IL7R-activating mutations identified in B-ALL and T-ALL patient leukemic cells facilitate cytokine-independent growth.
Abstract
Interleukin-7 receptor α (IL7R) is required for normal lymphoid development. Loss-of-function mutations in this gene cause autosomal recessive severe combined immune deficiency. Here, we describe somatic gain-of-function mutations in IL7R in pediatric B and T acute lymphoblastic leukemias. The mutations cause either a serine-to-cysteine substitution at amino acid 185 in the extracellular domain (4 patients) or in-frame insertions and deletions in the transmembrane domain (35 patients). In B cell precursor leukemias, the mutations were associated with the aberrant expression of cytokine receptor-like factor 2 (CRLF2), and the mutant IL-7R proteins formed a functional receptor with CRLF2 for thymic stromal lymphopoietin (TSLP). Biochemical and functional assays reveal that these IL7R mutations are activating mutations conferring cytokine-independent growth of progenitor lymphoid cells. A cysteine, included in all but three of the mutated IL-7R alleles, is essential for the constitutive activation of the receptor. This is the first demonstration of gain-of-function mutations of IL7R. Our current and recent observations of mutations in IL7R and CRLF2, respectively suggest that the addition of cysteine to the juxtamembranous domains is a general mechanism for mutational activation of type I cytokine receptors in leukemia.
IL-7R α (IL7R) is required for normal lymphoid development (Peschon et al., 1994). Loss-of-function mutations in this receptor cause severe combined immune deficiency (OMIM 608971; Puel et al., 1998). IL-7R heterodimerizes either with IL-2R γ (IL2RG) to form a receptor to IL-7 or with cytokine receptor-like factor 2 (CRLF2) in the receptor for thymic stromal lymphopoietin (TSLP; Noguchi et al., 1993; Liu et al., 2007). TSLP is a cytokine that mediates inflammation and allergy expressed on macrophages and T cells (Liu et al., 2007). Signaling from the TSLP receptor activates signal transducer and activator of transcription (STAT5) by phosphorylation of JAK1 and JAK2 through association with IL-7R and CRLF2, respectively (Rochman et al., 2010).
We and others have recently described the aberrant expression of CRLF2 in B cell precursor acute lymphoblastic leukemia (BCP-ALL) caused by genomic aberrations (Mullighan et al., 2009; Russell et al., 2009; Hertzberg et al., 2010; Yoda et al., 2010). These are either translocations into the immunoglobulin heavy locus or interstitial deletions creating chimeric P2RY8-CRLF2 transcripts. Additional mutations in the pathway, most commonly in JAK2 but occasionally in JAK1 or in CRLF2 itself, occur in approximately half of the patients. Overall, CRLF2 deregulation occurs in 5–10% of childhood ALL and in 60% of children with Down syndrome and BCP-ALL (DS-ALL; Izraeli, 2010) and is associated with a worse prognosis (Cario et al., 2010; Harvey et al., 2010; Yoda et al., 2010; Ensor et al., 2011). Importantly, these leukemias may be sensitive to JAK inhibitors, suggesting the potential for a targeted therapy.
IL7R is constitutively expressed in BCP-ALL, and the formation of a TSLP receptor is plausible in cases with aberrant expression of CRLF2 (Hertzberg et al., 2010). Receptor association is functional because primary BCP-ALL cells expressing CRLF2 respond to TSLP, but not to IL-7, by phosphorylation of STAT5 and RPS6 (Tasian et al., 2010). We therefore hypothesized that IL7R might be mutationally activated in ALL samples expressing CRLF2. In addition to confirmation of this hypothesis, we also report the identification of such mutations in 10% of T-ALLs, thereby demonstrating a general involvement of the IL-7R in ALL.
RESULTS AND DISCUSSION
We screened DNA derived from 133 bone marrow samples of BCP-ALLs, including 62 DS-ALLs, with aberrant expression of CRLF2 and additional 153 BCP-ALL not expressing CRLF2 including 21 DS-ALLs. Nine leukemias with IL7R mutations were identified. The rate of mutations in the CRLF2 group (8/133; 6%) was significantly higher than the rest of BCP-ALLs (1/153; 0.6%; P = 0.01, Fisher’s exact test; Table I).
Table I.
Patients with B cell precursor ALL and somatic mutations in IL7R
| ID | IL7R mutation DNA | IL-7R mutation protein | CRLF2 | JAK2 | Gender | Age at Dx | WBC/liter | Events |
| yr | ||||||||
| DS46 | c.819 Ins 12 | 243 InsPPCL | P2RY8-CRLF2 | mut | M | 4.9 | 30.5 × 109 | first CCR |
| DS92 | c.642 A>T | S185C | IGHα translocation | WT | M | 15.1 | 112.8 × 109 | first CCR |
| M90 | c.828 Ins7 Del T | 246 InsKCH | P2RY8-CRLF2 | WT | M | 1.7 | 123 × 109 | relapse |
| M112 | c.642 A>T | S185C | P2RY8-CRLF2 | mut | F | 14.3 | 9.7 × 109 | relapse |
| M117 | c.814 Ins13 Del A | 241 InsFSCGP | P2RY8-CRLF2 | mut | F | 10.3 | 7 × 109 | first CCR |
| M122 | c.642 A>T | S185C | P2RY8-CRLF2 | WT | M | 7.0 | 7.3 × 109 | relapse |
| M124 | c.820 Ins10 Del C | 244 InsCHL | High expression | WT | M | 4.9 | 8.9 × 109 | first CCR |
| M223 | c.642 A>T | S185C | P2RY8-CRLF2 | WT | M | 8.0 | 27 × 109 | first CCR |
| M61 | c.820 ins21 | 244 InsPPVCSVT | CRLF2 not expressed | WT | M | 10.7 | 152 × 109 | relapse |
DS, Down syndrome; M, male; F, female; mut, mutated at JAK2 R683; CCR, continuous complete remission; Dx, Diagnosis; Del, Deletion; Ins, Insertion; WBC, white blood cells count.
