Abstract
The expression of the breast cancer susceptibility protein BRCA2 is highly regulated in human breast, ovary, and pancreatic cells. BRCA2 is not expressed in the non-dividing cells, and expression is cell cycle stage-dependent and is elevated in the sporadic cancer cells. Mutational analysis of the upstream sequence of the human BRCA2 gene revealed an E2-box-containing silencer at the −701 to −921 position. The E2-box is essential for the cell-cycle stage-dependent activity of the silencer. We affinity-purified a 29-kDa silencer-binding protein (SBP) from the nuclear extracts of human breast cells BT-549 and MDA-MB-231. We explored whether the E2-box-binding repressor protein SLUG, which is of similar molecular size, is involved in the silencing process. Supershift assay with the purified SBP and anti-SLUG antibody revealed the identity of the SBP as SLUG. We found that silencer is inactive in the human breast cancer cells such as MDA-MB-468 and MCF-7 that do not express SLUG, further suggesting the involvement of SLUG in the BRCA2 gene silencing. Inducible expression of human SLUG in the dividing MDA-MB-468 cells reduced BRCA2 RNA levels with the activation of the silencer. Furthermore, small interfering RNA-mediated knockdown of SLUG mRNA in the BT-549 cells caused inhibition of the silencer function. Chromatin immunoprecipitation assays suggested that SLUG mediates its action by recruiting C-terminal-binding protein-1 (CtBP-1) and histone deacetylase-1 (HDAC-1) at the silencer E2-box. The general HDAC inhibitor, trichostatin A, inhibited the SLUG-mediated regulation of the silencer function. It thus appears that SLUG is a negative regulator for BRCA2 gene expression.
BRCA2 is a tumor suppressor protein with diverse functions (1–3). BRCA2 deficiency has been attributed as the cause for many cases of breast, ovarian, and pancreatic carcinoma (1–3). BRCA2 deficiency may be caused by the molecular defects in the BRCA2 gene or due to sporadic reasons. The BRCA2 gene is not expressed in non-dividing cells, and the rate of expression of this protein is increased with the rate of cell proliferation (4–6). This growth-dependent turn-on/turn-off mechanism of this gene is not well understood. Any number of environmental cues may influence the turn-on event and may initiate a molecular domino effect leading to DNA damage and oncogenesis. Although undue expression of BRCA2 protein in non-dividing cells may initiate apoptosis, the failure of the cell to produce the appropriate amount of BRCA2 corresponding to the growth rate of the cell may also be detrimental. Since the majority of the breast cancer cases are sporadic (1–6), such cancers may be initiated as a result of the transient absence of BRCA2 during the proliferative stages of the breast cells.
As alluded to above, the BRCA2 gene is stringently regulated during the cell cycle (7–11). To understand how BRCA2 gene expression is regulated in human mammary epithelial cells, the regulatory DNA sequence elements around the proximal upstream region of the gene are under extensive study (8–12). Examination of the minimal promoter sequence has revealed several canonical elements for the binding of transcription factors including an E-box, E2F, and Ets recognition motifs (9). Antibodies to candidate transcription factors used in supershift experiments revealed specific interactions between the BRCA2 promoter and the basic region/helix-loop-helix containing USF-1 and -2 proteins and Elf-1, an Ets domain protein. Myc-Max or Max-Max dimers were reported not to bind this E-box sequence (9). Analysis of the −144 to −59 region identified a putative NFκB binding site (10). Thus, NFκB and upstream stimulatory facter regulate BRCA2 expression through the BRCA2 promoter (10). P53 was found to repress the expression of BRCA2 promoter activity (11).
We previously reported a 221-bp silencer sequence located at 700 bp upstream of the BRCA2 gene transcription start site (12) (see Fig. 1A). We report here a mechanism for the negative regulation of the BRCA2 gene expression by this silencer. We show evidence that a unique E2-box sequence surrounded by Alu sequences is responsible for the function of the silencer. Our chromatin immunoprecipitation data suggest that the E2-box may mediate the silencing by recruiting the zinc finger suppressor protein SLUG, which most likely then recruits CtBP-1 and HDAC-11 to result in deacetylation of the acetylated histones at the BRCA2 gene promoter. Histone deacetylation then probably causes the inhibition of BRCA2 gene expression.
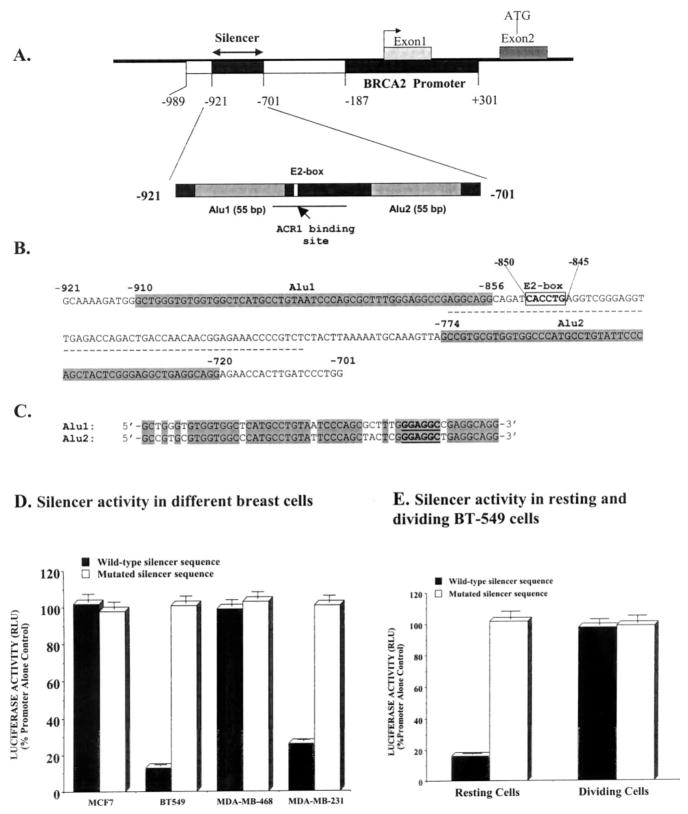
Fig. 1. Structure and function of human BRCA2 gene silencer.
