Abstract
Two new saponins, panajaponol (1) and pseudoginsenoside RT1 butyl ester (2), together with 35 known compounds (3–37), were isolated from the roots of Panax japonicus. The structures of 1 and 2 were elucidated on the basis of spectroscopic analysis and chemical methods. Furthermore, a LC-MS/MS method was developed for confirming 2, 3, and 8 as natural compounds containing a butyl ester group. This method should be useful for distinguishing between minor natural and artifactual compounds in Panax species. Moreover, compounds 3, 6, 8, 9, 11, 13, and 15 exhibited strong inhibition of superoxide anion generation and elastase release by human neutrophils in response to formyl-L-methionyl-L-leucyl-L-phenylalanine/cytochalasin B (fMLP/CB), with IC50 values ranging from 0.78 to 43.6 μM. In addition, 1 showed greater than two- to three-fold selective cytotoxic activity against KB and DU145 cancer cell lines.
The roots of Panax species (Araliaceae) are widely used in Chinese herbal medicine or as a food in Asian countries, because these plants are well-known for their beneficial properties. Pharmacological studies of triterpene saponins isolated from Panax species have reported anticancer,1,2 immunomodulatory,3,4 anti-inflammatory,5 antiallergic,6 neuroprotective,7 antihypertensive,8 and antidiabetic effects.9,10 The chemical constituents in Panax species are triterpenoid saponins that can be classified into two groups based on their sapogenin skeleton, the dammarane- and oleanane-types. The dammarane-type triterpenoid saponins are known as ginsenosides, including glycosides of (20S)-protopanaxadiol and (20S)-protopanaxatriol. Oleanolic acid-type ginsenosides seem to be typical constituents of ginseng species and are particularly characteristic for P. ginseng, P. pseudoginseng subsp. himalaicus (Himamayan ginseng), P. vietnamensis (Vietnamese ginseng), P. zingiberensis (ginger ginseng), and P. japonicus (Japanese ginseng or Zhujie-Shen),11,12

Panax japonicus C. A. Meyer var. major (Chinese name, “ZuTziSeng”) also contains many oleanolic acid saponins and dammarane saponins.13 It is an important herb prescribed in traditional Chinese medicine to treat diabetes (wasting-thirst),14 and exhibits expectorant, antitussive, hemostatic, sedative, and analgesic activities. Recently, we reported the isolation and structure elucidation of three new polyacetylenes, panaxjapynes A–C, from the active CHCl3-soluble fraction of the roots of P. japonicus var. major through bioassay-guided fractionation using an assay for inhibitory activity of α-glucosidase.15 As part of a continuing investigation on the bioactive constituents of this plant, we have now isolated two new compounds (1 and 2) together with 35 known compounds. Herein, we describe the detailed structure elucidation of the new compounds as well as their inhibitory effects on fMLP/CB-induced superoxide anion generation and elastase release in human neutrophils, their cytotoxic activity for cancer cells, and inhibitory effects on α-glucosidase. Moreover, we have confirmed that the butyl ester-containing compounds in this plant were indeed natural products using a LC-MS/MS method. This method was developed to distinguish between minor natural and artifactual compounds.
Results and Discussion
An ethanolic extract of the roots of P. japonicus var. major yielded two new compounds, panajaponol A (1) and pseudoginsenoside RT1 butyl ester (2), together with 35 known compounds, identified as taibaienoside I (3),16 chikusetsusaponin-IV (4),17 chikusetsusaponin-IV methyl ester (5),18 chikusetsusaponin-IVa (6),18 chikusetsusaponin-IVa methyl ester (7),19 chikusetsusaponin-IVa butyl ester (8),20 stipuleanoside R2 (9),21 chikusetsusaponin-V (10),22 chikusetsusaponin-Ib (11),23 pseudoginsenoside RT1 (12),24 pseudoginsenoside RT1 methyl ester (13),20 oleanolic acid 28-O-β-D-glucopyranoside (14),25 oleanolic acid (15),25 ginsenoside Rf (16),26 ginsenoside Rg1 (17),27 ginsenoside Rd (18),28 ginsenoside Rb1 (19),29 (20R)-ginsenoside Rg3 (20),30 (20S)-ginsenoside Rg3 (21),30 ginsenoside Rg5 (22),31 majonoside-R1 (23),24 β-sitosteroyl-β-D-glucoside (24),32 panaxjapynes A–C (25–27),15 (3R)-(−)-falcarinol (28),33 (3S,10S)-panaxydiol (29),34 (3S,9R,10R)-panaxytriol (30),35 (3S,9R,10R)-gensenoyne C (31),36 docosyl trans-ferulate (32),37 vanillin (33),38 syringaldehyde (34),39 3,4,5-trimethoxybenzoic acid (35),39 2,6-dimethoxyphenol (36),40 and 1β, 6α-dihydroxy-4(14)-eudesmene (37)41 by comparing their spectroscopic data with those reported in the literature.
