Abstract
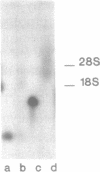
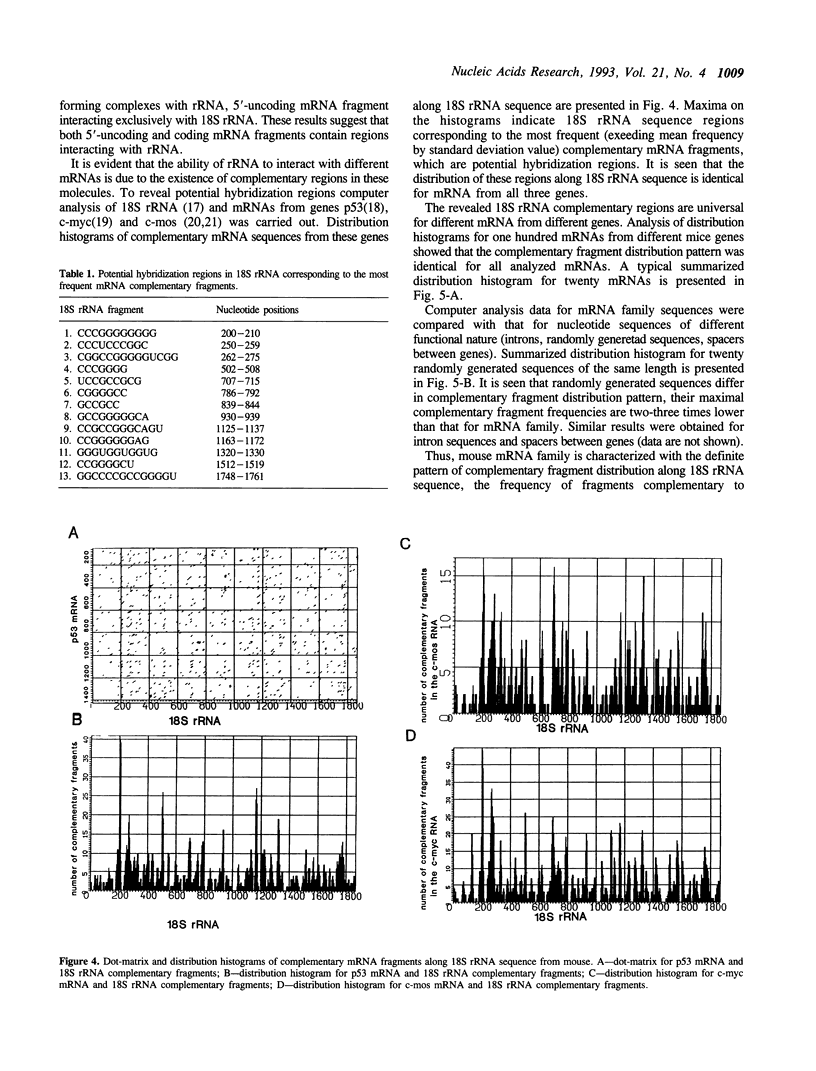
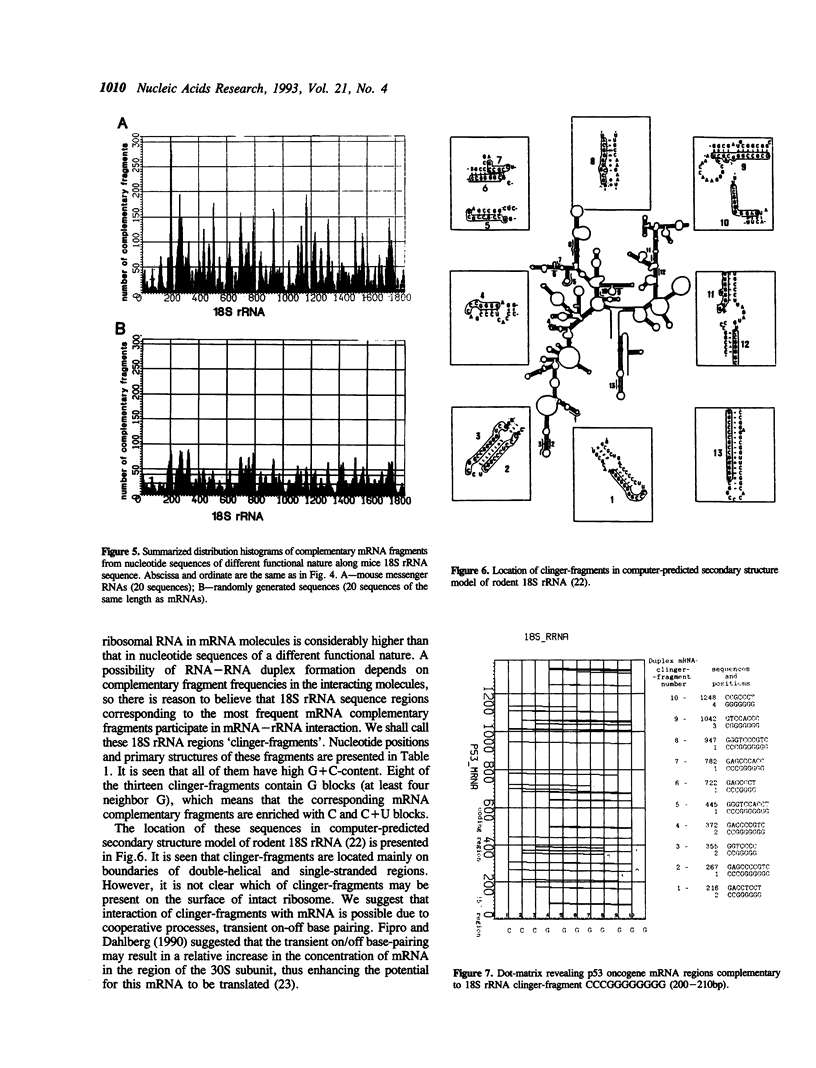
Intermolecular hybridization experiments show that murine 18S rRNA and 28S rRNA are capable of forming stable hybrid structures with mRNA from genes p53, c-myc and c-mos from the same species. Both 5'-uncoding and coding oncogene p53 mRNA regions contain fragments interacting with rRNA. Computer analysis revealed 18S rRNA fragments complementary to oligonucleotides frequently met in mRNA, which are potential hybridization regions (clinger-fragments). The distribution of clinger-fragments along 18S rRNA sequence is universal at least for one hundred murine mRNA sequences analyzed. Maximal frequencies of oligonucleotides complementary to 18S rRNA clinger-fragments are reliably (2-3 times) higher for mRNA than for intron sequences and randomly generated sequences. The results obtained suggest a possible role of clinger-fragments in translation processes as universal regions of mRNA binding.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chan Y. L., Gutell R., Noller H. F., Wool I. G. The nucleotide sequence of a rat 18 S ribosomal ribonucleic acid gene and a proposal for the secondary structure of 18 S ribosomal ribonucleic acid. J Biol Chem. 1984 Jan 10;259(1):224–230. [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Dahlberg A. E. The functional role of ribosomal RNA in protein synthesis. Cell. 1989 May 19;57(4):525–529. doi: 10.1016/0092-8674(89)90122-0. [DOI] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göringer H. U., Hijazi K. A., Murgola E. J., Dahlberg A. E. Mutations in 16S rRNA that affect UGA (stop codon)-directed translation termination. Proc Natl Acad Sci U S A. 1991 Aug 1;88(15):6603–6607. doi: 10.1073/pnas.88.15.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenbüchle O., Santer M., Steitz J. A., Mans R. J. Conservation of the primary structure at the 3' end of 18S rRNA from eucaryotic cells. Cell. 1978 Mar;13(3):551–563. doi: 10.1016/0092-8674(78)90328-8. [DOI] [PubMed] [Google Scholar]
- Hui A., de Boer H. A. Specialized ribosome system: preferential translation of a single mRNA species by a subpopulation of mutated ribosomes in Escherichia coli. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4762–4766. doi: 10.1073/pnas.84.14.4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob W. F., Santer M., Dahlberg A. E. A single base change in the Shine-Dalgarno region of 16S rRNA of Escherichia coli affects translation of many proteins. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4757–4761. doi: 10.1073/pnas.84.14.