A, a map of the promoter and the silencer at the upstream of the human BRCA2 gene. The putative ACR1 binding site at the silencer is indicated by a line. B, the nucleic acid sequence of the silencer showing the AluI, the AluII, and the E2-box. The 55-bp direct Alu repeats are in gray. The E2-box sequence (5′-CACCTG-3′) is boldfaced and boxed. The putative ACR1 binding site at the silencer is underscored. C, alignments of nucleotide sequences of the AluI/AluII. D, a Dual luciferase assay (Promega) to evaluate the function of the wild-type and E2-box-mutated silencer. Data are presented as mean (n = 12) ± S.E. The differences between the luciferase activities with the wild-type silencer sequence and the E2-box-mutated silencer sequence containing construct in BT-549 cells and in MDA-MB-231 cells are statistically significant (p < 0.0001). RLU, relative light units. E, activity of the silencer in the non-dividing (resting) and the dividing BT-549 cells. The differences between the luciferase activities with the wild-type silencer sequence and the E2-box-mutated silencer sequence-containing construct is statistically significant (p < 0.0005). The silencer DNA was mutated at the E2-box from 5′-CACCTG-3′ to 5′-AACCTA-3′.
MATERIALS AND METHODS
Cell Culture
We used a series of commercially available lines of human breast cells including HMEC (Clonetics, purchased through Fisher), MDA-MB-231, MCF-7, MDA-MB-468, and BT-549. All cells, other than the HMEC cells, were purchased from American Type Culture Collection (ATCC, Manassas, VA). HMEC cells were grown in medium purchased from Clonetics under their recommended conditions. Other cells were maintained and grown in ATCC recommended media and conditions, as described before (12, 13). Resting or non-dividing cells are defined as the cells that are in a 100% confluent state of growth in a culture flask or the cells that are arrested mostly at the G0 phase by serum starvation. Serum starvation of the cells to arrest them at the G0 phase was done for 60–80 h following established protocols (9). The population of non-dividing cells was determined by flow cytometry using propidium iodide staining methods (14). More than 88% of the cells used as non-dividing cells were at the G0/G1 phase. Dividing cells are the cells from S/G2 phase (>82%) of cell growth.
Promoter/Silencer Constructs and Luciferase Assay
Human BRCA2 promoter (−187 to +301) was amplified with the primer set PF1, 5′-GCGAGAAGAGAACACACA-3′/PR1, 5′-GCAGAGAAAAGGCAA-3′. The 497-bp fragment was initially cloned into pCRII-Topo vector (Invitrogen). The recombinant plasmid with the insert having the forward orientation with respect to the T7 RNA polymerase promoter was selected. The insert was cut out of the plasmid with BamHI/XhoI and subcloned into the BglII/XhoI site at the multiple cloning site of pGL3-Basic plasmid (Promega). This gave us the BRCA2 promoter vector, pGL3-P. The 221-bp silencer DNA (−701 to −921) was amplified using the primer set SF1, 5′-GCAAGATGGGCCGGGTGT-3′/SR1, 5′-CCA GGGTGTGGTTCTC-3′ and initially cloned into pCRII-Topo vector. The recombinant plasmid clone with insert in the reverse orientation with respect to T7 RNA polymerase promoter was selected. The silencer sequence was digested out of the recombinant plasmid with KpnI/XhoI and subcloned at the KpnI/XhoI sites of the pGL3-P vector. This gave us the promoter-silencer vector, pGL3-PS.
Transient transfections were performed in six-well plates using Lipofectamine 2000 transfection reagent (Invitrogen) with 1 μg of pGL3-P or pGL3-PS and 0.1 μg of pRL-TK-Renilla luciferase control vector (Promega). Protein lysates were prepared from the cells, and luciferase activities were measured as described previously (12). Firefly luciferase activity was normalized with respect to Renilla luciferase activity and presented as a ratio (relative light units). Protein contents of the extract, when needed, were determined using RC-DC reagents and protocol from Bio-Rad.
Site-directed Mutagenesis
PCR-based site-directed mutagenesis (the QuikChange site-directed mutagenesis kit, Stratagene, La Jolla, CA) technique was used for the generation of reporter gene constructs with E2-box mutations following the manufacturer’s instructions. The E2-box element was mutated from 5′-CACCTG-3′ to 5′-AACCTA-3′ (sense strand). Such mutations are known to ablate the function of the E2-box (15).
DNA Affinity Pull-down of Silencer-binding Proteins
We purified the silencer-binding proteins by DNA affinity chromatography following the protocol provided by Dynal Biotech, using biotin-tagged 221-bp BRCA2 silencer fragment employing Dynabeads M-280-streptavidin beads (Dynal Biotech). The biotin-labeled silencer fragment was obtained by PCR using 5′-biotin-labeled primers. The proteins in BT-549 or MDA-MB-231 cells were biosynthetically labeled with [35S]methionine, and the nuclear extract was isolated following standard protocols (14). The biotin-tagged silencer DNA was bound to Dynabeads M280 following the manufacturer’s instructions (Dynal Biotech). The nuclear protein was incubated for 1 h at 25 °C with biotinylated silencer DNA bound to Dynabeads M280 streptavidin in protein binding buffer (80 mM NaCl, 50 mM Tris-HCL, pH 7.5, 4% (v/v) glycerol, 5 mM MgCl2, 0.25 mg/ml poly(dI-dC), 2.5 mM EDTA, 30 μM mutant double-strand oligonucleotide, 1 mM phenylmethylsulfonyl fluoride, and 1 mM dithiothreitol). The magnetic beads were washed three times with protein binding buffer in 80 mM NaCl containing excess poly(dI-dC) and mutant (at the E2-box) duplex oligonucleotide competitor DNA. The fractions were eluted with elution buffer (0.5 M NaCl, 50 mM Tris-HCL, pH 7.5, 4%(v/v) glycerol, 5 mM MgCl2, 0.25 mg/ml poly(dI-dC), 2.5 mM EDTA, 30 μM mutant double-strand oligonucleotide, 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol) and were stored at −80 °C. Silencer-binding proteins (SBPs) were electrophoresed on a 10% Tris-glycine SDS-poly-acrylamide gel. The gel was fixed, dried, and exposed to x-ray film at −80 °C with intensifying screen.