Panajaponol A (1) was isolated as colorless powder ([α]D −14.1°) and showed a [M+Na]+ ion peak at m/z 839.4777 in its positive HRESIMS, corresponding to the molecular formula C42H72O15. This formula was corroborated from the 13C NMR spectrum, which showed 42 carbon resonances. The IR spectrum of 1 showed strong absorption bands at 3345, 1076, 1030 and 1640 cm−1 due to glucose and olefinic moieties, respectively. In the 1H NMR spectrum of 1, diagnostic signals were found for a sapogenin moiety with seven methyl groups at δ 2.07 (s), 1.99 (s), 1.45 (s), 1.34 (s), 1.15 (s), 0.94 (s), and 0.79 (s), an olefinic proton at δ 5.47 (m), and three oxymethine groups at δ 3.46 (d, J = 5.6 Hz), 4.34 (m), and 3.86 (m). In addition, the 1H NMR spectrum showed signals for two anomeric protons [δ4.91 (d, J = 7.6 Hz, H-1′) and δ5.91 (d, J = 7.2 Hz, H-1″)], which showed HMBC correlations to carbons at δ104.0 and δ103.9, respectively. Based on the coupling constants of the anomeric protons, both sugar substituents were identified as β-configured glucose units, which was confirmed by acid hydrolysis of 1 to give D-glucose, identified by HPLC analysis.42,43 The 1H and 13C NMR data (Table 1) of 1 were very similar to those of ginsenoside Rf (16), except that the C-27 signal was shifted downfield to δ 59.9, and the H-27 protons were present as a methylene group [δ 4.47 (m) and δ 4.54 (dd, J=12.0, 4.0 Hz)] in 1 rather than a methyl group in 16. These data indicate that the C-27 methyl group is oxygenated as a hydroxymethyl group in 1. HMBC experiments confirmed that H-27b (δ 4.54) correlated with C-24 (δ 128.1), C-25 (δ 135.5), and C-26 (δ 22.0). The oligoglycoside structure at position C-6 in 1 was determined from the HMBC spectrum, which showed proton-carbon correlations of H-1′ (δ 4.91)/C-6 (δ 79.9) and H-1″ (δ 5.91) with C-2′ (δ 79.6). The relative configurations at the ring junctions were determined from key NOE correlations observed between H-3 (δ 3.46)/H-5 (δ 1.38)/H-28 (δ 2.07), H-6 (δ 4.34)/H-18 (δ 1.15)/H-19 (δ 0.94), H-12 (δ 3.86)/H-9 (δ 1.50)/H-30 (δ 0.79), and H-17 (δ 2.25)/H-12 (δ 3.86)/H-21 (δ 1.34)/H-30 (δ0.79). Furthermore, the signals of H-26 (δ 1.99) and H-24 (δ 5.47) showed an NOE correlation, suggesting that the double bond has a Z-configuration. Thus, the structure of 1 was proposed as shown, and this compound has been named panajaponol A (1).
Table 1.
13C NMR Data of 1, 2, and 16 in Pyridine-d5 at 100 MHz
| no. |
1 δC (m) |
2 δC (m) |
16 δC (m) |
no. |
1 δC (m) |
2 δC (m) |
16 δC (m) |
|---|---|---|---|---|---|---|---|
| C-1 | 39.5 (t) | 38.8 (t) | 38.7 (t) | C-1′ | 104.0 (d) | 105.4 (d) | 105.3 (d) |
| C-2 | 27.8 (t) | 26.8 (t) | 26.2 (t) | C-2′ | 79.6 (d) | 83.4 (d) | 83.3 (d) |
| C-3 | 78.0 (d) | 89.4 (d) | 89.5 (d) | C-3′ | 76.1 (d) | 76.6 (d) | 76.6 (d) |
| C-4 | 40.2 (s) | 39.7 (s) | 40.0 (s) | C-4′ | 71.8 (d) | 72.8 (d) | 72.9 (d) |
| C-5 | 61.5 (d) | 55.9 (d) | 55.9 (d) | C-5′ | 79.9 (d) | 78.3 (d) | 78.3 (d) |
| C-6 | 79.9 (d) | 18.6 (t) | 17.5 (t) | C-6′ | 63.4 (t) | 170.0 (s) | 170.0 (s) |
| C-7 | 45.1 (t) | 33.2 (t) | 33.2 (t) | C-1″ | 103.9 (d) | 107.0 (d) | 107.0 (d) |
| C-8 | 41.2 (s) | 40.0 (s) | 40.0 (s) | C-2″ | 72.4 (d) | 77.0 (d) | 76.9 (d) |
| C-9 | 50.2 (d) | 48.1 (d) | 48.1 (d) | C-3″ | 78.0 (d) | 77.7 (d) | 77.7 (d) |
| C-10 | 39.5 (s) | 37.0 (s) | 37.0 (s) | C-4″ | 78.1 (d) | 71.2 (d) | 71.2 (d) |
| C-11 | 32.1 (t) | 23.7 (t) | 23.7 (t) | C-5″ | 78.5 (d) | 67.3 (t) | 67.2 (t) |
| C-12 | 71.1 (d) | 123.3 (d) | 123.3 (d) | C-6″ | 63.0 (t) | - | - |
| C-13 | 48.3 (d) | 144.2 (s) | 144.2 (s) | C-1‴ | - | 95.8 (d) | 95.8 (d) |
| C-14 | 51.7 (s) | 42.2 (s) | 42.2 (s) | C-2‴ | - | 74.2 (d) | 74.2 (d) |
| C-15 | 31.3 (t) | 30.8 (t) | 27.9 (t) | C-3‴ | - | 79.0 (d) | 79.2 (d) |
| C-16 | 26.8 (t) | 23.8 (t) | 23.7 (t) | C-4‴ | - | 71.2 (d) | 71.2 (d) |
| C-17 | 54.5 (d) | 47.1 (s) | 47.1 (s) | C-5‴ | - | 79.4 (d) | 79.4 (d) |
| C-18 | 17.5 (q) | 41.8 (d) | 41.8 (d) | C-6‴ | - | 62.3 (t) | 62.3 (t) |
| C-19 | 17.7 (q) | 46.3 (t) | 46.3 (t) | C-1″″ | - | 65.0 (t) | - |
