseases. To begin to test this possibility, we analyzed the time course of Ifng expression during Th1 differentiation. We observed high levels of Ifng transcripts at day 3 and a subsequent decrease in transcript levels by day 7 of Th1 differentiation (Fig. 7 D). Additionally, Ifng promoter-reporter activity was higher in day 3 versus day 7 primary Th1 cells (Fig. 7 E). Importantly, Bcl-6 was not associated with the Ifng promoter at day 3 when Ifng expression was approximately fivefold higher (Fig. 7 C). Therefore, the decrease in Ifng expression from day 3 to 7 correlates with the T-bet–dependent recruitment of Bcl-6 to the Ifng promoter specifically at this late stage of Th1 differentiation.
To rule out the possibility that the lower levels of Ifng transcripts in the late stages of Th1 differentiation are caused by a reduction in T-bet binding to the Ifng promoter, we performed a ChIP analysis on day 3 versus day 7 from WT and T-bet−/− Th1 cells. In contrast to the decrease observed in Ifng transcripts at day 7, there was actually an increase in the amount of T-bet bound to the promoter at day 7 (Fig. 7 F). Thus, the amount of T-bet bound to the Ifng promoter does not explain the decrease in transcription. Collectively, these data suggest the mechanism by which Bcl-6 represses T-bet–dependent promoter activity may be important for inhibiting both genes that are activated and repressed by T-bet in Th1 cells.
DISCUSSION
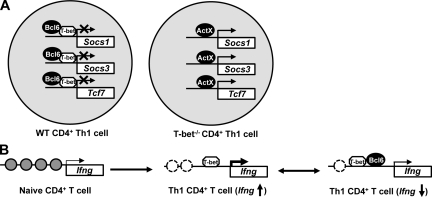
In this study, we have identified three novel gene targets that are negatively regulated by T-bet in Th1 cells. Interestingly, the mechanism by which T-bet functionally represses these genes involves a physical association with the Tfh cell lineage–defining factor, Bcl-6. Bcl-6 is targeted by T-bet to the promoters of Socs1, Socs3, and Tcf7 and, once associated, represses the transcriptional activation of these genes. Interestingly, Bcl-6 also antagonizes the T-bet–dependent activation of Ifng, a gene positively regulated by T-bet. Collectively, the data suggest that the interplay between T-bet and Bcl-6 is important for establishing appropriate gene expression patterns in Th1 cell development (Fig. 8).
Figure 8.
Models representing the mechanisms by which T-bet and Bcl-6 cooperate to regulate transcription. (A) Schematic portraying the differences in the regulation of Socs1, Socs3, and Tcf7 genes in WT versus T-bet–deficient CD4+ T cells. In WT Th1 cells, T-bet–dependent targeting of Bcl-6 to the Socs1, Socs3, and Tcf7 promoters results in transcriptional repression. Conversely, the absence of T-bet in the T-bet−/− Th1 cells results in a loss of Bcl-6 targeting to the Socs1, Socs3, and Tcf7 promoters and gene activation. (B) A schematic representation of Bcl-6 repression at the Ifng locus. In the naive CD4+ T cells, gray circles represent closed chromatin. By day 3 of Th1 differentiation, T-bet binds to the promoter and functionally induces chromatin remodeling, resulting in enhanced Ifng transcription. By day 7 of Th1 differentiation, the reduced level of Ifng transcription correlates with the T-bet–dependent recruitment of Bcl-6 to the Ifng promoter.
Recently, the transcriptional repressor Bcl-6 was identified as a key factor that promotes Tfh cell differentiation (Johnston et al., 2009; Nurieva et al., 2009; Yu et al., 2009a). In this study, we demonstrate that in differentiated Th1 cells, T-bet and Bcl-6 collaborate to repress the transcription of several genetic loci. Our findings, as well as others that demonstrate the simultaneous expression of multiple lineage-defining factors in a single Th cell type, now strongly suggest that the notion of one unique master regulator per Th cell type may not be entirely accurate (Zhou et al., 2009; O’Shea and Paul, 2010; Zhu and Paul, 2010b). Instead, the regulation of critical lineage-defining factors by each other and the interactions between them are clearly important aspects in determining Th cell fate. For example, it is known that Th1 cells express high levels of T-bet and low levels of Bcl-6, whereas the reverse scenario is found in Tfh cells (Johnston et al., 2009; Nurieva et al., 2009; Yu et al., 2009a). Our data suggest that when T-bet is in excess, it will target the available Bcl-6 to a subset of promoters that are preferentially repressed during the later stages of Th1 development. Furthermore, these data argue that a simple change in the expression levels of either of these two lineage-defining factors could result in dramatic changes in gene expression networks and possibly have implications for plasticity between the Th1 and Tfh cell types.