Expression of Recombinant SLUG in MDA-MB-468 Cells
Human SLUG coding sequence was amplified from RNA isolated from BT-549 cells using N-terminal primer (5′-GACGGATCCATGCCGCGCTCCTTCCTG-3′) and the C-terminal primer (5′-CGTCGACTCACTTATCGTCGTCATCCTTGTAATCGTGTGCTACACAGC-3′). The sequence-verified amplified cDNA (831 bp) was digested with BamHI/SalI and was cloned at the BamHI/SalI sites at the multiple cloning site of pRevTet-TRE plasmid (BD Biosciences). PT67 packaging cells were transfected with either the pRevTet-ON plasmid (BD Biosciences) or the pRevTet-TRE-SLUG plasmid to collect the recombinant retrovirus. PT67 cells were grown to ~50% confluency in 100-mm culture dishes and then transfected with 4 μg of plasmid DNA using Lipofectamine 2000 (Invitrogen). The culture medium was replaced at 24 h after transfection, and retrovirus-containing medium was collected at 48 h after transfection and filtered through a 0.45-μm filter. For long term storage, the retrovirus-containing medium was frozen in liquid nitrogen and stored at −80 °C. MDA-MB-468 cells were grown to ~40–50% confluency in 100-mm culture dishes and then transduced first by retrovirus containing pRevTet-ON plasmid by the addition of 1:1 retrovirus-containing medium and growth medium. The culture medium was replaced after 24 h with cell growth medium. Medium containing 1 mg/ml Geneticin was added to select stable cell population at 48 h. This Geneticin-resistant cell population was further transduced by pRevTRE-SLUG plasmid containing virus as described above, and the stable cell line was selected further with 500 μg/ml hygromycin B. For all the experiments, this double-resistant cell line was maintained in 100 μg/ml of Geneticin and hygromycin B. Expression of SLUG was induced by doxycycline (1 μg/ml).
Knock-down of SLUG Gene Expression in BT-549 Cells
The anti-human SLUG siRNAs were designed using the Invitrogen site software and custom-synthesized from Invitrogen. The nucleotide sequences of the siRNA pair (SR) and its respective control (SRC) are as follows (the number indicates the location of the sequence in the SLUG coding region): SR661 5′-GCAUUUGC AGACAGGUCAA-3′/5′-UUGAACUGUCUGCA AAUGC-3′; SRC661: 5′-GCAACGUACAGUG GUUCAA-3′/5′-UUGAACCACUGUACGUUGC-3′. We used Trans-messenger reagent kit (Qiagen) and protocol for the transfection of the BT-549 cells with the siRNAs. RNA was prepared by TRIZOL reagent and DNase I treated before RT-PCR analysis.
End Point RT-PCR Analysis
Total RNA was isolated from cells and analyzed for specific transcripts by RT-PCR analysis (13). β-Actin was used as a loading control. Total RNAs (5 μg) were treated with DNase I (Promega) and reverse-transcribed with Superscript II (Invitrogen). The resulting cDNA was used to carry out PCR amplification of BRCA2 (225 bp; 5′-GTACAGGAAACAAGCTTCTGA-3′ and 5′-GACTAACAGGTGGAGGTAAAG-3′); SLUG (807 bp; 5′-CCTGGTCAAGAAGCATTTCAACG-3′ and 5′-CCCCAAAGATGAGGAGTATCCG-3′); and β-actin (353 bp; 5′-GCTCGTCGTCGACAACGGCTC-3′ and 5′-CAAACATGATCTGGGTCATCTTCTC-3′).
Real-time RT-PCR Analysis
Quantitation of BRCA2 and SLUG transcripts were performed using a TaqMan assay (Applied Biosystems) as described before (13). 18 S rRNA was used as an endogenous control (product 4319413E). All PCR reactions were performed in triplicate for each sample and were repeated three times. All experimental data were expressed as the mean ± S.E. A one-way analysis of variance, a two-way repeated measure analysis of variance, and Student’s t test were used to determine the significance of the difference (16).
Western Blotting
SLUG and actin protein levels in cell lysates were analyzed by immunoblotting. We amplified the coding sequence of human SLUG and cloned in pET100D/Topo vector (Invitrogen), expressed the His6-tagged SLUG in Escherichia coli (DE3), and purified the recombinant protein using nickel-nitrilotriacetic acid agarose following the supplier’s instructions (Qiagen). Anti-SLUG antiserum was custom-developed (Antibody Solutions, Mountain View, CA) and purified. Proteins from cell lysate were analyzed by polyacrylamide gel electrophoresis and transferred to nitrocellulose membrane, as described before (13). Membranes were blotted with anti-SLUG antibody or anti-actin antibody (Sigma) and then incubated with horseradish peroxidase-conjugated secondary antibodies (Amersham Biosciences). Signals were visualized using the SuperSignal chemiluminescent detection system (Pierce).
Electerophoretic Mobility Shift Assay
Double-stranded DNA containing the 221-bp silencer fragment was labeled with [γ-32P]ATP and used in electrophoretic mobility shift assay (12). Double-stranded DNA probes were purified from the reaction mixture using a Bio-Gel P-100 column (Bio-Rad), incubated with whole cell extract from breast cells, and separated on 5% polyacrylamide gels as described previously (12, 17). Supershift assay using anti-SLUG antibodies (made in our laboratory) was also performed (17). Immuno-pull-down of SLUG to remove SLUG from the purified protein preparations was done using Dynabead-Protein A following supplier-provided protocol (Dynal Biotech, Brown Deer, WI).
Chromatin Immunoprecipitation Assays
To test the in vivo binding of SLUG, CtBP-1, and HDAC-1 to the silencer or the deacetylation of histones at the BRCA2 promoter during the silencing process, we performed chromatin immunoprecipitation assays (ChIP) using the standard protocol supplied by Upstate Biotechnology with their ChIP assay reagents (18, 19). For immunoprecipitation, we used anti-SLUG antibody developed in our laboratory as described above. Following this, other antibodies were procured from Upstate Biotechnology: anti-CtBP-1 (rabbit polyclonal IgG, catalog number 07-306); anti-HDAC-1, clone 2E10 (mouse monoclonal IgG, catalog number 05-614); anti-acetyl histone H3 (rabbit polyclonal, catalog number 06-599); and anti-acetyl histone H4 (rabbit polyclonal, catalog number 06-866). PCR was performed with 10 μl of DNA, and Taq DNA polymerase, using 5′-32P-labeled primers SF1/SR1 to amplify the 221 bp of the silencer or PF1/ PR1 to amplify the 497-bp BRCA2 gene promoter. The gel was dried and exposed to x-ray film to visualize the amplified band of expected size.