| C-20 | 73.1 (s) | 30.9 (s) | 30.8 (s) | C-2″″ | - | 30.8 (t) | - |
| C-21 | 27.0 (q) | 34.1 (t) | 33.2 (t) | C-3″″ | - | 19.3 (t) | - |
| C-22 | 36.2 (t) | 32.6 (t) | 30.8 (t) | C-4″″ | - | 13.8 (q) | - |
| C-23 | 22.6 (t) | 27.9 (q) | 27.9 (q) | OCH3 | - | - | 52.2 (q) |
| C-24 | 128.1 (d) | 16.4 (q) | 16.4 (q) | ||||
| C-25 | 135.5 (s) | 15.6 (q) | 15.6 (q) | ||||
| C-26 | 22.0 (q) | 17.5 (q) | 17.5 (q) | ||||
| C-27 | 59.9 (t) | 26.2 (q) | 26.8 (q) | ||||
| C-28 | 32.1 (q) | 176.5 (s) | 176.5 (s) | ||||
| C-29 | 16.9 (q) | 33.2 (q) | 33.2 (q) | ||||
| C-30 | 16.9 (q) | 23.7 (q) | 24.7 (q) |
Pseudosaponin RT1 butyl ester (2) was isolated as colorless powder with [α]D −18.2. The HRESIMS of 2 exhibited a sodiated molecular ion peak at m/z 1005.5405 [M+Na]+, consistent with the molecular formula, C51H82O18. The IR spectrum of 2 showed absorption bands at 3387, 1053, 1045, and 1744 cm−1, indicating the presence of an oliglycoside and ester carbonyl group. Acid hydrolysis of 2 gave D-glucose, D-xylose, and D-glucuronic acid, identified by HPLC analysis.42,43 The 1H NMR spectrum showed three anomeric proton signals at δ 6.33 (d, J = 8.0 Hz), 5.27 (d, J = 7.2 Hz), and 4.96 (d, J = 8.0 Hz), which were correlated to carbon resonances at δ 95.8 (C-1‴), 107.5 (C-1″), and 105.4 (C-1′) in the HMQC spectrum. Based on the coupling constants, all three sugar moieties have a β-configuration. The diagnostic 1H NMR spectrum signals for an oleanane-type aglycone included seven methyl groups at δ 1.26 (s), 1.26 (s), 1.09 (s), 1.08 (s), 0.90 (s), 0.87 (s), and 0.86 (s), an olefinic proton at δ 5.41 (m), and an oxymethine at δ 3.30 (dd, J = 9.6, 3.6 Hz). The NMR spectroscopic data of 2 were strikingly very similar to those of pseudosaponin RT1 methyl ester (13), except for the presence of signals corresponding to a butyl group [(δC 65.0, 30.8, 19.3, 13.8), correlated to δ 4.31 (m, H-1″″), 1.34 (m, H-2″″), 1.60 (m, H-3″″), 0.74 (t, J = 6.8 Hz, H-4″″)], in 2 rather than the methyl group in 13. This assignment was confirmed by HMBC correlations observed between H-1″″/C-6′ (δ 170.0) in the HMBC spectrum. The 1H and 13C NMR assignments for 2 were assigned unambiguously based on 1H-1H COSY, HMQC, and HMBC studies. Esterified derivatives have been reported previously in some Araliaceae species.16,19,20,44 Thus, the structure of 2 was determined as oleanolic acid 3-O-[β-D-xylopyranosyl-(1 → 2)-β-D-glucuronopyranoside-6-O-butyl ester]-28-O-β-D-glucopyranoside, and this compound has been named pseudosaponin RT1 butyl ester.
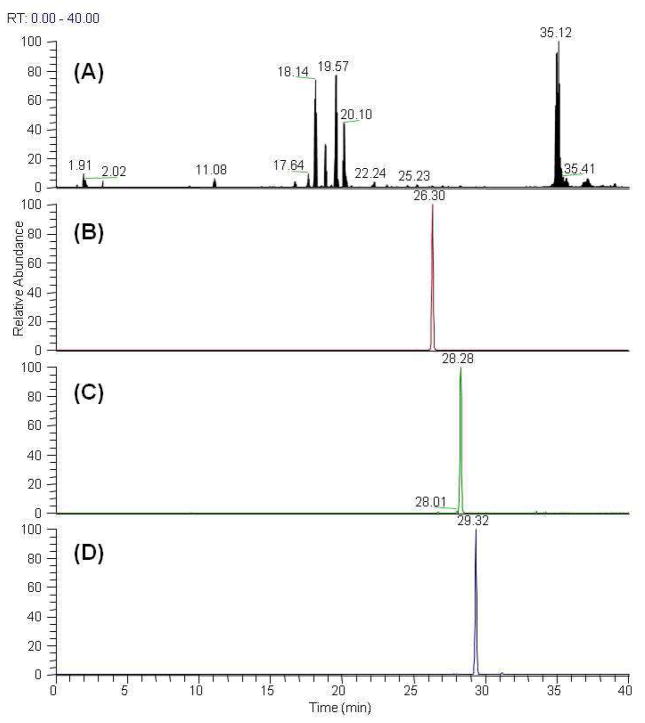
In order to confirm that butyl ester-containing compounds occur naturally in the plant rather than being artifactual compounds, 2, 3, and 8, which contain a butyl ester group, were analyzed using LC-MS/MS. Fragmentation of 2 produced ions at m/z 820 and 651 corresponding to [M-H-Glc]− and [M-H-Glc-Xyl-H2O]−, respectively (Figure 1B). In addition, the corresponding product ions of 3 and 8 are also shown in Figures 1A and 1C. The ethanolic root extraction (PJ) and n-BuOH soluble portion (PJB) were detected by using LC-MS/MS with optimized SRM parameters, and the chromatograms are shown in Figures 2 and 3. SRM signals for m/z 981.6→651.4, 820→669, and 849.5→687.4 corresponding to 2, 3, and 8 were detected simultaneously at 28.28, 26.30, and 29.32 min, respectively, during the LC-MS/MS run. Therefore, the butyl compounds were confirmed as natural occurring in this plant, and the LC-MS/MS method used could be applied to distinguish between natural and artifactual compounds in other Panax species.
Figure 1.
Product ion mass spectra of (A) taibaienoside I (3) (m/z 981.6), (B) pseudoginsenoside RT1 butyl ester (2) (m/z 981.6), and (C) chikusetsusaponin-IVa butyl ester (8) (m/z 849.5)
Figure 2.
Base peak intensity (BPI) chromatogram (A) and the SRM chromatograms of (B) taibaienoside I (3) (m/z 981.6 > m/z 669.4), (C) pseudoginsenoside RT1 butyl ester (2) (m/z 981.6 > m/z 651.4), and (D) chikusetsusaponin-IVa butyl ester (8) (m/z 849.5 > m/z 687.4) for PJ
Figure 3.