Two types of mutations in IL7R were identified (Fig. 1, A and B, and Table I). Replacement of serine with cysteine at position 185 at the extracellular domain in four patients and complex in-frame insertions and deletions resulted in the addition of 3–7 aa at the transmembrane domain in 5 patients. Whereas the inserted amino acids varied from patient to patient, cysteine was always included (Fig. 1 C).
Figure 1.
Somatic mutations of IL7R (NM_002185.2) in patients with BCP-ALL. (A) IL7R mutation localization. Amino acid numbers of the different domains are indicated. (B) Sequences of IL7R insertions and deletions mutations at the transmembrane domain. The inserted sequences are within brackets (All Ins-Del mutations were heterozygous, only the mutated allele is shown after allele separation). N, sites with both WT and mutant allele at the same position; Ins, insertion; Del, deletion. (C) Alignment of WT and mutated IL7R transmembrane domain sequences. Numbers show the positions of nucleotides and corresponding amino acids. The inserted amino acids are shown in red, with the cysteine residue in bold. (D) Expression of mutations: Examples of two mutated IL-7R sequences with S185C and c.814 Ins 13 Del A. The mutated allele is expressed in the RNA from diagnosis, but not in remission samples.
All mutations were heterozygous and somatic, as they were absent in remission bone marrows. The mutated mRNA was expressed (Fig. 1 D and Fig. S1). JAK2 mutations were present in three samples, whereas no mutations in JAK1 were identified. Four patients relapsed. Examination of DNA derived from matched diagnostic and relapse samples revealed clonal diversification. The same IL7R mutation was present in both diagnosis and relapse in patient M61. IL7R mutation was present only at relapse in patient M122, whereas CRLF2 expression was already noted at diagnosis, suggesting that IL7R mutation was a progression event. Conversely, in two patients, M90 and M112 CRLF2 abnormalities and IL7R mutations were present at diagnosis and not in relapse, suggesting that relapse arose from a different subclone. These findings are consistent with the recent studies on the marked clonal diversity of B- and T-ALLs (Rothman et al., 2005; Anderson et al., 2011; Clappier et al., 2011).
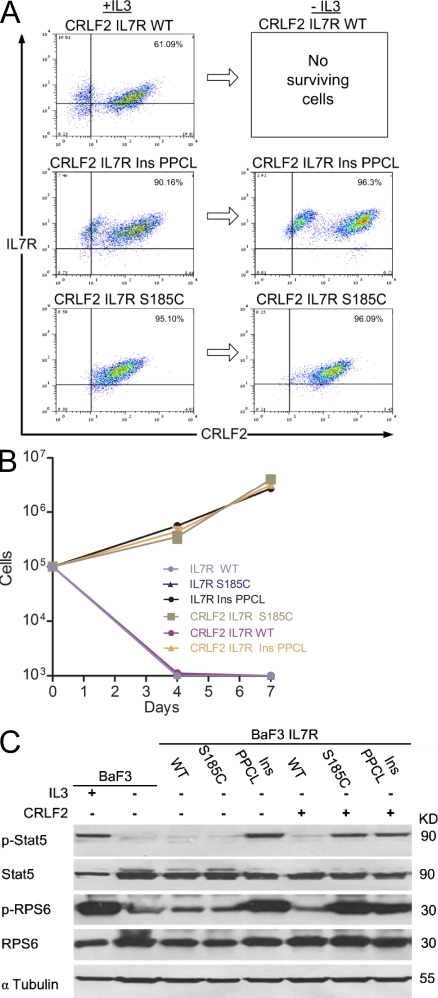
We next tested whether the somatic mutations in IL-7R are gain-of-function mutations that cooperate with CRLF2 to form a constitutively active TSLP receptor. We used the IL-3–dependent mouse pro–B cells BaF3 (not expressing endogenous TSLP receptor; Fig. S2) to generate cell lines that express either WT or mutated IL7R alone or together with CRLF2. We expressed IL-7R S185C and the c.819 Ins12 (CCCCCGTGCCTA) 243 insertion (Ins) PPCL (herein “InsPPCL”) representing the two types of mutations. All proteins were expressed at the cell membrane as demonstrated by flow cytometry (Fig. 2 A). 1 wk after IL-3 withdrawal, only cells transduced with CRLF2 and mutated IL-7R survived (Fig. 2, B and C). Closer examination revealed two populations of surviving BaF3 cells transduced with CRLF2 and IL-7R InsPPLC, suggesting that IL-7R InsPPLC provided survival advantage by itself. Indeed, growth assays of BaF3 cells transduced with the IL-7R InsPPLC construct in the absence of CRLF2 (Fig. 2 B) revealed robust cytokine-independent growth. In contrast, BaF3 cells expressing IL-7R S185C required CRLF2 co-expression for survival. Cells expressing only CRLF2, IL-7R S185C, or WT CRLF2 and WT IL-7R did not grow in the absence of cytokines.