In addition to Tfh cells, Bcl-6 has been implicated in the formation of both CD4+ and CD8+ memory T cells. Central memory T cells are characterized by reduced expression of Blimp1 as compared with effector memory T cells, which express high levels of Blimp1 (Kallies et al., 2009; Rutishauser et al., 2009). Importantly, Blimp1 and Bcl-6 negatively regulate each other’s expression, which results in an inverse correlation between the two factors. Thus, central memory T cells with low Blimp1 should express higher levels of Bcl-6 in comparison with effector memory T cells. Interestingly, the cytokine production from a reactivated central memory T cell is indeed subdued in comparison with that of an effector memory T cell response (Kaech and Wherry, 2007). Therefore, it is possible that the mechanisms described in this study could aid in tempering T-box protein–dependent cytokine production in central memory T cells.
It is also interesting to speculate about how the DNA binding–independent targeting of Bcl-6 to gene loci may differentially affect gene expression patterns in different cell types. To date, microarray datasets examining gene expression patterns between immune cell types that harbor high Bcl-6 expression levels contain few similarities (Johnston et al., 2009; Rutishauser et al., 2009). Our study demonstrates that some gene targets repressed by Bcl-6 are determined by its recruitment to T-bet binding sites rather than the DNA-binding capabilities of Bcl-6 itself. Therefore, if the mechanisms described in this study are conserved in other cells types, Bcl-6 could be targeted to genetic loci based on the predominance of a particular transcription factor, in this case, T-bet. This will then alter both T-bet and Bcl-6 target gene expression patterns to create a specialized profile for the cell.
The identification of Socs1, Socs3, and Tcf7 as genes that are negatively regulated by T-bet provides new insight into the gene expression network that is established during Th1 development. Importantly, the repression of the Socs family members Socs1 and Socs3 is necessary to create a functional Th1 cell. Numerous studies have shown that Socs1 and Socs3 repress the Th1 cell lineage and promote other Th cell types such as Th2 cells (Diehl et al., 2000; Egwuagu et al., 2002; Eyles et al., 2002; Seki et al., 2003). For Socs1, this is because of its role in inhibiting the IFN-γ and STAT1 signaling pathway, a signaling module critical for the development and function of a Th1 cell (Sakamoto et al., 1998; Alexander et al., 1999). Significantly, Socs1-deficient CD4+ T cells have a propensity to develop into Th1 cells, whereas overexpression of Socs1 inhibits the Th1 cell lineage (Diehl et al., 2000; Eyles et al., 2002). Similar to the role for Socs1 in antagonizing IFN-γ signaling, Socs3 plays an analogous role in disrupting STAT4 and IL-12 signaling, which is another pathway crucial for Th1 cell development (Seki et al., 2003; Yamamoto et al., 2003; Takatori et al., 2005). Furthermore, Socs3 is preferentially expressed in the CD4+ T cells of the Th2 cell lineage (Egwuagu et al., 2002). Importantly, the dysregulation of Socs1 and Socs3 has been linked to the onset of autoimmune diseases and some types of cancer, providing strong evidence for their biological significance in the proper regulation of the immune response (Cottet et al., 2001; Karlsen et al., 2001; He et al., 2003; Seki et al., 2003; Chong et al., 2004; Weniger et al., 2006). Collectively, the impact of Socs1 and Socs3 on immune cell function and its involvement in human disease highlights the importance in determining the mechanisms governing their proper regulation, including the role of T-bet and Bcl-6 in this process.