Base peak intensity (BPI) chromatogram (A) and the SRM chromatograms of (B) taibaienoside I (3) (m/z 981.6 > m/z 669.4), (C) pseudoginsenoside RT1 butyl ester (2) (m/z 981.6 > m/z 651.4), and (D) chikusetsusaponin-IVa butyl ester (8) (m/z 849.5 > m/z 687.5) for PJB
The isolated pure compounds 1–24 were evaluated for inhibitory effects on superoxide anion generation and elastase release by human neutrophils in response to fMLP/CB (Tables 2 and 3). Compounds 1 and 16 have very similar structural features, differing only at the C-27 position, where 1 has a hydroxymethyl group and 16 has a methyl group. However, with 20.3% (superoxide) and 42.0% (elastase) inhibition at 30 μM, 1 exhibited greater potency than 16 with 8.94% and 29.5% inhibition at the same concentration (Table 3). These results indicated that the presence of the hydroxymethyl group rather than methyl group enhanced inhibitory effects on superoxide anion generation and elastase release in human neutrophils. Compounds 4 and 5, with carboxylic acid and methyl ester functionalities, respectively, at the sugar C-6′ position, inhibited both O2•− generation and elastase release, and the potency increased significantly when the carboxylic acid group was esterified with a butyl group in 3 (IC50 2.24 μM for superoxide and 1.36 μM for elastase, Table 2). Compounds 6, 7, and 8 have a carboxylic acid, a methyl ester, and a butyl ester group, respectively, at the C-6′ position. Among them, 6 and 8 exhibited more potent inhibition than 7 in response to fMLP/CB-induced superoxide anion generation, while 7 was more potent than 6 and 8 against elastase release. The methyl ester analogue 13 showed significantly greater inhibition of both fMLP/CB induced O2•− generation and elastase release than butyl ester analogue 2. Compound 15, with a free carboxylic aid group at position C-28, showed more potent inhibition of both O2•− generation and elastase release (IC50 1.37 and 0.75 μM) than 14, esterified with glucose at this position.
Table 2.
Inhibitory Effects of 2, 3, 5–8, and 11–15 on Superoxide Anion Generation and Elastase Release by Human Neutrophils in Response to fMLP/CBa
| compound | superoxide | elastase |
|---|---|---|
| IC50 (μM)a | IC50 (μM)a | |
| 2 | 21.2 ± 3.61 | ---b, * |
| 3 | 2.24 ± 0.60 | 1.36 ± 0.12 |
| 5 | 19.7 ± 3.48c, ** | ---b, * |
| 6 | 2.88 ± 0.77 | 4.76 ± 1.71 |
| 7 | 13.6 ± 1.70d, ** | 1.97 ± 0.63 |
| 8 | 3.73 ± 1.22 | 7.28 ± 1.48 |
| 11 | 1.56 ± 0.18 | 1.87 ± 0.27 |
| 12 | 19.4 ± 4.29 | 44.5 ± 2.53 |
| 13 | 5.26 ± 0.30 | 7.75 ± 1.51 |
| 14 | ---b, *** | 9.64 ± 1.92 |
| 15 | 1.37 ± 0.19 | 0.75 ± 0.08 |
| DPIe | 1.02 ± 0.35 | N.T.f |
| PMSFe | N.T.f | 95.0 ± 25 |
Results are presented as mean ± S.E.M. (n = 3–4).
p < 0.05,
p < 0.01,
p < 0.001 compared with the control value.
50% inhibition was not reached at the maximum concentration tested (30 μM). For the indicated compounds, the percentages of inhibition at this concentration were 20.0%±5.69 (2), 27.5%±6.59 (5), and 43.6%±4.86 (14).
5 (30 μM) induced superoxide generation in the presence of cytochalasin B.
7 (30 μM) induced superoxide generation in the absence of fMLP/CB.
Diphenyleneiodonium (DPI, a NADPH oxidase inhibitor) and phenylmethylsulfonyl fluoride (PMSF, a serine protease inhibitior) were used as the positive controls in the generation of superoxide anion and release of elastase, respectively.
N.T.: not tested.
Table 3.
Inhibitory Effects of 1, 4, 9, 10, 16–19, 23, and 24 at 30 μM on Superoxide Anion Generation and Elastase Release by Human Neutrophils in Response to fMLP/CBa
| Compound | superoxide | elastase |
|---|---|---|
| inhibition % | inhibition % | |
| 1 | (20.3 ± 5.20)* | (42.0 ± 5.98)** |
| 4 | (14.6 ± 3.72)* | (27.6 ± 4.14)** |
| 9 | (0.78 ± 3.05) | (23.7 ± 6.39) |
| 10 | (14.2 ± 5.78) | (37.4 ± 5.60)** |
| 16 | (8.94 ± 0.96)*** | (29.5 ± 4.81)** |
| 17 | (14.1 ± 2.35)** | (19.0 ± 4.37)* |
| 18 | (−3.54 ± 3.46) | (2.88 ± 2.95) |
| 19 | (10.8 ± 1.37)** | (7.36 ± 3.72) |
| 23 | (7.40 ± 6.26) | (−4.95 ± 2.03) |
| 24 | (7.45 ± 2.20)* | (21.7 ± 1.63)*** |
Results are presented as mean ± S.E.M. (n = 3–4).
p < 0.05,
p < 0.01,
p < 0.001 compared with the control value.
In addition, the isolated compounds were evaluated for cytotoxicity against KB (nasopharyngeal), A549 (lung), HCT-8 (colon), and DU145 (prostate) human cancer cell lines. Compound 1 showed marginal cytotoxicity against KB and DU145 cell lines with GI50 values of 6.27 and 7.30 μg/mL, respectively. Interestingly, the lung and colon tumor-derived cell lines were significantly less susceptible, suggesting 1 may have some antitumor selectivity. The remaining compounds did not exhibit cytotoxicity.
In our previous report, the polyacetylenes 25, 27, 28, 29, and 30 strongly inhibited α-glucosidase activity.15 In this study, we found that saponins 14 and 15 also exhibited potent inhibition of α-glucosidase activity with IC50 values of 75.0 and 50.4 μM, respectively, compared with the positive control acarbose (IC50=677.97 μM).
In conclusion, it has been demonstrated in the present study that certain pure compounds found in the roots of P. japonicus var. major exhibit neutrophil pro-inflammatory activity, cytotoxicity against KB and DU145 cell lines, and inhibition of α-glucosidase activity. These results substantiate, in part, the past medicinal uses of this herb, and may provide a basis for potential therapeutic applications of the roots of P. japonicus var. major for anti-inflammatory, anticancer, or antidiabetes effects. In addition, the developed LC-MS/MS method could be applied to distinguish between natural and artifactual compounds in Panax species.
