Abstract
Epithelial cell transformation has been demonstrated in numerous animal models for the study of solid tumor biology. However, little evidence exists for human epithelial cell transformation without prior immortalization via genetic influences such as SV40 T-antigen, limiting our knowledge of the events that can transform naïve human epithelium. Here we describe a system developed in our lab to directly transform freshly-isolated primary human prostate epithelial cells without prior culture or immortalization. Prostate tissue is obtained from patients, and benign tissue is separated from cancer. Benign and cancer tissue are mechanically and enzymatically dissociated to single cells overnight, and immune cells and epithelial subsets are isolated based on differential expression of surface antigens. Epithelial progenitor cells are transduced with lentiviruses expressing oncogenes and combined with inductive stroma for in vivo studies. 8-16 weeks after transplantion into immune-deficient mice, the development of lesions histologically classified as benign prostate, prostatic intraepithelial neoplasia (PIN) and adenocarcinoma can be evaluated.
Keywords: Prostate cancer, Human regeneration, Human prostate, Flow cytometry, Lentivirus
INTRODUCTION
Understanding the genetic and cellular origins of human prostate cancer is a primary focus of interventional inquiry. Human prostate cancer is commonly regarded as a disease of luminal cell origin based on pathological evidence and immunohistochemical markers. Recent findings, however, complicate the picture and suggest that basal stem-like cells are a potential cell of origin1-3. The unique ability to self-renew suggests that tissue stem cells may serve as a likely origin of multiple cancers, making these cells intriguing targets for investigative research and therapeutic intervention4, 5. Therefore, delineating the epithelial hierarchy and purifying specific cell types from prostate and other tissues based on functional characteristics will help researchers to examine the cellular contexts and genetic influences undermining epithelial transformation.
Overview of the procedure
This protocol describes how to prepare, purify, and transform defined populations of primary human prostate epithelial cells and has been used recently to identify a cell of origin for human prostate cancer2. A sequential enzymatic digestion of primary tissue yields a total dissociated cell preparation containing all of the cell-types present in the original tissue, with the sole exception of red blood cells removed by a specific lysis step. We use Fluorescence Activated Cell Sorting (FACS) techniques to purify immune and epithelial cell subsets. This approach allows for the simultaneous isolation of distinct cell populations. Other methodologies to purify epithelial progenitor cells rely on differential centrifugation and/or preferential adhesion of specific cell types to extracellular matrices such as collagen6, 7. These protocols may be useful to enrich for epithelium or purify distinct subpopulations, but these methods leave other cells behind, whether they are stroma or stem cell-depleted epithelial populations. In contrast to magnetic bead separation, FACS provides precise control to purify cells that express high, intermediate, or low levels of multiple antigens. We have utilized this strategy for the purification of human prostate epithelial progenitor cells that form spheres8, tubules, and malignant structures resembling prostate cancer2 while also allowing for simultaneous isolation and discrimination of the remaining epithelial and non-epithelial subsets, including immune cells.
Purified epithelial cells are transduced with lentivirus to over-express different oncogenes, and transplanted into immune-deficient mice for regeneration of benign or malignant epithelium. This strategy is analogous to transplantation approaches used to study normal and malignant development in hematopoietic and mammary cells. The major advantage of this approach is the use of naïve adult human epithelial cells, which are the target cells for the disease. By directly transforming primary cells and transplanting them in vivo, this approach avoids potential artifacts introduced in tissue culture. While the current protocol focuses primarily on human prostate epithelium, recent literature in our lab has made use of modified versions of this protocol in both murine and human systems2, 8-11.