Figure 2.
IL7R mutations are gain-of-function mutations. (A) FACS analysis of BaF3 cells stably transduced with CRLF2 and either IL-7R WT, IL-7R S185C, or IL-7R InsPPCL in the presence of IL-3. The same cells were then grown without IL-3 for 1 wk and then analyzed again. (B) Cytokine withdrawal assay of BaF3 and BaF3-CRLF2 cells transduced with either IL-7R WT, IL-7R S185C, or IL-7R InsPPCL. Error bars represent SE. (C) Constitutive phosphorylation of Stat5 and ribosomal protein S6 (RPS6) in BaF3 and BaF3-CRLF2 cells expressing IL-7R mutants, after 5 h of cytokine deprivation. IL-3+ indicates cells harvested after 5 h of IL-3 deprivation followed by 20 min of IL-3 stimulation. All experiments were repeated four times.
Biochemical analysis of proteins extracted from BaF3 cell was consistent with the growth assays (Fig. 2 C). Both Stat5 and RPS6 were phosphorylated in the absence of cytokine in BaF3 cells transduced with the IL-7R InsPPCL alone, but not in cells expressing IL-7R S185C or WT IL-7R. Co-expression of CRLF2 with each of these mutated IL-7R proteins, but not with the WT IL-7R caused constitutive phosphorylation of Stat5 and RPS6. Together, the functional and biochemical assays demonstrate that the two types of somatic mutations in IL7R are activating mutations causing cytokine independent growth of mouse pro–B cells and constitutive activation of STAT and mTOR pathways. In accordance with the functional studies, the single patient in whom IL7R was mutated in the absence of CRLF2 expression (Table I, patient M61) had the insertion type of mutation with the CRLF2-independent activation phenotype.
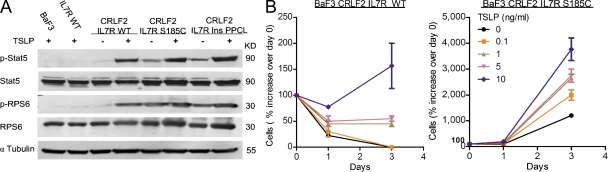
To see if CRLF2 and mutated IL-7R form a functional TSLP receptor, BaF3 cells expressing CRLF2 and either WT or mutated IL-7R were starved from IL-3 and treated with 100 ng/ml TSLP for 25 min. As noted in Fig. 2 C, cells expressing the mutated receptor exhibited Stat5 and RPS6 phosphorylation in the absence of cytokine. Yet all cells responded to TSLP with a marked increase in Stat5 and RPS6 phosphorylation (Fig. 3 A). Thus, CRLF2 and mutated IL-7R form a functional TSLP receptor. To test if the presence of mutated IL-7R sensitizes BaF3 cells to TSLP, we treated BaF3 transduced with CRLF2 and either WT IL-7R or IL-7R S185C with increasing doses of TSLP in the absence of IL-3 (Fig. 3 B). Cells expressing CRLF2–IL-7R S185C grew in the absence of cytokines, but also demonstrated higher sensitivity to TSLP. As little as 0.1 ng/ml TSLP doubled the growth rate, but had no effect on BaF3 cells transduced with CRLF2–IL-7R WT. Similarly, cells transduced with IL-7R InsPPCL also responded to TSLP, in addition to marked self-activation of this allele (Fig. S3).
Figure 3.
CRLF2 and mutated IL7R form a functional TSLP receptor and sensitize cells to TSLP. (A) BaF3 cells expressing CRLF2 and either WT or mutated IL-7R were starved of IL-3 for 5 h, and then treated or not with 100 ng/ml TSLP where indicated for 25 min. (B) BaF3-CRLF2 cells transduced with either WT IL-7R or IL-7R S185C were treated with increasing doses of TSLP in the absence of IL-3. The number of cells was normalized to day 0 (therefore relative growth was measured). Experiments were performed three times. Error bars indicate SE.
Loss-of-function mutations in IL-7R cause severe combined immunodeficiency characterized by the complete absence of T lymphocytes and the presence of B and NK cells (Puel et al., 1998). Both B- and T-ALLs express IL-7R and have been reported to respond to IL-7 (Touw et al., 1990). We have therefore hypothesized that gain-of-function mutations in IL-7R may also be detected in T-ALL. Accordingly, we screened 295 diagnostic childhood T-ALL samples treated on prospective BFM protocols (Kox et al., 2010) for IL7R mutations and identified 30 somatic mutations (10.5%). All the mutations were at the transmembrane domain encoded by exon 6, and all but one were in-frame insertions and deletions (Table II); 27 of the 30 cases included an insertion of cysteine. The patients with mutations were younger and tended to have higher white blood cells counts at diagnosis (Table S1). There was no association between the presence of the mutations and response to therapy, and a similar proportion (10%) relapsed. Notch mutations, which hallmark ∼50% of T-ALLs (Clappier et al., 2010; Kox et al., 2010), were detected in 64% of the IL7R-mutated leukemias compared with 48% of the T-ALL without IL7R mutations (P = 0.1, χ2 test).
Table II.