Experimental Section
General Experimental Procedures
Melting points were determined on a Yanagimoto MP-S3 micro-melting point apparatus. Optical rotations were measured using a JASCO DIP-370 polarimeter. IR spectra were recorded on a Shimazu FTIR Prestige-21 spectrometer. NMR spectra, including 1H, 13C, COSY, NOESY, HMBC, HMQC experiments, were recorded on Bruker AVANCE-500 and AMX-400, using tetramethylsilane (TMS) as internal standard, and all chemical shifts are reported in parts per million (ppm, δ). ESI and HRESI mass spectra were recorded on a Bruker APEX II mass spectrometer. Silica gel (E. Merck 70–230, 230–400 mesh) was used for column chromatography. RP-HPLC was carried out on Shimazu LC-8A and -10A VP systems equipped with CLASS-VP workstation (Shimadzu Co., Ltd., Japan) and SPD-10AV UV-Vis detector.
Plant Material
Dried roots of P. japonicus var. major were purchased from Lijiang and Yongsheng, Yunnan Province, People’s Republic of China, in September–December, 2005. The roots of P. japonicus var. major were 2–3 years growth. The material was identified by Prof. Xi-Wen Li, Kunming Institute of Botany. The voucher specimen has been deposited in Kunming Institute of Botany, Chinese Academy of Sciences, Kunming (KIB05102320) and the Department of Chemistry, National Cheng Kung University, Tainan, Taiwan (TSWu 2006010).
Extraction and Isolation
The dried roots of P. japonicus (4.6 kg) were pulverized and extracted with EtOH (5 × 10 L) under reflux. The filtrate was concentrated under reduced pressure to obtain crude EtOH extract (1212 g, PJ). This extract was suspended into H2O, and successively partitioned with CHCl3 and n-BuOH to obtain CHCl3-soluble brown syrup (77 g, PJC), a n-BuOH soluble portion (482 g, PJB), and a H2O layer. The isolation of compounds from the CHCl3 fraction was described in our previous report.15 The precipitate of the H2O portion was further chromatographed on a silica gel column eluted with CHCl3-EtOAc (3:1) to provide chikusetsusaponin-IV (4) (3.8 g). The n-BuOH portion was chromatographed on a Dianion HP-20 column eluted with H2O, followed by step gradients with MeOH to obtain seven fractions (Fr. 1–7). Fr.2 was chromatographed on a silica gel column eluted with CHCl3-MeOH (3:1) to give panajaponol A (1) (13.3 mg), chikusetsusaponin-Ib (11) (51.4 mg), oleanolic acid 28-O-β-D-glucopyranoside (14) (182.9 mg), and oleanolic acid (15) (40.9 mg). Fr.3 was also chromatographed on a silica gel column eluted with CHCl3-MeOH (3:1) to afford panajaponol A (1) (27.8 mg), chikusetsusaponin-IV (4) (778.4 mg), chikusetsusaponin-IV methyl ester (5) (12.5 mg), stipuleanoside R2 (9) (480.0 mg), pseudoginsenoside RT1 methyl ester (13) (19.1 mg), oleanolic acid 28-O-β-D-glucopyranoside (14) (43.4 mg), ginsenoside Rf (16) (324.5 mg), ginsenoside Rg1 (17) (2.3 mg), (20R)-ginsenoside Rg3 (20) (4.1 mg), and majonoside-R1 (23) (12.4 mg). Silica gel column chromatography of Fr. 4 eluted with EtOAc-MeOH (3:1) and further purification by reversed-phase HPLC provided chikusetsusaponin-IV (4) (420.5 mg), chikusetsusaponin-IV methyl ester (5) (21.8 mg), chikusetsusaponin-V (10) (607.9 mg), oleanolic acid 28-O-β-D-glucopyranoside (14) (130.5 mg), ginsenoside Rf (16) (110.5 mg), ginsenoside Rd (18) (3.1 g), and ginsenoside Rb1 (19) (302 mg). Fr. 5 was chromatographed on a silica gel column eluted with CHCl3-MeOH (3:1) and further purified by HPLC to afford taibaienoside I (3) (3.1 mg), chikusetsusaponin-IV (4) (214.9 mg), chikusetsusaponin-IV methyl ester (5) (60.4 mg), chikusetsusaponin-IVa methyl ester (7) (85.5 mg), chikusetsusaponin-IVa butyl ester (8) (2.3 mg), chikusetsusaponin-V (10) (16.3 mg), pseudoginsenoside RT1 methyl ester (13) (14.8 mg), oleanolic acid 28-O-β-D-glucopyranoside (14) (370.5 mg), ginsenoside Rd (18) (10.9 g), and ginsenoside Rb1 (19) (395.5 mg). Fr.6 was chromatographed on a silica gel using CHCl3-MeOH (4:1) as eluent and further purified by HPLC to give pseudoginsenoside RT1 butyl ester (2) (7.5 mg), taibaienoside I (3) (35.9 mg), chikusetsusaponin-IVa (6) (2.2 mg), chikusetsusaponin-IVa methyl ester (7) (94.8 mg), pseudoginsenoside RT1 (12) (1.1 mg), pseudoginsenoside RT1 methyl ester (13) (88.6 mg), ginsenoside Rd (18) (53.5 mg), (20S)-ginsenoside Rg3 (21) (1.3 mg), ginsenoside Rg5 (22) (3.5 mg), and β-sitosteryl-β-D-glucoside (24) (33.2 mg). β-Sitosteryl-β-D-glucoside (24) (423.1 mg) was also obtained from fr. 7, using silica gel column chromatography (eluent, CHCl3–MeOH, 9:1).