Model systems of prostate cancer
Several model systems are utilized for the study of prostate cancer and the specific pathways involved. Cell lines and xenografts12 represent the most wide-spread model in the field, and they provide valuable insight into fundamental pathways and mechanisms that support the survival and growth of prostate cancer cells. Since the majority of such cell lines are derived from late stage or metastatic cancer, this model system is useful for uncovering and testing biochemical mechanisms in vitro and in vivo, but is not sufficient to investigate the genetic and epigenetic alterations responsible for converting healthy tissue to a malignant state. The introduction of mouse models of prostate cancer, such as prostate specific deletion of PTEN, transgenic adenocarcinoma of the prostate (TRAMP), and c-Myc, has allowed researchers to further understand the pathways leading to disease initiation and progression in an oncogene or pathway-dependent fashion13-15. However, development of these murine models requires complex breeding strategies and multiple crosses that are time consuming, expensive and do not allow for parallel studies with human tissue. The approach described here allows researchers to introduce oncogenes, via lentivirus, into primary prostate epithelial cells for transplantation under the kidney capsule or skin of immune-deficient mice. This technique measures both the regenerative and tumorigenic capacity of isolated prostate cell-types9, 11. Although the prostate-regeneration model takes cells out of their natural microenvironment and requires organogenesis concomitant with tumorigenesis, the system is faster and cheaper than mouse breeding studies, allows for efficient testing of multiple oncogene combinations, and permits parallel studies in mouse and human tissues.
Diversity of potential applications and limitations
Elucidating the pathways important for prostate stem cell function, prostate cancer initiation, and castration-resistance is critical for moving forward. Since we can now routinely isolate basal and luminal cells from benign prostates, as well as cancer cells, we can interrogate these specific cell populations using an array of genomic and proteomic approaches, including, but not limited to, microarrays, deep sequencing, activity-based protein profiling, and kinase enrichment and mass spectrometry. However, without a human prostate-regeneration system, the functional relevance of these new pathways and targets cannot be tested. Hence, the ability to consistently introduce new genes of interest, discovered using existing genomic and proteomic technologies, into primary human benign cells will shed light on new avenues and opportunities for therapeutic development.
While we are capable of utilizing many different approaches to interrogate dissociated human prostate epithelium, we are limited by the frequency at which samples are obtained and the amount of tissue that is available for processing. This technique is designed to be utilized where prostate tissue can be readily transferred to the appropriate lab quickly for processing, such as at academic institutions attached to medical centers. If prostate tissue needs to be shipped or stored for long periods of time, the cell yield and viability may be greatly reduced.
Experimental design
In vivo regeneration
The subrenal regeneration assay, described in Box 4, is technically demanding and requires experience to perfect the procedure. Prior to using valuable human materials, we recommend practicing the technique with mouse prostate epithelium. If preferred, subcutaneous implantation of these cells may be performed, as described in Box 3. Using sorted cells, we have found that the rate of outgrowth formation is generally higher in the subcutaneous site compared to kidney capsule engraftment. The subcutaneous injection is an easier procedure and allows for monitoring of graft size through the skin. However, the reproducibility may be lower in the subcutaneous grafts due to the lack of blood supply, which is more abundant and consistent in the kidney capsule 16.
FACS machine accessibility
Sorting time should be arranged in advance at your institutional core facility to ensure that cells can be sorted on the same day that tissue is processed. Two types of controls are vital for accurate cell sorting, single color controls and fluorescence minus one (FMO). Control tubes should be set up such that cells are stained with one fluorescent antibody at a time. This will allow you to determine the bleed over for each fluorophore into each channel and set the appropriate voltage compensation. FMO is a strategy to leave one fluorescent antibody out at a time to ensure that positive staining is indeed due to that specific antibody. FMO analysis should be performed when testing an antibody or setting up the gates for the first time.
Functionality of Matrigel
Lot to lot variation exists in the functional ability of Matrigel to induce sphere formation in vitro and/or tubule formation in vivo. Testing of each specific lot prior to the experiment is crucial to avoid wasting valuable sample materials. We suggest plating approximately 5,000 or 10,000 enriched prostate basal cells in Matrigel to induce sphere formation as has been previously described10. A good lot of Matrigel will support robust sphere formation in as little as 5 days. Growth factor reduced Matrigel has been tested briefly in the murine system with no changes in effect. However, in the human system, growth factor reduced Matrigel has not been tested.