IL7R mutations in T-ALL
| ID | IL7R mutation DNA | IL-7R mutation protein |
| T1 | c.816 Ins 15 TTTTGTCGGAAGGAC | 243 Ins FCRKD |
| T2 | c.815 Ins 7 GAGATGC Del 1 A | 243 Ins RC |
| T3 | c.818Ins 10 CGTGCCCCCT Del 4 TAAC | 243 Ins PCPL |
| T4 | c.820 Ins 21 TGCCCGAGCAAGATTGCCCCA + point mutation c826 G->C | 244 Ins MPEQDCP +S246T |
| T5 | c.798 Ins 11 CCTCCTGGTGC Del 17 AGATGGATCCTATCTTA | 237 Ins ASWC |
| T6 | c.814 Ins 6 GCCCCC Del 6 TACTAA | 242 Ins CPP |
| T7 | c.817 Ins 11 GCTGCCCGTCC Del 2 TA | 243 Ins RCPS |
| T8 | c.815 Ins 16 CGACTGTATTGGGGGTC Del 1 A | 242 Ins FDCIGV |
| T9 | c.822 Ins 10 CACCGTGGGT Del 4 ATCA | 245 Ins HRGC |
| T10 | c.817 Ins 12 CCCTCTGTTCGG Del 3 TAA | 243 Ins PLCSA |
| T11 | c.849 Ins 9 GAGAGGCCG | 254 Ins GEA |
| T12 | c.817 Ins 19 CCATTTATCGGTGTGTCCT Del 4 TAAC | 243 Ins PIYRCVL |
| T13 | c.799 Ins 11 CCTCCTGGTGC Del 17 AGATGGATCCTATCTTA | 237 Ins ASWC |
| T14 | c.816 Ins 7 TGAGTGT Del 1 A | 242 Ins FEC |
| T15 | c.815 Ins 11 TACCTGCCCGT Del 5 ACTAA | 242 Ins FTCPS |
| T16 | c.816 Ins 11 TGCCCCTCTCC Del 2 CT | 243 Ins CPSP |
| T17 | c.850 Ins 6 AAAAAG Del 3 CTC | 254 Ins EKV |
| T18 | c.823 Ins 12 GTCATCAGCCCT Del 3 TCA | 245 Ins SHQPC |
| T19 | c.838 Ins 24 GTTCAACCATCAGCATTTTGAGTT | 250 Ins CSTISILS |
| T20 | c.832 Ins 7 GTCAAAG Del 13 TGAGTTTTTTCTC | 248 Ins CQ |
| T21 | c. 814 Ins 14 GTGGTATAAGGGAA Del 5 TACTA | 242 Ins CGIREI |
| T22 | c. 847 T>G | V253G |
| T23 | c. 817 Ins 6 GGCTGT | 243 Ins RC |
| T24 | c.817 Ins 6 GCTGTA | 244 Ins GC |
| T25 | c.809 Ins 13 GTGCCGTCCCCAT Del 7 TATCTTA | 241 Ins CRPH |
| T26 | c.816 Ins 7 GGCTGTA Del 1 C | 243 Ins GCI |
| T27 | c.821 Ins 12 GAGGCCTTGTGG | 245 Ins RPCG |
| T28 | c.821 Ins 15 ACTTCCCTGCGTCTAC | 245 Ins LPCVT |
| T29 | c.814 Ins 10 GCTGGATGAA Del 1 T | 242 Ins CWMK |
| T30 | c.820 Ins 13 AGAAATGCACAAA Del 1 C | 244 Ins KKCTN |
Del, Deletion; Ins, Insertion. The amino acid cysteine included in 27 of 30 mutations is shown in bold.
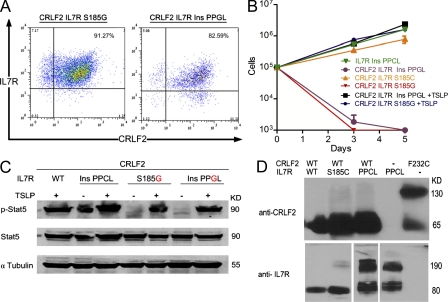
We and others (Hertzberg et al., 2010; Yoda et al., 2010) have recently reported an activating F232C mutation in CRLF2 that introduces cysteine in the juxtamembrane domain. All but three of the mutations observed in IL-7R included the addition of cysteine. To study whether the gain-of-function of the mutated IL-7R could be attributed to the addition of cysteine, we replaced the cysteines in the S815C and in the IL-7R InsPPCL alleles with glycine. These newly mutated receptors were expressed at the cell surface of transduced BaF3 cells (Fig. 4 A). Elimination of the cysteine abrogated the cytokine-independent growth (Fig. 4 B) and the constitutive phosphorylation of Stat5 (Fig. 4 C). Response to TSLP was not altered, confirming that “cysteine-lacking” mutated IL-7R formed a functional TSLP receptor with CRLF2 (Fig. 4 C). These results suggest that the presence of cysteine is critical for the gain-of-function phenotype both in the context of insertion of additional amino acids and as a point mutation.
Figure 4.