Our experiments also identified Tcf7 as a target of T-bet–mediated repression. Recently, the protein product of the Tcf7 gene, TCF-1, has been linked to the transcriptional regulation of GATA-3, the lineage-defining factor for the Th2 lineage (Yu et al., 2009b). TCF-1 promotes the development of the Th2 cell fate by promoting GATA-3 expression and repressing IFN-γ production. Therefore, the repression of Tcf7 by T-bet will inhibit the Th2 fate. To underscore the biological significance of this, Tcf7 has been linked to the onset of Type 1 diabetes, a disease which has also been associated with dysregulation of T-bet and Th1 cells (Noble et al., 2003; Erlich et al., 2009).
Interestingly, our data also suggest that Bcl-6 is directly involved in the regulation of the canonical Th1 cytokine, IFN-γ. Previous studies examining Tfh cells found that when Bcl-6 is in excess during Tfh cell development, Ifng expression is significantly impaired (Nurieva et al., 2009; Yu et al., 2009a). In addition, overexpression of Bcl-6 in primary T cells results in a modest decrease in Ifng transcription, whereas the absence of Bcl-6 results in a dramatic increase (Nurieva et al., 2009). These observations were previously attributed to an indirect mechanism related to the ability of Bcl-6 to repress Tbx21 (the gene encoding T-bet). However, the absolute levels of T-bet did not always correlate with Ifng expression in these experiments, and this explanation also could not account for an initial reduction in Ifng before the down-regulation of T-bet (Nurieva et al., 2009). Our data now extend these findings by identifying Ifng as a direct target of Bcl-6 repression. We demonstrate that Bcl-6 associates with the Ifng locus in a T-bet-dependent manner and inhibits its transcription at late time points in Th1 differentiation. Collectively, these data suggest that the interplay between T-bet and Bcl-6 may help to ensure that the expression of Ifng remains at a moderate level so that there is neither too little, which would impede the initiation of an appropriate immune response for pathogen clearance, nor too much, which could result in autoimmunity.
It is interesting to note the timing for the T-bet–dependent association of Bcl-6 with the Ifng promoter. Ifng transcript levels inversely correlate with the recruitment of Bcl-6 at later time points in Th1 differentiation. However, the environmental or intracellular cues that prompt the targeting of Bcl-6 to the Ifng locus are currently unknown. The answers to these questions await a better understanding of the additional regulatory signaling pathways and complexes involved in this process. A comprehensive understanding of these mechanisms will provide insight into Th1 and Tfh cell fate choices and the potential for plasticity between these two Th cell fates. Collectively, the study presented here provides evidence for a model of gene regulation in which two lineage-defining transcription factors collaborate to promote the development of the Th1 cell fate by repressing genes that contribute to the differentiation of other Th cell types, as well as regulating the expression of genes necessary for the proper functioning of the Th1 cell.
MATERIALS AND METHODS
Cell culture and transfection
Primary T cells.
Primary CD4+ T cells were isolated from the spleen and lymph nodes of WT and T-bet−/− mice using the Mag Cellect kit (R&D Systems) as previously described (Beima et al., 2006). After isolation, cells were grown on plate-bound α-CD3/α-CD28 and treated with Th1 polarizing cytokines: 5 µg/ml α–IL-4, 5 ng/ml IL-12, and 10 ng/ml IL-2 (National Cancer Institute preclinical repository). On day 3, cells were split and treated under Th1 conditions for an additional 3 d. All experiments involving mice were conducted in accordance with Institutional Animal Care and Use Committee approval. For transfections involving primary cells, the Amaxa nucleofection system (Lonza) was used. Manufacturer protocols were followed using mouse primary T cell solutions and program X-01.
EL4 T cells.
Mouse EL4 T cells (American Type Culture Collection) were grown in RPMI with 10% FBS and penicillin/streptomycin. Cells were transfected using the Amaxa nucleofection system using program 0-17 and solution V as previously described (Beima et al., 2006). After transfection, the cells were left untreated or treated overnight with PMA (Sigma-Aldrich) and ionomycin (P/I; Sigma-Aldrich) as indicated. Western blot analysis was performed to ensure equal protein expression.
Retroviral constructs and transduction
The T-bet–GFP-MSCV construct was generated by subcloning T-bet into the GFP-MSCV vector. T-bet–MSCV was transfected into Phoenix cells according to protocols from G. Nolan (Stanford University, Stanford, CA). Primary CD4+ T cells from T-bet−/− mice were spin infected with T-bet–GFP-MSCV or GFP-MSCV viral supernatant in the presence of 5 mg/ml polybrene at 37°C for 90 min at 2,000 rpm and then left overnight. After transduction, cells were cultured in Th1 polarizing conditions. GFP-positive cells were sorted on day 7, and RNA was harvested for analysis.