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins J. R., Rudge K., Redmond S., Wade-Evans A. Cloning and expression analysis of full length mouse cDNA sequences encoding the transformation associated protein p53. Nucleic Acids Res. 1984 Jul 25;12(14):5609–5626. doi: 10.1093/nar/12.14.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrach H., Diamond D., Wozney J. M., Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977 Oct 18;16(21):4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- Maizel J. V., Jr, Lenk R. P. Enhanced graphic matrix analysis of nucleic acid and protein sequences. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7665–7669. doi: 10.1073/pnas.78.12.7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min K. T., Kim M. H., Lee D. S. Search for the optimal sequence of the ribosome binding site by random oligonucleotide-directed mutagenesis. Nucleic Acids Res. 1988 Jun 10;16(11):5075–5088. doi: 10.1093/nar/16.11.5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynal F., Michot B., Bachellerie J. P. Complete nucleotide sequence of mouse 18 S rRNA gene: comparison with other available homologs. FEBS Lett. 1984 Feb 27;167(2):263–268. doi: 10.1016/0014-5793(84)80139-8. [DOI] [PubMed] [Google Scholar]
- Sargan D. R., Gregory S. P., Butterworth P. H. A possible novel interaction between the 3'-end of 18 S ribosomal RNA and the 5'-leader sequence of many eukaryotic messenger RNAs. FEBS Lett. 1982 Oct 18;147(2):133–136. doi: 10.1016/0014-5793(82)81026-0. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton L. W., Fahrlander P. D., Tesser P. M., Marcu K. B. Nucleotide sequence comparison of normal and translocated murine c-myc genes. Nature. 1984 Aug 2;310(5976):423–425. doi: 10.1038/310423a0. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N. Translation framing code and frame-monitoring mechanism as suggested by the analysis of mRNA and 16 S rRNA nucleotide sequences. J Mol Biol. 1987 Apr 20;194(4):643–652. doi: 10.1016/0022-2836(87)90241-5. [DOI] [PubMed] [Google Scholar]
- Van Beveren C., Galleshaw J. A., Jonas V., Berns A. J., Doolittle R. F., Donoghue D. J., Verma I. M. Nucleotide sequence and formation of the transforming gene of a mouse sarcoma virus. Nature. 1981 Jan 22;289(5795):258–262. doi: 10.1038/289258a0. [DOI] [PubMed] [Google Scholar]
- Van Beveren C., van Straaten F., Galleshaw J. A., Verma I. M. Nucleotide sequence of the genome of a murine sarcoma virus. Cell. 1981 Nov;27(1 Pt 2):97–108. doi: 10.1016/0092-8674(81)90364-0. [DOI] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Dahlberg A. E., Atkins J. F., Gesteland R. F. Reading frame switch caused by base-pair formation between the 3' end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO J. 1988 May;7(5):1503–1507. doi: 10.1002/j.1460-2075.1988.tb02969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]