Functional significance of the cysteine residue in mutated IL7R. (A) FACS analysis of BaF3-CRLF2 cells expressing either S185G or IL-7R InsPPGL. In these constructs, the cysteine was mutated to glycine. (B) Growth assay of BaF3 and BaF3-CRLF2 cells transduced with IL-7R WT, IL-7R S185G, or IL-7R InsPPGL in the presence or absence of 30 ng/ml TSLP and the absence of IL-3. Error bars indicate SE. (C) BaF3 and BaF3-CRLF2 cells expressing indicated IL-7R mutants were incubated without IL-3 for 5 h. Where indicated, 100 ng/ml TSLP was added for 25 min. (D) Homodimerization of IL-7R InsPPCL mutant under nonreducing conditions. BaF3 cells that stably express CRLF2 with IL-7R (WT or mutated), IL-7R InsPPCL alone, or CRLF2 F232C (that has been shown previously to homodimerize; Yoda et al., 2010), were grown in the presence of IL-3. Protein lysates were separated by gel electrophoresis in the presence of nonreducing conditions and immunoblotted with an anti-CRLF2 or anti–IL-7R antibody. All experiments were performed three times.
The experimental insertion of cysteine into the transmembrane domain of the erythropoietin receptor (Constantinescu et al., 2001; Lu et al., 2006) activated the receptor by causing ligand-independent receptor dimerization. Indeed, we observed marked homodimerization of the IL-7R InsPPCL in protein analysis under nonreducing conditions (Fig. 4 D). Interestingly, CRLF2 was not present in these dimers, which is consistent with the observation that this mutant protein does not require CRLF2 for its activation and with the presence of these mutations in T-ALLs in which CRLF2 is not highly expressed. Our current and recent observations (Chapiro et al., 2010; Hertzberg et al., 2010; Yoda et al., 2010) of mutations in IL-7R and CRLF2, respectively, suggest that the addition of cysteine to the juxtamembranous domain is a general mechanism for mutational activation of type I cytokine receptors in leukemias.
These discoveries are a prime example of the interconnection of development and leukemia (Izraeli, 2004). Although loss-of-function mutations in IL-7R perturb lymphoid development, the novel gain-of-function mutations described here are associated with both BCP- and T-ALLs. Activating mutations of IL-7R are present in leukemias of the same lineage, for which it is developmentally required (T cells), or in B cell precursor leukemias that aberrantly express a T cell and monocytic receptor (CRLF2). The broad significance of this pathway in leukemia is further suggested by the aberrant deregulation of CRLF2–JAK–STAT pathway in BCP-ALL (Bercovich et al., 2008; Constantinescu et al., 2008; Kearney et al., 2009; Mullighan et al., 2009; Russell et al., 2009; Hertzberg et al., 2010; Yoda et al., 2010) and by these newly described IL7R mutations and the previously reported JAK1 activating mutations in T-ALL (Flex et al., 2008; Hornakova et al., 2010). Similar to mutational activations of other growth factor receptors in cancer (Gazdar, 2009), our observations have potential implications for targeted therapy.
MATERIALS AND METHODS
Patient samples.
DNA and RNA were derived from leukemic bone marrow of children with ALL enrolled in a prospective BFM (Berlin Frankfurt Munster) and AIEOP (Associazione Italiana Ematologia Oncologia Pediatrica Group) ALL 2000 study (Conter et al., 2010). The T-ALL and the DS-ALL cohorts were previously described (Hertzberg et al., 2010; Kox et al., 2010). All specimens were collected with informed consent and approval of local and national ethic committees. Samples were anonymized for the study. The study was approved by the Israeli Health Ministry Ethic committee, approval # 920070771.
CRLF2 analysis.
CRLF2 levels were analyzed by the TaqMan Gene Expression Assay Hs00913509_s1 (Applied Biosystems), as we have previously reported (Cario et al., 2010; Hertzberg et al., 2010). The presence of the fusion transcript P2RY8-CRLF2 was analyzed by RT-PCR, as we have previously reported (Cario et al., 2010; Hertzberg et al., 2010), using primers designed in the first exon of P2RY8 (5′-GGACAGATGGAACTGGAAGG-3′) and the third exon of CRLF2 (5′-GTCCCATTCCTGATGGAGAA-3′). PCR product was ∼511 bp. Immunoglobulin heavy chain locus (IGHα) translocations were identified by fluorescence in situ hybridization (FISH) on interphase nuclei using the Vysis LSI IGHα Dual Color Break-Apart Rearrangement Probe (Abbott Molecular). The nuclei were counterstained with DAPI (4′, 6-diamino-2-phenylindol). Results were recorded using a fluorescence microscope (DMRB; Leica) fitted with a 100×/1.30 oil objective, charge-coupled device camera, and digital imaging software from MetaSystems (Isis FISH imaging system).
For the purpose of this study, the CRLF2-positive group consisted of BCP-ALL specimens that were either positive for the P2RY8-CRLF2 fusion or to the IGHα CRLF2 translocation or had CRLF2 RNA expression levels >20 fold of the median value of CRLF2 expression of 555 unselected BCP-ALLs (Cario et al., 2010). The CRLF2-negative group consisted of BCP-ALL specimens negative for the genomic rearrangements and with expression level at the median and below. JAK2 and CRLF2 mutations were identified as previously described (Hertzberg et al., 2010)
IL7R mutation analysis.