RNA and RT-PCR
RNA was isolated using the Nucleospin (Macherey-Nagel) RNA purification protocol. For quantitation, cDNA was prepared using the First Strand Superscript II Synthesis System protocol (Invitrogen). cDNA was then used as a template for quantitative PCR (qPCR) reactions using the qPCR Sybr Green mix (Thermo Fisher Scientific) and gene-specific primers (Table S1). Data obtained were normalized to Actb levels for each sample.
ChIP assay
The ChIP assay was performed as described previously (Beima et al., 2006; Lewis et al., 2007; Miller et al., 2008). Antibodies to T-bet (H-210) and Bcl-6 (C-19) were obtained from Santa Cruz Biotechnology, Inc., whereas the antibody to the H3K4me2 mark was obtained from Millipore. In brief, chromatin was harvested from primary WT and T-bet−/− CD4+ T cells stimulated under Th1 skewing conditions. Chromatin was incubated overnight with the indicated antibody. After a series of washes, DNA was isolated, and qPCR was conducted (for primers see Table S1). Samples were normalized to a standardized total input DNA control and nonspecific antibody control (IgG).
Restriction enzyme accessibility assay
This protocol has been described previously (Weinmann et al., 1999; Oestreich et al., 2006; Miller et al., 2010). In brief, nuclei from 5 × 106 cells were isolated and treated with the restriction enzyme BanII for 2.5 min at 37°C. After digestion, DNA was purified, and samples were ligated to a BanII-specific linker using T4 DNA ligase (New England Biolabs, Inc.). Nested ligation-mediated PCR was performed with primers specific for both the linker and the promoter of the gene analyzed (Table S1). DNA input was controlled for by amplifying Actb, which is independent of the restriction enzyme digestion.
Promoter-reporter assay
The Socs1, Socs3, and Tcf7 promoters were cloned into the pGL3 luciferase reporter construct (Promega). Socs1 mutant promoter-reporters were generated by either PCR, in the case of the truncation mutants, or QuikChange (Agilent Technologies) for the T-bet and Bcl-6 DNA-binding element mutant constructs. The Ifng promoter-reporter construct has been described previously (Shnyreva et al., 2004). EL4 cells were transfected with the indicated promoter-reporter constructs in combination with T-bet, Bcl-6, Bcl-6 mutant V5-tagged expression vectors, or an empty vector control as indicated. A TK-renilla control plasmid was used to normalize transfection efficiency. Cell aliquots were collected for analysis 16–22 h after transfection. For luciferase analysis, the cells were treated and analyzed according to the Dual-Luciferase Reporter assay protocol (Promega). The siRNA experiments included a siBcl6 smart-pool (Thermo Fisher Scientific) and siGFP (Invitrogen) and were performed as described previously (Miller et al., 2008, 2010).
Co-IP
The co-IP assay was performed as described previously (Miller et al., 2008, 2010). A V5 antibody (Invitrogen) and a T-bet–specific antibody (H-210) were used for the experiments in the EL4 cells. For endogenous protein expression in the primary Th1 cells, an antibody for Bcl-6 (C-19) was used for IP, with the presence of T-bet detected using the 4B10 antibody (Santa Cruz Biotechnology, Inc.).
Online supplemental material
Fig. S1 uses a chromatin accessibility assay to show that the T-bet–dependent repression of Socs1, Socs3, and Tcf7 is independent of the chromatin environment. Fig. S2 demonstrates that a T-bet mutant deficient in chromatin-remodeling capabilities is unable to activate the Socs1, Socs3, and Tcf7 promoters. Fig. S3 is a schematic representation of the Socs1, Socs3, and Tcf7 promoters displaying the locations of the T-bet and Bcl-6 binding sites. Fig. S4 is a promoter-reporter assay demonstrating an increase in Tcf7 and Ifng promoter activity when Bcl-6 levels are reduced. Fig. S5 is a Western blot analysis of the expression levels for the WT Bcl-6 and Bcl6DBmut constructs. Fig. S6 demonstrates that the overexpression of two unique Bcl6DBmut constructs represses Socs1 and Tcf7 promoter-reporter activity but not that of Gzmb. Fig. S7 shows that there is no alteration of Socs1 promoter activity in T-bet−/− primary Th1 cells when T-bet binding sites are mutated. Fig. S8 demonstrates that the expression levels of Bcl-6 are similar in WT and T-bet−/− primary Th1 cells. Fig. S9 is a promoter-reporter assay demonstrating the requirement for both the BTB/POZ and C-terminal zinc finger domains in the repression of Tcf7 promoter activity by Bcl-6. Table S1 lists the primer sequences used in this study. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20102144/DC1.