RESULTS
Human BRCA2 Gene Silencer Is Differentially Active in Various Breast Cells and at Different Cell Growth Stages
The 221-bp silencer sequence at the upstream (−701 to −921) of the human BRCA2 gene has several interesting structural features. It has two Alu sequences (AluI and AluII) each of 55 bp separated by 81-bp inter-Alu sequences (Fig. 1, A and B). This inter-Alu sequence houses an E2-box sequence (5′-CACCTG-3′) as well as a 63-bp patch overlapping the E2-box that has >83% identity with the footprint of a RNA polymerase III transcriptional regulator ACR1 (20) (Fig. 1, A and B). The Alu sequences have ~82% identity at the nucleotide level (Fig. 1C). We have shown previously that the entire 221-bp sequences are essential for the function of the silencer (12). We further verified that this silencer is not only active with heterologous promoter, such as SV40 promoter (12), but also active with its endogenous BRCA2 gene promoter (Fig. 1D). This functionality of the silencer is not only demonstrated in a transient transfection assay but also in stably transfected BT-549 cells (data not shown). One of the interesting features of this silencer is that it is active in certain breast cancer cells (Fig. 1D) and only in the non-dividing stages (G0/G1 phases) of the cell (Fig. 1E). Thus, the activity of the silencer is 8–10-fold higher in resting or predominantly G0-arrested BT-549 cells than in the dividing cells (Fig. 1E). The cell cycle stage-dependent activity of the silencer may be relevant because BRCA2 mRNA and protein are expressed significantly only in the dividing cells with optimum levels in the S/G2 phase cells (9, 10). The silencer is also active in the non-dividing HMEC cells (data not shown), suggesting that the activity of the silencer may not be specific for certain cancer cells only.
The E2-box in the Silencer Is Essential for Its Function
Since the E2-box is involved in many transcriptional repression processes, we checked whether the E2-box sequence in the BRCA2 silencer is necessary for its function. When the E2-box sequence was mutated from 5′-CACCTG-3′ to 5′-AACCTA-3′, the function of the silencer is abolished in the cells where it is functional (Fig. 1, D and E). The silencer is not active in the MCF-7 and the MDA-MB-468 cells. Thus, there are no significant differences in the activity of the promoter whether or not wild-type or mutated silencer is present (Fig. 1D). On the other hand, the silencer is significantly active in the BT-549 and MDA-MB-231 cells (Fig. 1D). Mutation of the E2-box of the silencer ablated its activity (Fig. 1D) in these cells. The essentiality of the E2-box for the silencer function was context-dependent. Thus, replacement of any of the Alu elements from the silencer with unrelated sequence of similar size ablated the function of the silencer although the E2-box remained unaltered (data not shown).
We previously reported that the silencer DNA forms protein-DNA complexes with the nuclear proteins from MDA-MB-231 cells (12). These complexes were not formed with nuclear proteins from the breast cells where the silencer is inactive. Oligonucleotide sequence containing the wild-type E2-box could successfully compete the binding of the protein to the silencer DNA probe, but that containing the mutated E2-box could not (12). All these data strongly suggest that the E2-box sequence within its context in the BRCA2 silencer is necessary for the binding of proteins to it and thus for its function. We also have verified the essentiality of the E2-box in BT-549 cells stably transfected with the wild type or the mutated silencer-containing luciferase constructs (data not shown).
Silencer Binds with SLUG from Breast Cell Nuclear Extract
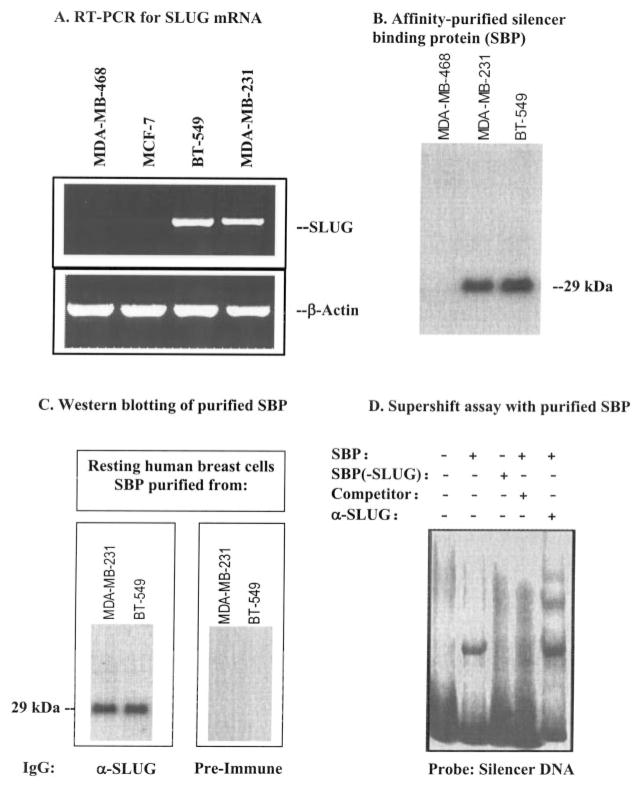
Since the E2-box of the silencer is essential for the binding of the nuclear proteins as well as for the function of the silencer, our hypothesis was that the E2-box-binding proteins regulate the function of the silencer. We had noted that the silencer failed to function in the human breast cancer cells where the E2-box binding repressor protein SLUG is not expressed. For example, it has been reported that SLUG mRNAs are below detection levels in the MCF-7 and MDA-MB-468 cells, whereas the BT-549 and MDA-MB-231 cells express plenty of SLUG mRNA (15). Our RT-PCR data further verified these observations (Fig. 2A). We experimented with the nuclear extracts from different human breast cells to verify whether it indeed binds to SLUG. These pull-down experiments show the purification of a 29-kDa protein from the nuclear extracts of the cells where the silencer is functional, e.g. BT-549 and MDA-MB-231, whereas it did not purify any such protein from MDA-MB-468 cells where the silencer is not active (Fig. 2B). Immunoblot analysis with anti-SLUG antibodies revealed that the affinity-purified protein band contains SLUG (Fig. 2C). An electrophoretic mobility shift assay with the purified SBP showed that the protein specifically binds to the wild-type silencer DNA but not the E2-box-mutated silencer DNA (Fig. 2D). The formation of the protein-DNA complex was competed out with unlabeled wild-type silencer DNA. There was a super-shift of the retarded protein-DNA band with anti-SLUG antibody (Fig. 2D), indicating the presence of SLUG in the protein-DNA complex.