We used intronic primers of human IL7R sequence (available from GenBank/EMBL/DDBJ under accession no. NM_002185.2; Table S2) to amplify exons 1–8 of the gene with PCR. Fast start Taq DNA polymerase (Roche) was used with the following thermal cycling conditions: 1 cycle of 94°C for 5 min, followed by 5 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C 30 s, followed by 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C 30 s, followed by 1 cycle of 72°C for 7 min. We then analyzed the fragments by denaturing high-performance liquid chromatography as described before (Bercovich et al., 2008; Hertzberg et al., 2010).
Fragments with abnormal chromatography patterns were sequenced directly by the primer used for PCR, or cloned into TA vector pCR2.1 (Invitrogen). DNA from plasmid clones was extracted by AccuPrep Spin Miniprep kit (Bioneer) and sequenced.
Site-directed mutagenesis.
The human IL7R cloned into the MSCViresGFP retrovirus (encoding GFP) was used as a template for the generation of mutations by site-directed mutagenesis (QuikChange II XL; Stratagene). The plasmid sequences were verified by sequencing.
BaF3 assays.
Parental BaF3 cells and those expressing human CRLF2 receptor (BaF3/CRLF2; Hertzberg et al., 2010) were transduced with the retroviruses containing WT or mutant IL7R. Transduced cells were sorted by flow cytometer 2–4 d later. PE anti–human CRLF2 (322806; BioLegend) and Alexa Fluor 647 anti–human CD127 (IL-7R, 317606; BioLegend) antibodies were used. Cytokine withdrawal assays were performed as previously described (Hertzberg et al., 2010)
Western blotting.
For Western blot analyses, 10% polyacrylamide linear gels were used. BaF3 cells were harvested after 5 h of IL-3 deprivation, and then stimulated with either 10 ng/ml IL-3 or 100 ng/ml TSLP for 25 min. The antibodies used were anti–phospho-STAT5 Tyr 694 (Epitomics), anti-STAT5, anti-S6 ribosomal protein and anti-phospho-S6 ribosomal protein Ser 235/6 (Cell Signaling Technology).
For Western blot analyses of disulfide-bonded dimmers, 8% polyacrylamide linear gels were used. For nonreducing conditions, proteins were boiled in SDS buffer without 2-mercaptoethanol. Antibodies used were anti–human IL-7R (R&D Systems) and anti–human CRLF2 (BioLegend).
Online supplemental material.
Fig. S1 shows expression of mutated IL7R. Fig. S2 shows FACS analysis of parental BaF3 cells in the presence of IL-3. Fig. S3 shows CRLF2 and IL7R Ins PPCL respond to TSLP. Table S1 shows the clinical characteristics of T-ALL patients with IL7R mutations. Table S2 shows primers used for genomic amplification of IL7R exons. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20110580/DC1.
Acknowledgments
We thank M. Happich, B. Fedders, Drs. S. Korem and C. Eckert for their assistance. The IL7R vector was a gift from D. Weinstock.
This study was funded by grants from the Israel Science Foundation Legacy program, Waxman foundation and Children for Leukaemia UK (S. Izraeli), Israel Trade Ministry (D. Bercovich), German Ministry for Education and Research (BMBF) in the context of the NGFN Leukemia Net and the Tour der Hoffnung (A. Kulozik), Deutsche Krebshilfe, and the Madeleine-Schickedanz-Kinder-krebsstiftung (G. Cario and M. Schrappe), EU NMP4-SL-2009-228971 (GteK), Associazione Italiana per la Ricerca sul Cancro (AIRC), Fondazione Cariplo, and Ministero dell’Istruzione dell’Università e della Ricerca (MIUR; A. Biondi and G. Cazzaniga).
This work was performed in partial fulfillment of the requirements for a PhD degree of Chen Shochat, Noa Tal and Ithamar Ganmore, Sackler Faculty of Medicine, Tel Aviv University, Israel. This is a study of the international BFM study group.
The authors have no competing financial interests.
Footnotes
Abbreviations used:
- ALL
- acute lymphoblastic leukemia
- BCP-ALL
- B cell precursor acute lymphoblastic leukemia
- CCR
- continuous complete remission
- CRLF2
- cytokine receptor-like factor 2
- DS
- Down syndrome
- DS-ALL
- Down syndrome with BCP-ALL
- IGHα
- immunoglobulin heavy locus
- IL2RG
- IL-2 receptor γ
- RPS6
- ribosomal protein S6
- STAT5
- signal transducer and activator of transcription 5
- TSLP
- thymic stromal lymphopoietin
References
- Anderson K., Lutz C., van Delft F.W., Bateman C.M., Guo Y., Colman S.M., Kempski H., Moorman A.V., Titley I., Swansbury J., et al. 2011. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 469:356–361 10.1038/nature09650 [DOI] [PubMed] [Google Scholar]
- Bercovich D., Ganmore I., Scott L.M., Wainreb G., Birger Y., Elimelech A., Shochat C., Cazzaniga G., Biondi A., Basso G., et al. 2008. Mutations of JAK2 in acute lymphoblastic leukaemias associated with Down’s syndrome. Lancet. 372:1484–1492 10.1016/S0140-6736(08)61341-0 [DOI] [PubMed] [Google Scholar]