Acknowledgments
We would like to thank Sarah Mohn and Margaret Brassil for technical assistance as well as Sara Miller for help with the restriction enzyme assay and helpful discussions. We also would like to thank the National Cancer Institute preclinical repository for the IL-2 and anti–IL-4.
The project described was supported by grants from the National Institute of Allergy and Infectious Diseases (NIAID; AI061061 and AI07272) and the American Cancer Society (RSG-09-045-01-DDC) to A.S. Weinmann. K.J. Oestreich was supported by a postdoctoral training grant from the NIAID (NRSA 5T32 AI 07411). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIAID.
The authors declare that they have no financial conflicts of interest.
Footnotes
Abbreviations used:
- ChIP
- chromatin IP
- H3K4me2
- histone 3-lysine 4 dimethyl
- IP
- immunoprecipitation
- P/I
- PMA and ionomycin
- qPCR
- quantitative PCR
- siRNA
- small interfering RNA
- Tfh cell
- T follicular helper cell
References
- Alexander W.S., Starr R., Fenner J.E., Scott C.L., Handman E., Sprigg N.S., Corbin J.E., Cornish A.L., Darwiche R., Owczarek C.M., et al. 1999. SOCS1 is a critical inhibitor of interferon gamma signaling and prevents the potentially fatal neonatal actions of this cytokine. Cell. 98:597–608 10.1016/S0092-8674(00)80047-1 [DOI] [PubMed] [Google Scholar]
- Basso K., Dalla-Favera R. 2010. BCL6: master regulator of the germinal center reaction and key oncogene in B cell lymphomagenesis. Adv. Immunol. 105:193–210 10.1016/S0065-2776(10)05007-8 [DOI] [PubMed] [Google Scholar]
- Beima K.M., Miazgowicz M.M., Lewis M.D., Yan P.S., Huang T.H., Weinmann A.S. 2006. T-bet binding to newly identified target gene promoters is cell type-independent but results in variable context-dependent functional effects. J. Biol. Chem. 281:11992–12000 10.1074/jbc.M513613200 [DOI] [PubMed] [Google Scholar]
- Chong M.M., Chen Y., Darwiche R., Dudek N.L., Irawaty W., Santamaria P., Allison J., Kay T.W., Thomas H.E. 2004. Suppressor of cytokine signaling-1 overexpression protects pancreatic beta cells from CD8+ T cell-mediated autoimmune destruction. J. Immunol. 172:5714–5721 [DOI] [PubMed] [Google Scholar]
- Cottet S., Dupraz P., Hamburger F., Dolci W., Jaquet M., Thorens B. 2001. SOCS-1 protein prevents Janus Kinase/STAT-dependent inhibition of beta cell insulin gene transcription and secretion in response to interferon-gamma. J. Biol. Chem. 276:25862–25870 10.1074/jbc.M103235200 [DOI] [PubMed] [Google Scholar]
- Diehl S., Anguita J., Hoffmeyer A., Zapton T., Ihle J.N., Fikrig E., Rincón M. 2000. Inhibition of Th1 differentiation by IL-6 is mediated by SOCS1. Immunity. 13:805–815 10.1016/S1074-7613(00)00078-9 [DOI] [PubMed] [Google Scholar]
- Djuretic I.M., Levanon D., Negreanu V., Groner Y., Rao A., Ansel K.M. 2007. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat. Immunol. 8:145–153 10.1038/ni1424 [DOI] [PubMed] [Google Scholar]
- Egwuagu C.E., Yu C.R., Zhang M., Mahdi R.M., Kim S.J., Gery I. 2002. Suppressors of cytokine signaling proteins are differentially expressed in Th1 and Th2 cells: implications for Th cell lineage commitment and maintenance. J. Immunol. 168:3181–3187 [DOI] [PubMed] [Google Scholar]
- Erlich H.A., Valdes A.M., Julier C., Mirel D., Noble J.A. 2009. Evidence for association of the TCF7 locus with type I diabetes. Genes Immun. 10:S54–S59 10.1038/gene.2009.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyles J.L., Metcalf D., Grusby M.J., Hilton D.J., Starr R. 2002. Negative regulation of interleukin-12 signaling by suppressor of cytokine signaling-1. J. Biol. Chem. 277:43735–43740 10.1074/jbc.M208586200 [DOI] [PubMed] [Google Scholar]