Importance of urogenital sinus mesenchyme (UGSM)
UGSM cells are derived from the developing urogenital sinus, the site of prostate gland formation. This fetal stroma creates a highly inductive environment by secreting specific growth factors important for prostate tubule formation and development. In contrast, stromal cells isolated from the adult prostate provide growth factors that are primarily involved in maintaining tissue homeostasis. In the absence of UGSM, the prostate epithelial cells are prone to form undifferentiated cord-like structures, rather than well-differentiated tubules. Mouse UGSM is sufficient to support human prostate-regeneration, however prostate development may be enhanced with the addition of species-specific factors through the use of parallel human fetal stroma. Since human fetal stroma is difficult to obtain, we are currently testing mixtures of inductive mouse UGSM with human stromal cells, such as immortalized or primary benign and carcinoma-associated fibroblasts7, to support human prostate epithelial growth and regeneration.
An alternative approach to using syringes for cell dissociation
The use of syringes with unscreened human material is very dangerous and represents a potential biohazard. To avoid the use of sharps during cell dissociation (see Step #12), the plunger from a 10 cc syringe can be used to mash the tissue onto either a 10 cm tissue culture dish or through a mesh strainer. Alternatively, the digested tissue can be dissociated by pipeting up and down using a plastic micro pipet tip. These alternate methods of dissociation reduce potential hazards but are less efficient at recovering epithelial cells.
Antibody dilutions
It is highly recommended to empirically determine the antibody concentration needed for efficient labeling of the cells. The dilutions outlined in this protocol are recommended as a starting point in this process.
MATERIALS
REAGENTS
Prostate tissue: prepared from surgery through the Pathology department, as described in Box 1.
UGSM, prepared as described in Box 2.
NOD-SCID-IL2Rγnul1 (NSG) mice Obtained from Jackson Laboratories and housed and bred under the supervision and regulations of the Division of Laboratory Animal Medicine at the University of California, Los Angeles ! CAUTION All experiments involving live rodents must conform to national and institutional standards and regulations.
DMEM (Invitrogen, cat. no. 31800-089)
Glutamine (Fisher Scientific, cat. no. BP379-100)
Penicillin-Streptomycin (Omega Scientific, cat. no. PS-20)
Collagenase, type I (Invitrogen, cat. no. 17018-029)
Dispase (Invitrogen, cat. no. 17105-041)
10x PBS (Omega Scientific, cat. no. PB-10)
Fetal bovine serum (Omega Scientific, cat. no. FB-01)
Nu Serum (BD Biosciences, cat. no. 35504) ▲CRITICAL This serum was determined to be optimal for the growth of UGSM cells.
Human recombinant Insulin (Invitrogen, cat. no. 12585-014)
Fungizone Amphotericin B (Invitrogen, cat. no. 15290-018)
RBC lysis buffer (BioLegend, cat. no. 420301)
Trypsin/EDTA 0.05% (Invitrogen, cat. no. 25300-054)
10% Trypsin (Invitrogen, cat. no. 15090046)
DNase I (Roche, cat. no. 10104159001)
PrEGM (Clonetics, cat. no. CC-3165) complete with nine supplements to be stored at -20 °C, see REAGENT SETUP
Matrigel (BD Biosciences, cat. no 354234) ▲CRITICAL Needs to be thawed at 4 °C according to manufacturer's specifications; each lot should be tested empirically as they may vary from lot to lot in their ability to support epithelial growth.
Collagen (BD Biosciences, cat. no. 354236)
1N NaOH (Sigma, cat. no. S2770) ! CAUTION Corrosive agent; handle with care and appropriate personal protective equipment (PPE).