- Cario G., Zimmermann M., Romey R., Gesk S., Vater I., Harbott J., Schrauder A., Moericke A., Izraeli S., Akasaka T., et al. 2010. Presence of the P2RY8-CRLF2 rearrangement is associated with a poor prognosis in non-high-risk precursor B-cell acute lymphoblastic leukemia in children treated according to the ALL-BFM 2000 protocol. Blood. 115:5393–5397 10.1182/blood-2009-11-256131 [DOI] [PubMed] [Google Scholar]
- Chapiro E., Russell L., Lainey E., Kaltenbach S., Ragu C., Della-Valle V., Hanssens K., Macintyre E.A., Radford-Weiss I., Delabesse E., et al. 2010. Activating mutation in the TSLPR gene in B-cell precursor lymphoblastic leukemia. Leukemia. 24:642–645 10.1038/leu.2009.231 [DOI] [PubMed] [Google Scholar]
- Clappier E., Collette S., Grardel N., Girard S., Suarez L., Brunie G., Kaltenbach S., Yakouben K., Mazingue F., Robert A., et al. ; EORTC-CLG 2010. NOTCH1 and FBXW7 mutations have a favorable impact on early response to treatment, but not on outcome, in children with T-cell acute lymphoblastic leukemia (T-ALL) treated on EORTC trials 58881 and 58951. Leukemia. 24:2023–2031 10.1038/leu.2010.205 [DOI] [PubMed] [Google Scholar]
- Clappier E., Gerby B., Sigaux F., Delord M., Touzri F., Hernandez L., Ballerini P., Baruchel A., Pflumio F., Soulier J. 2011. Clonal selection in xenografted human T cell acute lymphoblastic leukemia recapitulates gain of malignancy at relapse. J. Exp. Med. 208:653–661 10.1084/jem.20110105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinescu S.N., Keren T., Socolovsky M., Nam H., Henis Y.I., Lodish H.F. 2001. Ligand-independent oligomerization of cell-surface erythropoietin receptor is mediated by the transmembrane domain. Proc. Natl. Acad. Sci. USA. 98:4379–4384 10.1073/pnas.081069198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinescu S.N., Girardot M., Pecquet C. 2008. Mining for JAK-STAT mutations in cancer. Trends Biochem. Sci. 33:122–131 10.1016/j.tibs.2007.12.002 [DOI] [PubMed] [Google Scholar]
- Conter V., Bartram C.R., Valsecchi M.G., Schrauder A., Panzer-Grümayer R., Möricke A., Aricò M., Zimmermann M., Mann G., De Rossi G., et al. 2010. Molecular response to treatment redefines all prognostic factors in children and adolescents with B-cell precursor acute lymphoblastic leukemia: results in 3184 patients of the AIEOP-BFM ALL 2000 study. Blood. 115:3206–3214 10.1182/blood-2009-10-248146 [DOI] [PubMed] [Google Scholar]
- Ensor H.M., Schwab C., Russell L.J., Richards S.M., Morrison H., Masic D., Jones L., Kinsey S.E., Vora A.J., Mitchell C.D., et al. 2011. Demographic, clinical, and outcome features of children with acute lymphoblastic leukemia and CRLF2 deregulation: results from the MRC ALL97 clinical trial. Blood. 117:2129–2136 10.1182/blood-2010-07-297135 [DOI] [PubMed] [Google Scholar]
- Flex E., Petrangeli V., Stella L., Chiaretti S., Hornakova T., Knoops L., Ariola C., Fodale V., Clappier E., Paoloni F., et al. 2008. Somatically acquired JAK1 mutations in adult acute lymphoblastic leukemia. J. Exp. Med. 205:751–758 10.1084/jem.20072182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdar A.F. 2009. Personalized medicine and inhibition of EGFR signaling in lung cancer. N. Engl. J. Med. 361:1018–1020 10.1056/NEJMe0905763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey R.C., Mullighan C.G., Chen I.M., Wharton W., Mikhail F.M., Carroll A.J., Kang H., Liu W., Dobbin K.K., Smith M.A., et al. 2010. Rearrangement of CRLF2 is associated with mutation of JAK kinases, alteration of IKZF1, Hispanic/Latino ethnicity, and a poor outcome in pediatric B-progenitor acute lymphoblastic leukemia. Blood. 115:5312–5321 10.1182/blood-2009-09-245944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzberg L., Vendramini E., Ganmore I., Cazzaniga G., Schmitz M., Chalker J., Shiloh R., Iacobucci I., Shochat C., Zeligson S., et al. 2010. Down syndrome acute lymphoblastic leukemia, a highly heterogeneous disease in which aberrant expression of CRLF2 is associated with mutated JAK2: a report from the International BFM Study Group. Blood. 115:1006–1017 10.1182/blood-2009-08-235408 [DOI] [PubMed] [Google Scholar]
- Hornakova T., Chiaretti S., Lemaire M.M., Foa R., Ben Abdelali R., Asnafi V., Tartaglia M., Renauld J.C., Knoops L. 2010. ALL-associated JAK1 mutations confer hypersensitivity to the anti-proliferative effect of type I interferon. Blood. 115:3287–3295 [DOI] [PubMed] [Google Scholar]
- Izraeli S. 2004. Leukaemia — a developmental perspective. Br. J. Haematol. 126:3–10 10.1111/j.1365-2141.2004.04986.x [DOI] [PubMed] [Google Scholar]