- Finotto S., Neurath M.F., Glickman J.N., Qin S., Lehr H.A., Green F.H., Ackerman K., Haley K., Galle P.R., Szabo S.J., et al. 2002. Development of spontaneous airway changes consistent with human asthma in mice lacking T-bet. Science. 295:336–338 10.1126/science.1065544 [DOI] [PubMed] [Google Scholar]
- Harada M., Nakashima K., Hirota T., Shimizu M., Doi S., Fujita K., Shirakawa T., Enomoto T., Yoshikawa M., Moriyama H., et al. 2007. Functional polymorphism in the suppressor of cytokine signaling 1 gene associated with adult asthma. Am. J. Respir. Cell Mol. Biol. 36:491–496 10.1165/rcmb.2006-0090OC [DOI] [PubMed] [Google Scholar]
- He B., You L., Uematsu K., Zang K., Xu Z., Lee A.Y., Costello J.F., McCormick F., Jablons D.M. 2003. SOCS-3 is frequently silenced by hypermethylation and suppresses cell growth in human lung cancer. Proc. Natl. Acad. Sci. USA. 100:14133–14138 10.1073/pnas.2232790100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori S., Nomura T., Sakaguchi S. 2003. Control of regulatory T cell development by the transcription factor Foxp3. Science. 299:1057–1061 10.1126/science.1079490 [DOI] [PubMed] [Google Scholar]
- Hwang E.S., Szabo S.J., Schwartzberg P.L., Glimcher L.H. 2005. T helper cell fate specified by kinase-mediated interaction of T-bet with GATA-3. Science. 307:430–433 10.1126/science.1103336 [DOI] [PubMed] [Google Scholar]
- Ivanov I.I., McKenzie B.S., Zhou L., Tadokoro C.E., Lepelley A., Lafaille J.J., Cua D.J., Littman D.R. 2006. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 126:1121–1133 10.1016/j.cell.2006.07.035 [DOI] [PubMed] [Google Scholar]
- Jenner R.G., Townsend M.J., Jackson I., Sun K., Bouwman R.D., Young R.A., Glimcher L.H., Lord G.M. 2009. The transcription factors T-bet and GATA-3 control alternative pathways of T-cell differentiation through a shared set of target genes. Proc. Natl. Acad. Sci. USA. 106:17876–17881 10.1073/pnas.0909357106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R.J., Poholek A.C., DiToro D., Yusuf I., Eto D., Barnett B., Dent A.L., Craft J., Crotty S. 2009. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 325:1006–1010 10.1126/science.1175870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S.M., Wherry E.J. 2007. Heterogeneity and cell-fate decisions in effector and memory CD8+ T cell differentiation during viral infection. Immunity. 27:393–405 10.1016/j.immuni.2007.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies A., Xin A., Belz G.T., Nutt S.L. 2009. Blimp-1 transcription factor is required for the differentiation of effector CD8(+) T cells and memory responses. Immunity. 31:283–295 10.1016/j.immuni.2009.06.021 [DOI] [PubMed] [Google Scholar]
- Karlsen A.E., Rønn S.G., Lindberg K., Johannesen J., Galsgaard E.D., Pociot F., Nielsen J.H., Mandrup-Poulsen T., Nerup J., Billestrup N. 2001. Suppressor of cytokine signaling 3 (SOCS-3) protects beta -cells against interleukin-1beta - and interferon-gamma -mediated toxicity. Proc. Natl. Acad. Sci. USA. 98:12191–12196 10.1073/pnas.211445998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis M.D., Miller S.A., Miazgowicz M.M., Beima K.M., Weinmann A.S. 2007. T-bet’s ability to regulate individual target genes requires the conserved T-box domain to recruit histone methyltransferase activity and a separate family member-specific transactivation domain. Mol. Cell. Biol. 27:8510–8521 10.1128/MCB.01615-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascle X., Albagli O., Lemercier C. 2003. Point mutations in BCL6 DNA-binding domain reveal distinct roles for the six zinc fingers. Biochem. Biophys. Res. Commun. 300:391–396 10.1016/S0006-291X(02)02873-5 [DOI] [PubMed] [Google Scholar]