CD49f antibodies: PE conjugated (eBiosciences, cat. no. 12-0495-83, clone GoH3, 0.2 mg ml-1), Alexa Fluor 647 conjugated (BioLegend, cat. no. 313610, clone GoH3, concentration N/A)
Trop2 antibodies: (R&D Systems, APC conjugated- cat. no. FAB650A, FITC conjugated- cat. no. FAB650F, PE conjugated- cat. no. FAB650P, all colors are clone 77220, 10 μg ml-1)
Biotin conjugated anti-CD24 (StemCell Technologies, cat. no. 10231, clone 32D12, concentration N/A)
APC-eFluor 780 conjugated anti CD45 (eBiosciences, cat. no. 47-0459-42, clone HI30, 3 μg ml-1)
PE-Cy7 conjugated anti CD8 (eBiosciences, cat. no. 25-0088-42, clone RPA-T8, 3 μg ml-1)
FITC conjugated Streptavidin (BD Biosciences, cat. no. 554060, clone N/A, 500 μg ml-1)
Testosterone pellet, 12.5 mg 90 day release (Innovative Research of America, cat. no. NA-151)
Third generation lentiviral vectors (a generous gift from Inder Verma, Salk Institute for Biological Studies, La Jolla, CA and described in more detail in Refs. 17, 18; ref. 18 also includes a protocol for preparing and tittering lentivirus.) ! CAUTION Biosafety level II+ and appropriate PPE is required for working with lentivirus. Please conform to all national and institutional regulations prior to handling lentivirus.
Lentiviral packaging vectors: pVSVg, pMDL, pRev (Invitrogen, cat. no. K4975-00)
Polybrene (Hexadimethrine bromide) (Sigma, cat. no. H9268)
Trypan blue stain 0.4% (Invitrogen, cat. no. 15250-061)
EQUIPMENT
Tungsten Carbide Scissors 15 cm (Fine Sciences Tools)
Ice bucket (Fisher)
18-, 20-, 22-G needles (Kendall)
10-cc syringes (BD Biosciences)
1-cc insulin syringes (BD Biosciences)
Syringe attachable 22 μm pore size filters (Millipore)
Nylon mesh filter, 40 and 100 μm pore size (BD Biosciences)
Reichert bright line hemacytometer (Hausser Scientific)
Alcohol and iodine prep swabs
6-0 Coated vicryl sutures (Ethicon)
Wound clip applier, clips, and remover (Fisher Scientific, cat. nos. 01-804, 01-804-5, and 01-804-15, respectively)
Cell culture centrifuge (Beckman Coulter)
Tissue culture hood approved for use of lentivirus and human cell work (SteriGuard)
Tissue culture water bath (Thermo Scientific)
CO2 incubator set to 5% or 8% CO2 and 37°C (SteriCult)
Adams™ Nutator Mixer (BD Biosciences)
FACS Aria cell sorter (Becton Dickinson or similar)
Personal protective equipment (PPE) is comprised of gloves (CURAD), an appropriate P95 or N95 Respirator (3M) and a knee length laboratory coat (Cardinal Health)
REAGENT SETUP
DMEM complete digestion solution
Add 50 ml Fetal Bovine Serum (FBS), Glutamine to a final concentration of 4 mM, Penicillin-G/Streptomycin solution (Penicillin: final concentration of 100 units ml-1, Streptomycin: final concentration of 100 μg ml-1), to 440 ml DMEM media. Pass media through 0.22 μm filter to sterilize. Store at 4 °C for up to 2 months.
Collagenase/Dispase digestion solution
Dissolve collagenase type I and Dispase in DMEM complete digestion solution to a final concentration of 1 mg ml-1 for each enzyme. Add Fungizone (final concentration of 2.5 μg ml-1). Pass the media through 0.22 μm filter to sterilize. It is highly recommended to make this solution fresh each time. However, any excess can be stored at -20 °C for up to 2 weeks.