Fig. 2. DNA affinity purification of the SBP from the nuclear extracts from BT-549 and MDA-MB-231 cells and its characterization as SLUG.
A, RT-PCR data showing the expression of SLUG in the BT-549 and MDA-MB-231 cells and not in the MDA-MB-468 and MCF-7 cells. β-Actin was used as a reference control. B, an autoradiogram showing the [35S]methionine-labeled SBP of 29 kDa from resting MDA-MB-231 and BT-549 cells. A parallel experiment was done with the nuclear extract from the resting MDA-MB-468 cells, and no SBP band was detected. C, a Western blot showing the binding of the affinity-purified protein with immunoglobulins raised against recombinant human SLUG protein. Proteins were isolated from the nuclear extracts of unlabeled resting cells similarly as in B, separated by SDS-PAGE, and immunoblotted with anti-SLUG (α-SLUG) purified IgG. IgG similarly purified from rabbit preimmune antiserum was used as a control with a duplicate blot. D, a supershift assay showing the binding of SLUG to the silencer DNA. 32P-labeled wild-type 221-bp silencer DNA was used as probe. The probe was incubated with SBP purified from the nuclear extracts of the unlabeled resting BT-549 cells. The removal of SLUG from the purified SBP preparation by immuno-pull-down (SBP(−SLUG)) inhibited the mobility shift of the probe (lane 3 from the left). To determine specificity of the binding, purified SBP was preincubated with 50-fold excess of unlabeled wild-type 221-bp silencer DNA (lane 4 from the left); lane 5, same as lane 2 except that anti-SLUG antibodies were added to the protein-DNA complex for supershift assay. Multiple supershifted bands were formed.
The Function of the Silencer Is Directly Correlated with the Level of Silencer Protein in the Cell
To test whether the inability of the 221-bp silencer sequence to function in the context of MDA-MB-468 cells is indeed due to the absence of SLUG, we overexpressed SLUG from a doxycycline-inducible retroviral promoter and evaluated whether the silencer become functional in the presence of the inducer. Northern analysis of the SLUG mRNA levels in the uninduced and induced recombinant MBA-MB-468 cells indicated that there is a 7–10-fold increase in the mRNA (2.2 kilonucleotide) level after doxycycline induction (Fig. 3A). The basal level of SLUG mRNA is also increased (Fig. 3A), perhaps because of the “leakiness” of the system. Western blot analysis with the nuclear proteins from the induced or uninduced MDA-MB-468 cells also showed the increase of the SLUG protein level in the induced cells (Fig. 3B). The extent of SLUG expression was also confirmed by real time RT-PCR analysis (Fig. 3C).
Fig. 3. Activation of BRCA2 gene silencer in MDA-MB-468 cells by the overexpression of the SLUG protein.
A, a Northern autoradiogram showing overexpression of SLUG in the doxycycline-induced (Dox +) cells. An ~0.8 kb DNA containing full-length human SLUG coding sequence was used as probe. The size of SLUG mRNA is 2.2 kilo nucleotide. 28 S rRNA is shown as a loading control. B, a Western blot showing induced expression of SLUG protein in the recombinant MDA-MB-468 cells. C, real-time RT-PCR data showing an increase in the SLUG mRNA level and a decrease in the BRCA2 mRNA level in the presence of doxycycline. D, an electrophoretic mobility shift assay (EMSA) showing the doxycycline-inducible binding of protein in recombinant MDA-MB-468 cells. Nuclear proteins (NE) were isolated from wild-type (*) or recombinant (Dox − or Dox +) MDA-MB-468 cells. The probe was 32P-labeled 221-bp wild-type silencer DNA. E, the functional activity of the silencer in the uninduced (No Dox) or doxycycline-induced (With Dox) recombinant MDA-MB-468 cells. Cells were transfected with pGL3-P, pGL3-PS, or pGL3-PS* (which has the E2-box-mutated silencer sequence instead of the wild-type silencer) plasmid DNA. Data are presented as mean (n = 12) ± S.E. The differences between the luciferase activities in the cells (transfected with pGL3-PS construct) treated with and without doxycycline were statistically significant (p < 0.0001). RLU, relative light units.
Interestingly, the real-time RT-PCR experiment also revealed that the induction of SLUG mRNA in the recombinant non-dividing MDA-MB-468 cells is associated with the significant decrease in the BRCA2 mRNA levels (Fig. 3C). Although the proteins of the nuclear extract from the control wild-type MDA-MB-468 cells do not bind to the silencer DNA in vitro, there was a significant mobility shift of the silencer probe with the proteins of nuclear extract from the induced recombinant MDA-MB-468 cells (Fig. 3D). The protein-DNA complex that is formed with the nuclear extract isolated from doxycycline-induced recombinant MDA-MB-468 cells contained SLUG, as was revealed by supershift assay using anti-SLUG antibodies (data not shown). We thus established that doxycycline induced the expression of functional SLUG protein in the stable recombinant MDA-MB-468 cells. Additional findings provide evidence that the inhibition of BRCA2 mRNA level in the SLUG-overexpressing recombinant MDA-MB-468 cells is due to the activation of the BRCA2 silencer in these cells. Although there is no noteworthy change in the activity of the BRCA2 gene promoter itself in the induced or uninduced cells, its activity was significantly reduced in the presence of doxycycline only when the wild-type silencer sequence was present in the construct but not when the construct contained the E2-box-mutated silencer sequence (Fig. 3E). Likewise, induced overexpression of SLUG in recombinant BT-549 cells increased silencer activity several fold, and in parallel, decreased the expression of BRCA2 transcripts, as revealed by real-time RT-PCR analysis (data not shown).