Panajaponol A (1)
Colorless powder; mp 208–210 °C; [α]D25 −14.1 (c 0.046, MeOH); IR (KBr) νmax 3345, 2936, 2878, 1640, 1076, 1030 cm−1; 1H NMR (pyridine-d5, 400 MHz) δ 5.91 (1H, d, J = 7.2 Hz, H-1″), 5.47 (1H, t, J = 6.8 Hz, H-24), 4.91 (1H, d, J = 7.6 Hz, H-1′), 4.54 (1H, dd, J = 12.0, 4.0 Hz, H-27b), 4.47 (1H, m, H-27a), 4.34 (1H, m, H-6), 3.86 (1H, m, H-12), 3.46 (1H, d, J = 5.6 Hz, H-3), 2.70 (1H, m, H-23b), 2.42 (1H, m, H-7b), 2.39 (1H, m, H-23a), 2.25 (1H, m, H-17), 2.07 (1H, m, H-11b), 2.07 (3H, s, H-28), 2.03 (1H, m, H-22b), 1.99 (3H, s, H-26), 1.96 (1H, m, H-7a), 1.81 (2H, m, H-2), 1.78 (1H, m, H-16b), 1.50 (1H, m, H-22b), 1.60 (1H, m, H-1b), 1.58 (1H, m, H-15b), 1.67 (1H, m, H-11b), 1.50 (1H, m, H-9), 1.45 (3H, s, H-29), 1.39 (1H, m, H-16a), 1.38 (1H, m, H-5), 1.34 (3H, s, H-21), 1.23 (1H, m, H-13), 1.15 (3H, s, H-18), 1.09 (1H, m, H-15a), 0.98 (1H, m, H-1a), 0.94 (3H, s, H-19), 0.79 (3H, s, H-30); 13C NMR data see Table 1; ESIMS m/z 839 ([M+Na]+); HRESIMS m/z 839.4777 [M+Na]+ (calcd for C42H72O15Na, 839.4769).
Pseudoginsenoside RT1 Butyl Ester (2)
Colorless powder; mp 202–204 °C; [α]D25 −18.2 (c 0.048, MeOH); IR (KBr) νmax 3387, 2943, 1744, 1173, 1053, 1045 cm−1; 1H NMR (pyridine-d5, 400 MHz) δ 6.33 (1H, d, J = 8.0 Hz, H-1‴), 5.41 (1H, m, H-12), 5.27 (1H, d, J = 7.2 Hz, H-1″), 4.96 (1H, d, J = 8.0 Hz, H-1′), 4.31 (2H, m, H-1″″), 3.30 (dd, J = 9.6, 3.6 Hz, H-3), 3.16 (1H, m, H-18), 2.10 (1H, m, H-2b), 2.09 (1H, m, H-11b), 1.86 (1H, m, H-2a), 1.62 (1H, m, H-9), 1.46 (2H, m, H-6), 1.43 (2H, m, H-7), 1.40 (2H, m, H-1), 0.71 (1H, m, H-5), 1.97 (1H, m, H-11a), 1.89 (2H, m, H-16), 1.79 (1H, m, H-22b), 1.75 (1H, m, H-19b), 1.71 (1H, m, H-22a), 1.60 (2H, m, H-3″″), 1.53 (2H, m, H-15), 1.34 (2H, m, H-2″″), 1.26 (1H, m, H-19a), 1.26 (3H, s, H-27), 1.26 (3H, s, H-23), 1.09 (3H, s, H-26), 1.08 (3H, s, H-24), 1.07 (2H, m, H-21), 0.90 (3H, s, H-29), 0.87 (3H, s, H-30), 0.86 (3H, s, H-25), 0.74 (3H, t, J=6.8 Hz, H-4″″); 13C NMR data see Table 1; ESIMS m/z 1005 ([M+Na]+); HRESIMS m/z 1005.5405 [M+Na]+ (calcd for C51H82O18Na: 1005.5399).
Chromatography and Mass Spectrometry
A LTQ Velos mass spectrometer (Thermo Scientific, San Jose, CA, USA) equipped with Accela HPLC (Thermo Scientific, San Jose, CA, USA) using a BioBasic C18 column (150 × 2.1 mm, 5 μm, Thermo Scientific, San Jose, CA, USA) was used for LC-MS experiments. The HPLC instrument was programmed to form a linear gradient from 90% solvent A (H2O+0.1% formic acid) to 55% solvent B (MeCN+0.1% formic acid) over 30 min at a flow rate of 250 μL/min. The MS system was operated in negative ion mode with capillary temperature at 250 °C, spray needle voltage at −3.5 kV, normalized collision energy at 35%, and sheath gas flow and auxiliary gas flow at 50 and 10 arbitrary units, respectively.
Acid Hydrolysis of 1 and Determination of Sugar Configuration
A solution of 1 (1 mg) in 1.0 M HCl/p-dioxane (1:1 v/v, 1.0 mL) was heated at 80 °C for 4 h. After cooling, the solution was neutralized with Amberlite IRA-400 (OH− form), and the resin was removed by filtration. The filtrate was extracted with EtOAc. The aqueous layer was dried in vacuo, and the residue was dissolved in pyridine (0.1 mL) containing L-cysteine methyl ester hydrochloride (0.5 mg) and heated at 60 °C for 1 h. A 0.1 mL solution of o-torylisothiocyanate (1 μL) in pyridine was added to the mixture at room temperature and kept for 1 h. The reaction mixture was directly analyzed by reversed-phase HPLC. The peak (tR at 38.30 min) coincided with a derivative of D-glucose, as compared with standard D-glucose with tR at 38.12 min.
Acid Hydrolysis of 2 and Determination of Sugar Configuration
A solution of 2 (1 mg) was hydrolyzed in HCl 1.0 M/p-dioxane (1:1 v/v, 1.0 mL) as described above for 1. The peaks (tR at 25.939, 39.850 and 41.801 min) coincided with derivatives of D-glucuronic acid, D-glucose, and D-xylose. Derivatives obtained for standard D-glucuronic acid, D-glucose, and D-xylose gave peaks at tR at 25.970, 39.384, and 41.701 min, respectively.
Preparation of Human Neutrophils
Blood was taken from healthy human donors (20–32 years old) by venipuncture, using a protocol approved by the Institutional Review Board at Chang Gung Memorial Hospital. Neutrophils were isolated using a standard method of dextran sedimentation prior to centrifugation in a Ficoll Hypaque gradient and the hypotonic lysis of erythrocytes. Purified neutrophils that contained >98% viable cells, as determined by the trypan blue exclusion method, were resuspended in calcium (Ca2+)-free HBSS buffer at pH 7.4, and maintained at 4 °C before use.