PrEGM
PrEBM basal media is supplied with 9 pre-aliquoted supplements (BPE, INSULIN, HC, GA-1000, RETINOIC ACID, TRANSFERRIN, T3, EPINEPHRINE, rhEGF). Upon receiving shipment, PrEBM media is stored at 4 °C and supplements are stored at -20 °C. Thaw 9 frozen supplements, and add all to PrEBM basal media. Filter to sterilize. Store PrEGM in dark bottle for up to 3 weeks at 4 °C or divide into 50 ml aliquots and freeze at -20 °C for long term storage of up to 2 months.
1X RBC lysis buffer
Combine 9 ml distilled water with 1 ml 10x RBC lysis buffer. Pipet to mix. Make fresh each time.
1X PBS
Dilute 50 ml 10x PBS in 450 ml distilled water and filter sterilize. Store at 25 °C for up to 12 months.
DNase I digestion solution
Dissolve 1 mg recombinant DNase I in 50 ml DMEM complete digestion solution and filter sterilize. Make fresh each time.
Collagen
Combine 250 μl collagen, 5.8 μl 1N NaOH, 28.4 μl 10x PBS. Pipet gently to mix. Storage on ice for up to 15 minutes will ensure for proper mixing and neutralization of components. Make fresh each time.
UGSM Medium
Add 25 ml FBS (5%), 25 mL NuSerum IV (5%), Glutamine to a final concentration of 4 mM, Penicillin-G/Streptomycin solution (Penicillin: final concentration of 100 units ml-1, Streptomycin: final concentration of 100 μg ml-1), insulin to a final concentration of 25 μl ml-1 to DMEM basal media. Store at 4 °C for up to 1 month.
1% Trypsin
This is for UGSM Preparation in Box 2. Dilute the 10% Trypsin (Invitrogen) to 1% (vol/vol) in sterile 1x PBS. Prepare a fresh, small aliquot each time. Do not store.
1:4 Trypan Blue Mix
Dilute 10 ml 0.4% Trypan Blue into 30 ml 1x PBS for a total of 40 ml.
PROCEDURE
Weighing and preparing tissue for overnight enzymatic digestion ● TIMING 1 h
-
1
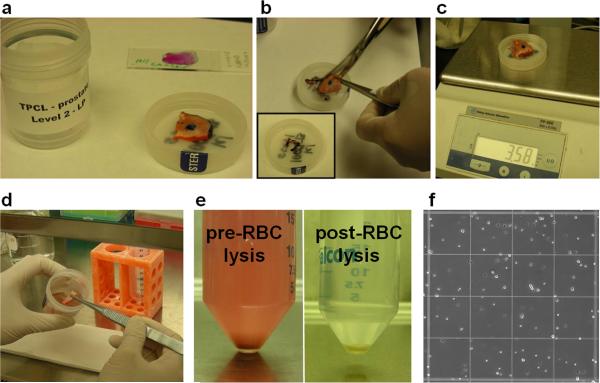
Obtain tissue samples from Pathology (Fig. 1a). Trim marked regions of tissue using scissors (Fig. 1b), and measure the weight of unmarked tissue in grams (Fig. 1c). Transfer tissue to a 100 mm × 20 mm tissue culture dish (Fig. 1d).
Figure 1. Tissue preparation to single cells.
(a) Tissue is marked up by the Pathology core laboratories. (b) Marked edges of the tissue are removed manually using scissors (inset shows trimmed edges), (c) and the remaining unmarked tissue is weighed. (d) Tissue is transferred to a tissue culture dish to be cut into small fragments. (e) After overnight digestion, samples contain red blood cells (left) that can be lysed to leave remaining cells (right). (f) Single dissociated cells can be counted by hemacytometer.
! CAUTION All work with primary human tissue needs to be done in an approved tissue culture hood with proper attire including PPE.
-
2
Use scissors to cut tissue into small chunks. Add a small amount of PBS to keep tissue together and continue to cut until chunks are small enough to pass through a 5 ml pipet. Transfer tissue chunks/PBS to 50 ml conical tube using a 5 ml pipet and wash the tissue culture dish with additional PBS as needed.