To further verify the reciprocal relationship between SLUG gene expression and the function of BRCA2 silencer in human breast cells, we knocked down SLUG expression in the BT-549 cells using SLUG siRNA. RT-PCR analysis of the SLUG mRNA levels in the control siRNA-treated or SLUG siRNA-treated BT-549 cells showed a decrease in the SLUG mRNA level in the latter (Fig. 4A). The concomitant decrease in the SLUG protein level in the SLUG siRNA-treated cells was evident from the Western blot data (Fig. 4B). Functional analysis of the BRCA2 silencer sequence in SLUG siRNA-treated non-dividing BT-549 cells showed a significant decrease in the level of the silencer activity (Fig. 4C) and of BRCA2 mRNA in the BT-549 cells (Fig. 4D). Taken together, the data provide strong evidence that the transcription factor SLUG regulates expression of the human BRCA2 gene via this newly identified silencer sequence at −701 to −921 nucleotides upstream of the BRCA2 gene start site.
Fig. 4. Inhibition of BRCA2 gene silencer function by the knockdown of SLUG in the non-dividing (resting) BT-549 cells.
A, RT-PCR data showing a decrease in the SLUG transcript level in SLUG knocked down (SLUG siRNA) resting BT-549 cells over that in the control siRNA-treated cells. B, a Western blot showing a decrease in the SLUG protein level in SLUG knocked down (SLUG siRNA) BT-549 cells over that in the control siRNA-treated cells. C, inhibition of the function of the BRCA2 gene silencer in the SLUG knocked down (siRNA) resting BT-549 cells. Data are presented as mean (n = 12) ± S.E. The differences between the luciferase activities in the cells (transfected with the pGL3-PS construct) treated with control siRNA and SLUG siRNA were statistically significant (p < 0.0005). RLU, relative light units. D, RT-PCR data showing a severalfold increase in the BRCA2 mRNA level in the anti-SLUG siRNA-treated resting BT-549 cells over that in the control siRNA-treated cells. A parallel evaluation of β-actin mRNA was used as a loading control for the RT-PCR assays.
SLUG Recruits CtBP-1 and HDAC-1 at the Silencer
SLUG is known to repress the expression of a gene by binding to the E2-box and then recruiting the co-repressor protein CtBP-1 and eventually the histone deacetylase HDAC-1 (21, 22). To explore whether SLUG is indeed recruited at the BRCA2 gene silencer inside the cell nucleus, we performed a chromatin immunoprecipitation assay. This analysis revealed that not only SLUG but also the co-repressor CtBP-1 and the histone modifier HDAC-1 are recruited at the silencer in SLUG-expressing MDA-MB-231 and BT-549 cells, perhaps to form the silencing complex (Fig. 5A). Importantly, CtBP-1 and HDAC-1 are not recruited in the nucleus in MDA-MB-468 cells lacking SLUG gene expression (Fig. 5A). The consequence of the recruitment of HDAC-1 at the promoter is the deacetylation of the acetylated histones on the DNA (23, 24). We tested whether the induction of SLUG expression in the recombinant MDA-MB-468 cells alters the abundance of acetylated histones at the BRCA2 promoter. Our ChIP assay data suggest that indeed HDAC-1 is recruited at the silencer and may be involved in the deacetylation of acetylated histones at the BRCA2 promoter (Fig. 5B).
Fig. 5. Recruitment of HDAC-1 at the silencer by SLUG is correlated with decrease in acetyl histones at the BRCA2 promoter.
A, a ChIP assay showing the in vivo binding of SLUG, CtBP-1, and HDAC-1 at the silencer in the non-dividing MDA-MB-231 and BT-549 cells but not in the non-dividing MDA-MB-468 cells. There was no amplification of the silencer DNA in the transfection when no antibody or preimmune rabbit serum was used (data not shown). The lowest panel is an amplification of input DNA prior to immunoprecipitation. B, ChIP assay showing the levels of acetyl histones H3 and H4 at the BRCA2 promoter in BT-549 cells. Cells were treated without (−TSA) or with (+TSA) trichostatin A (100 nM) for 24 h before ChIP assay. The lowest panel is an amplification of input DNA prior to immunoprecipitation. There is no change in the amplification of input DNA in all the cases. C, functional evaluation of the activity of the BRCA2 gene silencer in non-dividing BT-549, MDA-MB-231, and MDA-MB-468 cells in the presence or absence of the HDAC inhibitor, trichostatin A (100 nM). Data are presented as mean ± S.E. All experiments were done six times in triplicate. The differences between the luciferase activities in the cells treated with the solvent dimethyl sulfoxide (DMSO) or trichostatin A were statistically significant (p < 0.0001). RLU, relative light units.
We further verified the involvement of HDAC in the SLUG-induced histone deacetylation at the BRCA2 promoter of the recombinant MDA-MB-468 cells using the general HDAC inhibitor trichostatin A (TSA). TSA is a specific inhibitor of multiple HDACs (23, 24). It is believed to enhance transcription through inhibiting HDACs and thus in turn cause increased acetylated histones in the promoter region. The ChIP assay data evaluating the abundance of acetylated histones at the BRCA2 gene promoter with or without treatment with TSA (100 nM) is shown in Fig. 5B. In the absence of inducer (doxycycline), very little SLUG protein was present, and the promoter is bound to acetylated histones H3 and H4, among others. The acetyl histone content did not change with the addition of TSA (Fig. 5B). On the other hand, the induction of SLUG expression was associated with the decrease of acetylated histones H3 and H4. TSA reversed the decrease of the acetyl histone content of the BRCA2 promoter, further suggesting the involvement of HDAC in the process (Fig. 5B).
We also performed ChIP assays with the recombinant MDA-MB-468 cells transfected with the pGL3-PS* construct, which has the E2-box-mutated silencer sequence instead of the wild-type silencer (Fig. 3E). As expected, we found amplification of the silencer DNA from the SLUG antibody pull-down chromatin fragments only when SLUG was induced by doxycycline (data not shown). Although the E2-box was mutated in the plasmid construct, there are two wild-type copies of the silencer in the cell that gave us the ChIP assay data. To verify whether the PCR-amplified products had any mutated silencer sequence, we cloned the PCR products into pCRII-Topo vector and randomly sequenced the inserts of 12 clones. None of those clones showed any mutation, suggesting that the mutated silencer did not bind to SLUG in vivo. On the other hand, 30% of the clones we obtained from the PCR product from input DNA template had the mutated silencer sequence, as expected.