- Izraeli S. 2010. Similar yet different. Blood. 116:1019–1020 10.1182/blood-2010-05-285197 [DOI] [PubMed] [Google Scholar]
- Kearney L., Gonzalez De Castro D., Yeung J., Procter J., Horsley S.W., Eguchi-Ishimae M., Bateman C.M., Anderson K., Chaplin T., Young B.D., et al. 2009. Specific JAK2 mutation (JAK2R683) and multiple gene deletions in Down syndrome acute lymphoblastic leukemia. Blood. 113:646–648 10.1182/blood-2008-08-170928 [DOI] [PubMed] [Google Scholar]
- Kox C., Zimmermann M., Stanulla M., Leible S., Schrappe M., Ludwig W.D., Koehler R., Tolle G., Bandapalli O.R., Breit S., et al. 2010. The favorable effect of activating NOTCH1 receptor mutations on long-term outcome in T-ALL patients treated on the ALL-BFM 2000 protocol can be separated from FBXW7 loss of function. Leukemia. 24:2005–2013 10.1038/leu.2010.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.J., Soumelis V., Watanabe N., Ito T., Wang Y.H., Malefyt Rde.W., Omori M., Zhou B., Ziegler S.F. 2007. TSLP: an epithelial cell cytokine that regulates T cell differentiation by conditioning dendritic cell maturation. Annu. Rev. Immunol. 25:193–219 10.1146/annurev.immunol.25.022106.141718 [DOI] [PubMed] [Google Scholar]
- Lu X., Gross A.W., Lodish H.F. 2006. Active conformation of the erythropoietin receptor: random and cysteine-scanning mutagenesis of the extracellular juxtamembrane and transmembrane domains. J. Biol. Chem. 281:7002–7011 10.1074/jbc.M512638200 [DOI] [PubMed] [Google Scholar]
- Mullighan C.G., Collins-Underwood J.R., Phillips L.A., Loudin M.G., Liu W., Zhang J., Ma J., Coustan-Smith E., Harvey R.C., Willman C.L., et al. 2009. Rearrangement of CRLF2 in B-progenitor- and Down syndrome-associated acute lymphoblastic leukemia. Nat. Genet. 41:1243–1246 10.1038/ng.469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi M., Nakamura Y., Russell S.M., Ziegler S.F., Tsang M., Cao X., Leonard W.J. 1993. Interleukin-2 receptor gamma chain: a functional component of the interleukin-7 receptor. Science. 262:1877–1880 10.1126/science.8266077 [DOI] [PubMed] [Google Scholar]
- Peschon J.J., Morrissey P.J., Grabstein K.H., Ramsdell F.J., Maraskovsky E., Gliniak B.C., Park L.S., Ziegler S.F., Williams D.E., Ware C.B., et al. 1994. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J. Exp. Med. 180:1955–1960 10.1084/jem.180.5.1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puel A., Ziegler S.F., Buckley R.H., Leonard W.J. 1998. Defective IL7R expression in T(-)B(+)NK(+) severe combined immunodeficiency. Nat. Genet. 20:394–397 10.1038/3877 [DOI] [PubMed] [Google Scholar]
- Rochman Y., Kashyap M., Robinson G.W., Sakamoto K., Gomez-Rodriguez J., Wagner K.U., Leonard W.J. 2010. Thymic stromal lymphopoietin-mediated STAT5 phosphorylation via kinases JAK1 and JAK2 reveals a key difference from IL-7-induced signaling. Proc. Natl. Acad. Sci. USA. 107:19455–19460 10.1073/pnas.1008271107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman R., Trakhtenbrot L., Bielorai B., Izraeli S., Ishoev G., Amariglio N., Rechavi G., Toren A. 2005. Co-existence of multiple subclones in TEL-AML1 at diagnosis of acute lymphoblastic leukaemia in association with submicroscopic deletion of AML1. Br. J. Haematol. 129:491–498 10.1111/j.1365-2141.2005.05479.x [DOI] [PubMed] [Google Scholar]
- Russell L.J., Capasso M., Vater I., Akasaka T., Bernard O.A., Calasanz M.J., Chandrasekaran T., Chapiro E., Gesk S., Griffiths M., et al. 2009. Deregulated expression of cytokine receptor gene, CRLF2, is involved in lymphoid transformation in B-cell precursor acute lymphoblastic leukemia. Blood. 114:2688–2698 [DOI] [PubMed] [Google Scholar]
- Tasian S.K., Doral M.Y., Wood B.L., Borowitz M.J., Collins-Underwood J.R., Harvey R.C., Sakamoto K.M., Willman C.L., Hunger S.P., Mullighan C.G., Loh M.L. 2010. Thymic stromal lymphopoietin stimulation of pediatric acute lymphoblastic leukemias with CRLF2 alterations induces JAK/STAT and PI3K phosphosignaling. Blood. 116:182–183 [Google Scholar]
- Touw I., Pouwels K., van Agthoven T., van Gurp R., Budel L., Hoogerbrugge H., Delwel R., Goodwin R., Namen A., Löwenberg B. 1990. Interleukin-7 is a growth factor of precursor B and T acute lymphoblastic leukemia. Blood. 75:2097–2101 [PubMed] [Google Scholar]
- Yoda A., Yoda Y., Chiaretti S., Bar-Natan M., Mani K., Rodig S.J., West N., Xiao Y., Brown J.R., Mitsiades C., et al. 2010. Functional screening identifies CRLF2 in precursor B-cell acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. USA. 107:252–257 10.1073/pnas.0911726107 [DOI] [PMC free article] [PubMed] [Google Scholar]