-
3
Spin down tissue at 1800 rpm (754 xg) for 5 min at 4 °C. Aspirate supernatant and resuspend pellet in 30 ml PBS. Repeat spin to wash.
-
4
Aspirate supernatant and resuspend pellet in 20-30 ml DMEM complete digestion solution. Spin down tissue at 1800 rpm (754 xg) for 5 min at 4 °C.
-
5
While centrifuging, prepare collagenase/dispase digestion solution (See reagent setup). Calculate total volume needed (10 ml per gram of tissue and up to 50 ml for 5 grams in a single 50 ml conical tube).
-
6
Aspirate supernatant from tissue in step 4 and resuspend in collagenase/dispase digestion solution. For greater than 5 grams of starting tissue, the digestion process will need to be carried out in multiple 50 ml conical tubes. ■ PAUSE POINT If samples are processed earlier in the day, then tissue can be kept on ice or at 4 °C until the end of the day in DMEM complete digestion solution, prior to overnight digestion at 37 °C in collagenase/dispase digestion solution.
Overnight enzymatic digestion of tissue ● TIMING 8-12 h
-
7
Digest tissue chunks in 37 °C incubator overnight with rocking action on an Adams™ Nutator Mixer. ▲CRITICAL STEP Length of digestion depends on size of tissue sample and timing of tissue processing. Overnight digestion has been optimized for tissue that is processed at the end of the day, for samples that weigh approximately 1-5 grams. For smaller samples, digestion time should be decreased.
Preparation of single cells ● TIMING 1-2 h
-
8
The next morning, spin down cells/digestion mix at 1800 rpm (754 xg) for 5 min at 4 °C.
? TROUBLESHOOTING
-
9
Aspirate the supernatant, wash cells 2 times in 25 ml PBS and spin down at 1800 rpm (754 xg) for 5 min at 4 °C.
-
10
Aspirate supernatant, resuspend cells in 5 ml 0.05% Trypsin/EDTA, incubate for 5 min in a 37 °C degree water bath with occasional shaking to ensure that the enzyme can access all of the tissue.
-
11
During incubation, prepare DNase I digestion solution. After 5 minutes in water bath, add 15 ml DNase I digestion solution to trypsinized cells and mix. Spin down cells at 1800 rpm (754 xg) for 5 min at 4 °C. ▲CRITICAL STEP In the absence of DNase I, the trypsinized tissue will stick together and make it very difficult to go through the syringe in step 12.
-
12
Aspirate supernatant, resuspend in 10 ml DMEM complete digestion solution. Draw cells/media up and down through 18 gauge needle attached to a 10 cc syringe up to 5 times. Repeat with 20 and 22 gauge needles if possible. ! CAUTION The use of sharps around unscreened human biological material is very dangerous. Please take extra care when using syringes in this manner. An alternative approach to using sharps is discussed in the experimental design section of the introduction.
? TROUBLESHOOTING
-
13
Filter cells/media through 100 μm cell strainer and wash the conical tube and filter with an additional 10 ml DMEM complete digestion solution. Spin down cells at 1800 rpm (754 xg) for 5 min at 4 °C.
? TROUBLESHOOTING
-
14
During spin, prepare 1x RBC lysis buffer (See reagent setup).
-
15
Aspirate supernatant and resuspend cell pellet in 5 ml 1x RBC lysis buffer. Keep cells/lysis buffer on ice for 5 minutes with occasional shaking. After 5 minutes, add 25 ml PBS to conical tube and spin down cells at 1800 rpm (754 xg) for 5 min at 4 °C. ▲CRITICAL STEP If collection of red blood cells is desired, step 15 should be avoided. In the absence of RBC lysis, cell sorting time may be significantly increased due to the large number of red blood cells (Fig. 1e).
-
16
Aspirate supernatant and resuspend cell pellet in 10 ml DMEM complete digestion solution. Filter cells through 40 μm cell strainer. Wash the tube and strainer with an additional 10 ml DMEM complete digestion solution. Spin down cells at 1800 rpm (754 xg) for 5 min at 4 °C.