- Miller S.A., Huang A.C., Miazgowicz M.M., Brassil M.M., Weinmann A.S. 2008. Coordinated but physically separable interaction with H3K27-demethylase and H3K4-methyltransferase activities are required for T-box protein-mediated activation of developmental gene expression. Genes Dev. 22:2980–2993 10.1101/gad.1689708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S.A., Mohn S.E., Weinmann A.S. 2010. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol. Cell. 40:594–605 10.1016/j.molcel.2010.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K.M., Reiner S.L. 2002. The lineage decisions of helper T cells. Nat. Rev. Immunol. 2:933–944 10.1038/nri954 [DOI] [PubMed] [Google Scholar]
- Noble J.A., White A.M., Lazzeroni L.C., Valdes A.M., Mirel D.B., Reynolds R., Grupe A., Aud D., Peltz G., Erlich H.A. 2003. A polymorphism in the TCF7 gene, C883A, is associated with type 1 diabetes. Diabetes. 52:1579–1582 10.2337/diabetes.52.6.1579 [DOI] [PubMed] [Google Scholar]
- Nurieva R.I., Chung Y., Martinez G.J., Yang X.O., Tanaka S., Matskevitch T.D., Wang Y.H., Dong C. 2009. Bcl6 mediates the development of T follicular helper cells. Science. 325:1001–1005 10.1126/science.1176676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea J.J., Paul W.E. 2010. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 327:1098–1102 10.1126/science.1178334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich K.J., Cobb R.M., Pierce S., Chen J., Ferrier P., Oltz E.M. 2006. Regulation of TCRbeta gene assembly by a promoter/enhancer holocomplex. Immunity. 24:381–391 10.1016/j.immuni.2006.02.009 [DOI] [PubMed] [Google Scholar]
- Oldenhove G., Bouladoux N., Wohlfert E.A., Hall J.A., Chou D., Dos Santos L., O’Brien S., Blank R., Lamb E., Natarajan S., et al. 2009. Decrease of Foxp3+ Treg cell number and acquisition of effector cell phenotype during lethal infection. Immunity. 31:772–786 10.1016/j.immuni.2009.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer D.C., Restifo N.P. 2009. Suppressors of cytokine signaling (SOCS) in T cell differentiation, maturation, and function. Trends Immunol. 30:592–602 10.1016/j.it.2009.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutishauser R.L., Martins G.A., Kalachikov S., Chandele A., Parish I.A., Meffre E., Jacob J., Calame K., Kaech S.M. 2009. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 31:296–308 10.1016/j.immuni.2009.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto H., Yasukawa H., Masuhara M., Tanimura S., Sasaki A., Yuge K., Ohtsubo M., Ohtsuka A., Fujita T., Ohta T., et al. 1998. A Janus kinase inhibitor, JAB, is an interferon-gamma-inducible gene and confers resistance to interferons. Blood. 92:1668–1676 [PubMed] [Google Scholar]
- Seki Y., Inoue H., Nagata N., Hayashi K., Fukuyama S., Matsumoto K., Komine O., Hamano S., Himeno K., Inagaki-Ohara K., et al. 2003. SOCS-3 regulates onset and maintenance of T(H)2-mediated allergic responses. Nat. Med. 9:1047–1054 10.1038/nm896 [DOI] [PubMed] [Google Scholar]
- Shnyreva M., Weaver W.M., Blanchette M., Taylor S.L., Tompa M., Fitzpatrick D.R., Wilson C.B. 2004. Evolutionarily conserved sequence elements that positively regulate IFN-gamma expression in T cells. Proc. Natl. Acad. Sci. USA. 101:12622–12627 10.1073/pnas.0400849101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo S.J., Kim S.T., Costa G.L., Zhang X., Fathman C.G., Glimcher L.H. 2000. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 100:655–669 10.1016/S0092-8674(00)80702-3 [DOI] [PubMed] [Google Scholar]