To determine whether HDAC activity is functionally involved in the mediation of the silencer activity, TSA was added to the silencer assays (Fig. 5C). TSA inhibited the repression of BRCA2 promoter activity by the silencer in both BT-549 and MDA-MB-231 cells, whereas there was no effect on the promoter activity by the silencer sequence in presence of TSA in MDA-MB-468 cells (Fig. 5C).
DISCUSSION
Studies on BRCA2 indicate a pivotal role of this protein in the biology of most of the eukaryotic cells including mammalian cells. In human mammary epithelial cells, the expression of BRCA2 protein appears to be regulated by a series of genetic and epigenetic mechanisms that are often duplicate and complementary of one another. The present study provides evidence for yet another mechanism for the regulation of BRCA2 gene expression. During the course of our study of the transcription of the human BRCA2 gene, we have discovered an Alu-containing epigenetic silencer that negatively regulates the function of the BRCA2 gene promoter only in the resting cells and only when the cells express the zinc finger transcription factor SLUG.
We paid particular attention to the role of SLUG in BRCA2 gene expression because SLUG superfamily members bind specifically to DNA that contains the E2-box sequence 5′-CACCTG-3′ (21, 22). It is implicated as one of the determinants of tumor cell invasiveness (25, 26). Members of this superfamily of transcriptional regulators are implicated in the formation of mesoderm and neural crest, as well as in the pathological progression of epithelial tumors (21, 22). Overexpression of SLUG protein in epithelial cells leads to loss of expression of key cell-cell adhesion molecules, such as E-cadherin, claudins, and occludin (25–27). SLUG contains a highly conserved region at the C terminus of the protein containing 4–6 zinc fingers of the C2H2 type (21, 22). The zinc fingers mediate sequence-specific interactions with DNA. The N terminus of SLUG contains the evolutionarily conserved SNAG (for Snail/Gfi) domain (21, 22) that may mediate the transcriptional repression function of SLUG. It is generally believed that SLUG recruits a co-repressor, possibly CtBP-1, which in turn recruits HDAC, and that leads to gene repression (25, 26).
We previously reported differential activity of the BRCA2 gene silencer in various human breast cell lines (12). Verification of SLUG content of those cells revealed a strong direct correlation between the SLUG gene expression in a cell and the activity of the silencer in that cell. MCF-7 cells do not express SLUG, and thus, the silencer is not functional in these cells. With a laboratory stock of MCF-7 cells, we initially found activity of the silencer with those cells (12). With a fresh procurement of MCF-7 cells from ATCC, we later revealed that the silencer is indeed inactive in the MCF-7 cells. Inactivity of the silencer in the MCF-7 cells may also explain why this negative regulatory element was not identified previously by other more elaborate studies on the human BRCA2 gene promoter (9, 10) as those studies mainly used the MCF-7 cells.
One of the interesting features of the human BRCA2 silencer is the requirement of the Alu repeat for the suppression (12). Many Alu sequences are transcribed by RNA polymerase III, and this transcription is regulated by many RNA polymerase III transcription factors (20). One of these factors is an ~23-kDa Alu-co-repressor protein ACR1 (20). Although the human BRCA2 silencer has the Alu repeat, it lacks consensus B-box sequence (5′-G(A/T)TCRANNC-3′). Thus, this silencer may not be transcribed by RNA polymerase III. On the other hand, the inter-Alu sequence contains the E2-box and the overlapping ACR1 binding site. ACR1 protein may still bind to it and mediate the masking of the E2-box from being bound by SLUG repressor protein. Apart from ACR1, the polymerase II transcription factor YY1 may also bind to the same sequence of the Alu element (20, 28) and thus, has the potential to add to the desilencing mechanism.
It is noteworthy that many other silencer sequences of several human genes were found to have Alu repeats (29). They include the silencers of WT1, keratin 18, erythropoietin, θ-globin, γ-chain Fc/T cell receptor, and ε-globin genes. The mechanisms by which these silencers operate are not clear. How Alu sequences aid the function of the BRCA2 gene silencer is yet to be determined. One of the possible mechanisms could be the specific recruitment of SLUG at the E2-box by some yet to be understood modality.
Although we have evidenced here that SLUG specifically binds to the BRCA2 gene silencer to mediate the silencing effect, many other proteins have the potential for binding to the E2-box sequence. They include many proteins belonging to the class of basic helix-loop-helix transcription factors, other members of the Snail family of proteins, and the ZEB family of repressor proteins (21, 22, 30). All these E2-binders thus have the potential to bind the E2-box of the silencer and compete with SLUG.
The most straightforward model of the SLUG requirement for BRCA2 gene expression would be that SLUG binding to the silencer E2-box of the BRCA2 gene in the resting growth stage of the breast cells results in the formation of a silencing complex that represses the expression of the gene by histone deacetylation at the promoter region (Fig. 6). Although we do not have any evidence at present, proteins such as ACR1 or YY1 may sequester the E2-box in the silencer, in the dividing cells, preventing the binding of SLUG and derepressing BRCA2 gene expression. Our study does not exclude the possibilities of participation of other co-repressor proteins or HDAC isozymes in the silencing process. CtBP-1 is also shown to recruit histone methyltransferase to mediate transcriptional silencing (31). We have not yet verified that possibility for the regulation of BRCA2 gene expression. Identification of all the members of the silencing complex thus will require further purification of the complete silencing complex and the characterization of each component.
Fig. 6. Probable model for the silencing of BRCA2 gene expression at the non-dividing BT-549 cells.

In the non-dividing cells, the SLUG protein, perhaps with some specific assistance from the surrounding Alu sequences, is recruited at the E2-sequence of the silencer. It then in turn helps in recruiting CtBP-1 and HDAC-1 at the silencer. Recruited HDAC-1 deacetylates the acetylated histones at the BRCA2 promoter and thus silences the expression of the gene.
Repression of the BRCA2 gene at the resting phase of the cell is important because accumulation of the BRCA2 in the cell may potentially lead to cell death. On the other hand, desilencing of the BRCA2 gene must occur when the cell is dividing to maintain appropriate control of cell growth rate. The critical regulatory role of BRCA2 thus explains why multiple complementary mechanisms, including SLUG expression and SLUG function, are critical for the control of its expression. Immortalized breast cancer cells that do not express SLUG may have developed alternate mechanisms to regulate the expression of their BRCA2 gene at the non-dividing stage because unregulated expression of BRCA2 should be lethal in any cell. Further evaluation of the role of the Alu sequences in the recruitment and/or function of SLUG at the E2-box of the silencer may reveal a very unique mechanism of BRCA2 gene expression regulation at the epigenetic level and help us to understand the possible causes of some of the sporadic breast and ovarian cancer cases.