Measurement of O2•− Generation
The assay of O2•− generation was based on the SOD-inhibitable reduction of ferricytochrome c. In brief, after supplementation with 0.5 mg/mL ferricytochrome c and 1 mM Ca2+, neutrophils were equilibrated at 37 °C for 2 min and incubated with drugs for 5 min. Cells were activated with 100 nM FMLP for 10 min. When FMLP was used as a stimulant, CB (1 μg/mL) was incubated for 3 min before activation by the peptide (fMLP/CB). Changes in absorbance with the reduction of ferricytochrome c at 550 nm were monitored continuously in a double-beam, six-cell positioner spectrophotometer with constant stirring (Hitachi U-3010, Tokyo, Japan). Calculations are based on the differences in the reactions with and without SOD (100 U/mL) divided by the extinction coefficient for the reduction of ferricytochrome c (ε = 21.1/mM/10 mm).
Measurement of Elastase Release
Degranulation of azurophilic granules was determined by elastase release, as described previously. Experiments were performed using MeO-Suc-Ala-Ala-Pro-Val-p-nitroanilide as the elastase substrate. Briefly, after supplementation with MeO-Suc-Ala-Ala-Pro-Val-p-nitroanilide (100 μM), neutrophils (5 × 105/mL) were equilibrated at 37 °C for 2 min and incubated with test compound for 5 min. Cells were activated by 100 nM FMLP and 0.5 μg/mL CB. Changes in absorbance at 405 nm were continuously monitored to assay elastase release. The results are expressed as a percentage of the initial rate of elastase release in the fMLP/CB-activated, drug-free control system.
In Vitro Cytotoxicity Bioassays
All stock cultures were grown in T-25 flasks. Freshly trypsinized cell suspensions were seeded in 96-well microtiter plates at densities of 5000 cells per well with compounds added from DMSO-diluted stock. After three days in culture, attached cells were fixed with cold 10% trichloroacetic acid and then stained with 0.4% sulforhodamine B. The absorbency at 562 nm was measured using a microplate reader after the bound dye was solubilized. The mean EC50 is the concentration of agent that reduces cell growth by 50% under the experimental conditions and is the average from at least three independent determinations that were reproducible and statistically significant. The following human tumor cell lines were used in the assay: nasopharyngeal (KB), human lung (A549), colon (HCT-8), and prostate (DU145) cancer cell lines. All cell lines were obtained from the Lineberger Cancer Center (UNC-CH) or from ATCC (Rockville, MD) and cultured in RPMI-1640 medium supplemented with 25 mM HEPES, 0.25% sodium bicarbonate, 10% fetal bovine serum, and 100 μg/mL kanamycin.
α-Glucosidase Inhibitory Assay
The α-glucosidase inhibitory activity was determined by a partial modification of the procedure reported by Matsui et al.45 Briefly, each well of the 96-well plates contained 150 μL of 2 mM 4-nitrophenyl α-D-glucopyranoside (PNP-G) in 100 mM potassium phosphate buffer (pH 7.0) and 20 μL of sample in DMSO. The reaction was initiated by the addition of 150 μL of the enzyme solution (32 mU/mL from bakers yeast purchased from Sigma Chemical Co.). The plates were incubated at 37 °C for 20 min. The absorbance of 4-nitrophenol released from PNP-G at 400 nm was measured using a μ Quant universal microplate spectrophotometer. The increased absorbance (ΔA) was compared with that of the control (DMSO in place of sample solution) to calculate the inhibition.
The concentration of inhibitors required for inhibiting 50% of α-glucosidase activity under the assay conditions was defined as the IC50 value.
Supplementary Material
Acknowledgments
The authors are grateful for financial support from the National Science Council of Republic of China awarded to T.-S. Wu. This study was supported in part by Taiwan Department of Health Cancer Research Center of Excellence (DOH99-TD-C-111-005). Thanks are also due in part to support from NIH grant CA17625 awarded to K.-H. Lee.
Footnotes
Supporting Information Available: 1H, 13C, COSY, NOESY, HMQC, and HMBC NMR spectra for compounds 1 and 2.
References and Notes
- 1.Li QF, Shi SL, Liu QR, Tang J, Song J, Liang Y. Int J Biochem Cell Biol. 2008;40:1918–1929. doi: 10.1016/j.biocel.2008.01.031. [DOI] [PubMed] [Google Scholar]
- 2.Wang W, Zhao Y, Rayburn ER, Hill DL, Wang H, Zhang R. Cancer Chemother Pharmacol. 2007;59:589–601. doi: 10.1007/s00280-006-0300-z. [DOI] [PubMed] [Google Scholar]
- 3.Lee SK, Wong CK, Poon PM, Ip PS, Che CT, Fung KP, Leung PC, Lam CW. Phytother Res. 2006;20:883–888. doi: 10.1002/ptr.1955. [DOI] [PubMed] [Google Scholar]
- 4.Lee JH, Lee JH, Lee YM, Kim PN, Jeong CS. Food Chem Toxicol. 2008;46:3749–3752. doi: 10.1016/j.fct.2008.09.055. [DOI] [PubMed] [Google Scholar]
- 5.Park EK, Choo MK, Han MJ, Kim DH. Int Arch Allergy Immunol. 2004;113:113–120. doi: 10.1159/000076383. [DOI] [PubMed] [Google Scholar]
- 6.Park EK, Choo MK, Kim EJ, Han MJ, Kim DH. Biol Pharm Bull. 2003;26:1581–1584. doi: 10.1248/bpb.26.1581. [DOI] [PubMed] [Google Scholar]