- Szabo S.J., Sullivan B.M., Stemmann C., Satoskar A.R., Sleckman B.P., Glimcher L.H. 2002. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science. 295:338–342 10.1126/science.1065543 [DOI] [PubMed] [Google Scholar]
- Takatori H., Nakajima H., Kagami S., Hirose K., Suto A., Suzuki K., Kubo M., Yoshimura A., Saito Y., Iwamoto I. 2005. Stat5a inhibits IL-12-induced Th1 cell differentiation through the induction of suppressor of cytokine signaling 3 expression. J. Immunol. 174:4105–4112 [DOI] [PubMed] [Google Scholar]
- Wei G., Wei L., Zhu J., Zang C., Hu-Li J., Yao Z., Cui K., Kanno Y., Roh T.Y., Watford W.T., et al. 2009. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 30:155–167 10.1016/j.immuni.2008.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann A.S., Plevy S.E., Smale S.T. 1999. Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity. 11:665–675 10.1016/S1074-7613(00)80141-7 [DOI] [PubMed] [Google Scholar]
- Weniger M.A., Melzner I., Menz C.K., Wegener S., Bucur A.J., Dorsch K., Mattfeldt T., Barth T.F., Möller P. 2006. Mutations of the tumor suppressor gene SOCS-1 in classical Hodgkin lymphoma are frequent and associated with nuclear phospho-STAT5 accumulation. Oncogene. 25:2679–2684 10.1038/sj.onc.1209151 [DOI] [PubMed] [Google Scholar]
- Yamamoto K., Yamaguchi M., Miyasaka N., Miura O. 2003. SOCS-3 inhibits IL-12-induced STAT4 activation by binding through its SH2 domain to the STAT4 docking site in the IL-12 receptor beta2 subunit. Biochem. Biophys. Res. Commun. 310:1188–1193 10.1016/j.bbrc.2003.09.140 [DOI] [PubMed] [Google Scholar]
- Yoshida K., Sakamoto A., Yamashita K., Arguni E., Horigome S., Arima M., Hatano M., Seki N., Ichikawa T., Tokuhisa T. 2006. Bcl6 controls granzyme B expression in effector CD8+ T cells. Eur. J. Immunol. 36:3146–3156 10.1002/eji.200636165 [DOI] [PubMed] [Google Scholar]
- Yu D., Rao S., Tsai L.M., Lee S.K., He Y., Sutcliffe E.L., Srivastava M., Linterman M., Zheng L., Simpson N., et al. 2009a. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 31:457–468 10.1016/j.immuni.2009.07.002 [DOI] [PubMed] [Google Scholar]
- Yu Q., Sharma A., Oh S.Y., Moon H.G., Hossain M.Z., Salay T.M., Leeds K.E., Du H., Wu B., Waterman M.L., et al. 2009b. T cell factor 1 initiates the T helper type 2 fate by inducing the transcription factor GATA-3 and repressing interferon-gamma. Nat. Immunol. 10:992–999 10.1038/ni.1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W., Flavell R.A. 1997. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 89:587–596 10.1016/S0092-8674(00)80240-8 [DOI] [PubMed] [Google Scholar]
- Zhou L., Chong M.M., Littman D.R. 2009. Plasticity of CD4+ T cell lineage differentiation. Immunity. 30:646–655 10.1016/j.immuni.2009.05.001 [DOI] [PubMed] [Google Scholar]
- Zhu J., Paul W.E. 2010a. CD4+ T cell plasticity-Th2 cells join the crowd. Immunity. 32:11–13 10.1016/j.immuni.2010.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Paul W.E. 2010b. Heterogeneity and plasticity of T helper cells. Cell Res. 20:4–12 10.1038/cr.2009.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Yamane H., Paul W.E. 2010. Differentiation of effector CD4 T cell populations. Annu. Rev. Immunol. 28:445–489 10.1146/annurev-immunol-030409-101212 [DOI] [PMC free article] [PubMed] [Google Scholar]