Acknowledgments
We thank Drs. Lynn Matrisian and J. Ochieng for the suggestions during the course of this study and Dr. Lee Limbird for reading the manuscript.
Footnotes
This work was supported by the Department of Defense Grant DAMD17-00-1-0341 and the Meharry Medical College/Vanderbilt Ingram Cancer Center Cancer Partnership Grant 1U54CA091408-010003 from NCI, National Institutes of Health (to G. C.).
The abbreviations used are: HDAC-1, histone deacetylase-1; CtBP-1, C-terminal-binding protein-1; ACR1, Alu-co-repressor 1; HMEC, human mammary epithelial cells; siRNA, small interfering RNA; Dox, doxycycline; RT, reverse transcriptase; SBP, silencer-binding protein; ChIP, chromatin immunoprecipitation; TSA, trichostatin A.
References
- 1.Turner N, Tutt A, Ashworth A. Nat Rev Cancer. 2004;4:814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 2.Scully R, Xie A. J Mammary Gland Biol Neoplasia. 2004;9:237–246. doi: 10.1023/B:JOMG.0000048771.93906.3e. [DOI] [PubMed] [Google Scholar]
- 3.Keen JC, Davidson NE. Cancer. 2003;97:825–833. doi: 10.1002/cncr.11126. [DOI] [PubMed] [Google Scholar]
- 4.Chodosh LA. J Mammary Gland Biol Neoplasia. 1998;3:389–402. doi: 10.1023/a:1018784031651. [DOI] [PubMed] [Google Scholar]
- 5.Venkitaraman AR. Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 6.Narod SA, Foulkes WD. Nat Rev Cancer. 2004;4:665–676. doi: 10.1038/nrc1431. [DOI] [PubMed] [Google Scholar]
- 7.Bertwistle D, Swift S, Marston NJ, Jackson LE, Crossland S, Crompton MR, Marshall CJ, Ashworth A. Cancer Res. 1997;57:5485–5488. [PubMed] [Google Scholar]
- 8.Su LK, Wang SC, Qi Y, Luo W, Hung MC, Lin SH. Oncogene. 1998;17:2377–2381. doi: 10.1038/sj.onc.1202162. [DOI] [PubMed] [Google Scholar]
- 9.Davis PL, Miron A, Andersen LM, Iglehart JD, Marks JR. Oncogene. 1999;18:6000–6012. doi: 10.1038/sj.onc.1202990. [DOI] [PubMed] [Google Scholar]
- 10.Wu K, Jiang SW, Thangaraju M, Wu G, Couch FJ. J Biol Chem. 2000;275:35548–35556. doi: 10.1074/jbc.M004390200. [DOI] [PubMed] [Google Scholar]
- 11.Wu K, Jiang SW, Couch FJ. J Biol Chem. 2003;278:15652–15660. doi: 10.1074/jbc.M211297200. [DOI] [PubMed] [Google Scholar]
- 12.Sharan C, Hamilton N, Parl AK, Singh PK, Chaudhuri G. Biochem Biophys Res Commun. 1999;265:285–290. doi: 10.1006/bbrc.1999.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tripathi MK, Chaudhuri G. Biochem Biophys Res Commun. 2005;328:43–48. doi: 10.1016/j.bbrc.2004.12.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spector DL, Goldman RD, Leinwand LA. Cells: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. [Google Scholar]
- 15.Hajra KM, Chen DY, Fearon ER. Cancer Res. 2002;62:1613–1618. [PubMed] [Google Scholar]
- 16.Campbell MJ, Machin D. Medical Statistics: A Commonsense Approach. 2. John Wiley & Sons, Inc; New York: 1994. [Google Scholar]
- 17.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- 18.Boyd KE, Farnham PJ. Mol Cell Biol. 1999;19:8393–8399. doi: 10.1128/mcb.19.12.8393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orlando V. Trends Biochem Sci. 2000;25:99–104. doi: 10.1016/s0968-0004(99)01535-2. [DOI] [PubMed] [Google Scholar]
- 20.Kropotov A, Sedova V, Ivanov V, Sazeeva N, Tomilin A, Krutilina R, Oei SL, Griesenbeck J, Buchlow G, Tomilin N. Eur J Biochem. 1999;260:336–346. doi: 10.1046/j.1432-1327.1999.00162.x. [DOI] [PubMed] [Google Scholar]
- 21.Hemavathy K, Ashraf SI, Ip YT. Gene (Amst) 2000;257:1–12. doi: 10.1016/s0378-1119(00)00371-1. [DOI] [PubMed] [Google Scholar]
- 22.Nieto MA. Nat Rev Mol Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 23.Jenuwein T, Allis CD. Science. 2001;293:1074–1079. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 24.Finnin MS, Donigian JR, Cohen A, Richon VM, Rifkind RA, Marks PA, Breslow R, Pavletich NP. Nature. 1999;401:188–193. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- 25.Kang Y, Massagué J. Cell. 2004;118:277–279. doi: 10.1016/j.cell.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 26.Come C, Arnoux V, Bibeau F, Savagner P. J Mammary Gland Biol Neoplasia. 2004;9:183–193. doi: 10.1023/B:JOMG.0000037161.91969.de. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kajita M, McClinic KN, Wade PA. Mol Cell Biol. 2004;24:7559–7566. doi: 10.1128/MCB.24.17.7559-7566.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Humphrey GW, Englander EW, Howard BH. Gene Expr. 1996;6:151–168. [PMC free article] [PubMed] [Google Scholar]
- 29.Hewitt SM, Fraizer GC, Saunders GF. J Biol Chem. 1995;270:17908–17912. doi: 10.1074/jbc.270.30.17908. [DOI] [PubMed] [Google Scholar]
- 30.Zwijsen A, van Grunsven LA, Bosman EA, Collart C, Nelles L, Umans L, Van de Putte T, Wuytens G, Huylebroeck D, Verschueren K. Mol Cell Endocrinol. 2001;180:13–24. doi: 10.1016/s0303-7207(01)00505-6. [DOI] [PubMed] [Google Scholar]
- 31.Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y, Shi Y. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]