- 7.Rudakewich M, Ba F, Benishin CG. Planta Med. 2001;67:533–537. doi: 10.1055/s-2001-16488. [DOI] [PubMed] [Google Scholar]
- 8.Zhang G, Liu A, Zhou Y, San X, Jin T, Jin Y. J Ethnopharmacol. 2008;15:441–448. doi: 10.1016/j.jep.2007.10.026. [DOI] [PubMed] [Google Scholar]
- 9.Kim K, Kim HY. J Ethnopharmacol. 2008;120:190–195. doi: 10.1016/j.jep.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 10.Park MW, Ha J, Chung SH. Biol Pharm Bull. 2008;31:748–751. doi: 10.1248/bpb.31.748. [DOI] [PubMed] [Google Scholar]
- 11.Christensen LP. Adv Food Nutr Res. 2009;55:1–99. doi: 10.1016/S1043-4526(08)00401-4. [DOI] [PubMed] [Google Scholar]
- 12.Morita T, Kasai R, Kohda H, Tanaka O, Zhou J, Yang TR. Chem Pharm Bull. 1983;31:3205–3209. [Google Scholar]
- 13.Morita T, Kasai R, Tanaka O, Zhou J, Yang T, Shoji J. Chem Pharm Bull. 1982;30:4341–4346. [Google Scholar]
- 14.State Administration of Traditional Chinese Medicine. Zhong-hua-ben-cao. Vol. 5. Shanghai Science and Technology Press; Shanghai: 1999. pp. 836–839. [Google Scholar]
- 15.Chan HH, Sun HD, Reddy MVB, Wu TS. Phytochemistry. 2010;71:1360–1364. doi: 10.1016/j.phytochem.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 16.Tang H, Yi Y, Wang Z, Jiang Y, Li Y. Yao Xue Xue Bao. 1997;32:685–691. [PubMed] [Google Scholar]
- 17.Fujioka N, Kohda H, Yamasaki K, Kasai R, Shoyama Y, Nishioka I. Phytochemistry. 1989;28:1855–1858. doi: 10.1055/s-2006-962105. [DOI] [PubMed] [Google Scholar]
- 18.Nie RL, Morita T, Kasai R, Wu CY, Tanaka O. Planta Med. 1984;50:322–327. doi: 10.1055/s-2007-969721. [DOI] [PubMed] [Google Scholar]
- 19.Sakai S, Katsumata M, Satoh Y, Nagasao M, Miyakoshi M, Ida Y, Shoji J. Phytochemistry. 1994;35:1319–1324. doi: 10.1016/s0031-9422(00)94846-5. [DOI] [PubMed] [Google Scholar]
- 20.Hu M, Ogawa K, Sashida Y, Xiao PG. Phytochemistry. 1995;39:179–184. [Google Scholar]
- 21.Yang C, Jiang Z, Zhou J, Kasai R, Tanaka O. Acta Bot Yunnan. 1985;7:103–108. [Google Scholar]
- 22.Huan VD, Yamamura S, Ohtani K, Kasai R, Yamasaki K, Nham NT, Chau HM. Phytochemistry. 1998;47:451–458. doi: 10.1016/s0031-9422(97)00618-3. [DOI] [PubMed] [Google Scholar]
- 23.Yu SS, Yu DQ, Liang XT. J Nat Prod. 1994;57:978–982. doi: 10.1021/np50109a016. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka O, Morita T, Kasai R, Kinouchi J, Sanada S, Ida Y, Shoji J. Chem Pharm Bull. 1985;33:2323–2330. [Google Scholar]
- 25.Elujoba AA, Fell AF, Linley PA, Maitland DM. Phytochemistry. 1990;29:3281–3285. [Google Scholar]
- 26.Zhou J, Wu M, Taniyasu S, Besso H, Tanaka O, Saruwatari Y, Fuwa T. Chem Pharm Bull. 1981;29:2844–2850. [Google Scholar]
- 27.Ko SR, Choi KJ, Suzuki K, Suzuki Y. Chem Pharm Bull. 2003;51:404–408. doi: 10.1248/cpb.51.404. [DOI] [PubMed] [Google Scholar]
- 28.Ma WG, Mizutani M, Malterud KE, Lu SL, Ducrey B, Tahara S. Phytochemistry. 1999;52:1133–1138. [Google Scholar]
- 29.Du Q, Jerz G, Waibel R, Winterhalter P. J Chromatogr A. 2003;1008:173–180. doi: 10.1016/s0021-9673(03)00988-9. [DOI] [PubMed] [Google Scholar]
- 30.Teng R, Ang C, McManus D, Armstrong D, Mau S, Antony B. Helv Chim Acta. 2004;87:1860–1872. [Google Scholar]
- 31.Kim S, Park J, Ryu J, Lee Y, Kim T, Kim J, Beak N. Arch Pharm Res. 1996;19:551–553. [Google Scholar]
- 32.Basent P, Kadota S, Terashima S, Shimizu M, Namba T. Chem Pharm Bull. 1993;41:1238–1243. doi: 10.1248/cpb.41.1238. [DOI] [PubMed] [Google Scholar]
- 33.Zheng G, Lu W, Aisa HA, Cai J. Tetrahedron Lett. 1999;40:2181–2182. [Google Scholar]
- 34.Hirakura K, Morita M, Nakajima K, Ikeya Y, Mitsuhashi H. Phytochemistry. 1992;31:899–903. [Google Scholar]
- 35.Kobayashi M, Mahmud T, Umezome T, Wang W, Murakami N, Kitagawa I. Tetrahedron. 1997;53:1569115700. [Google Scholar]
- 36.Hirakura K, Morita M, Nakajima K, Ikeya Y, Mitsuhashi H. Phytochemistry. 1991;30:3327–3333. [Google Scholar]
- 37.Kuo YH, Chen WC. J Chin Chem Soc. 1999;46:819–824. [Google Scholar]
- 38.Ito J, Chang FR, Wang HK, Park YK, Ikegaki M, Kilgore N, Lee KH. J Nat Prod. 2001;64:1278–1281. doi: 10.1021/np010211x. [DOI] [PubMed] [Google Scholar]
- 39.Chen CY, Chang FR, Teng CM, Wu YC. J Chin Chem Soc. 1997;46:77–78. [Google Scholar]
- 40.Goda Y, Shibuya M, Sankawa U. Chem Pharm Bull. 1987;35:2675–2677. doi: 10.1248/cpb.35.2675. [DOI] [PubMed] [Google Scholar]
- 41.Hu JF, Bai SP, Jia ZJ. Phytochemistry. 1996;43:815–817. [Google Scholar]
- 42.Nakamura S, Sugamoto S, Matsuda H, Yoshikawa M. Chem Pharm Bull. 2007;55:1342–1348. doi: 10.1248/cpb.55.1342. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka T, Nakashima T, Ueda T, Tomii K, Koun I. Chem Pharm Bull. 2007;55:899–901. doi: 10.1248/cpb.55.899. [DOI] [PubMed] [Google Scholar]
- 44.Srivastava SK, Jain DC. Phytochemistry. 1989;28:644–647. [Google Scholar]
- 45.Matsui T, Yoshimoto C, Osajima K, Oki T. Biosci Biotech Biochem. 1996;60:2019–2022. doi: 10.1271/bbb.60.2019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.