-
17
Aspirate supernatant and resuspend cell pellet in 1-3 ml PrEGM complete media. Count cells by hemacytometer to determine the yield (Fig. 1f). Approximately 5 million total cells can regularly be collected per gram of starting tissue. ■ PAUSE POINT Dissociated cells can be stored on ice or at 4 °C for several hours prior to sorting, depending on the availability of the FACS machine.
? TROUBLESHOOTING
Staining cells for FACS ● TIMING 1 h
-
18
Label appropriate number of FACS tubes (1 tube for unstained cells, 1 separate tube per single fluorescent antibody control, 1 tube for “all” antibodies)
-
19
Aliquot approximately 1-5 × 104 cells in 100 μl PrEGM media per tube for unstained control and single antibody controls, used to set the compensation for sorting experiments. Add the appropriate amount of antibody as indicated in Table 1 for single antibody controls.
-
20
Prepare remaining cells in the tube labeled “all” at a concentration of no more than 1 × 107 cells per ml of PrEGM. Add appropriate amounts of antibody as indicated in Table 1 for sorted sample.
-
21
Incubate tubes on ice with a cover to block light for 30 min with occasional shaking. ▲CRITICAL STEP Conjugated fluorophores are sensitive to ambient light. Mixing is essential to ensure exposure of antibody to all cells.
-
22
Spin down cells at 1800 rpm (754 xg) for 5 min at 4 °C.
-
23
Aspirate supernatant, resuspend in 1 ml PrEGM media to wash, and spin down cells at 1800 rpm (754 xg) for 5 min at 4 °C.
-
24
If secondary antibody (FITC conjugated streptavidin) is required, resuspend cells used for single antibody control in 100 μl PrEGM, and resuspend cells used for “all” in the appropriate volume (no more than 1 × 107 cells per ml) of PrEGM. If secondary antibody is not required, resuspend single color antibody samples in 300 μl PrEGM and keep on ice.
-
25
Incubate cells with secondary antibody on ice with a cover for 15 min with occasional shaking. ▲CRITICAL STEP Conjugated fluorophores are sensitive to ambient light. Mixing is essential to ensure exposure of antibody to all cells.
-
26
Spin down cells at 1800 rpm (754 xg) for 5 min at 4 °C.
-
27
Aspirate supernatant, resuspend cells in 1 ml PrEGM media to wash, spin down cells at 1800 rpm (754 xg) for 5 min at 4 °C.
-
28
Aspirate supernatant, resuspend single antibody control samples in 300 μl PrEGM. Resuspend “all” sample in 1-2 ml PrEGM. Filter “all” sample through a 40 μm cell strainer. Wash tube and strainer with an additional 1 ml PrEGM. Combine 2-3 ml of cells/PrEGM, transfer to a new FACS tube for sorting. Keep all tubes on ice. ▲CRITICAL STEP Cells tend to clump after spinning down and can clog the fluidics of the cell sorter without filtering the sample first.
Table 1.
Antibodies used to identify and isolate distinct epithelial and immune cell subsets
| Antigen | Conjugate (Company) | Volume (μl) for Single Antibody Controls (1 × 10E4 cells) | Volume (μl) for Sorted Sample (per 1 × 10E6 cells/100 (μl) |
|---|---|---|---|
| CD49f | PE (eBiosciences) | 1 | 3 |
| Alexa647 (BioLegend) | 1 | 3 | |
| Trop2 | APC (R&D) | 3 | 10 |
| CD24 | Biotin (StemCell Technologies) | 5 | 15 |
| Streptavidin | FITC (BD) | 1 | 5 |
| CD45 | APC-eFluor780 (eBiosciences) | 1 | 5 |
| CD8 | PE-cy7 (eBiosciences) | 1 | 5 |
Isolation of distinct epithelial and non-epithelial cell subsets by FACS ● TIMING 2-4 h
-
29
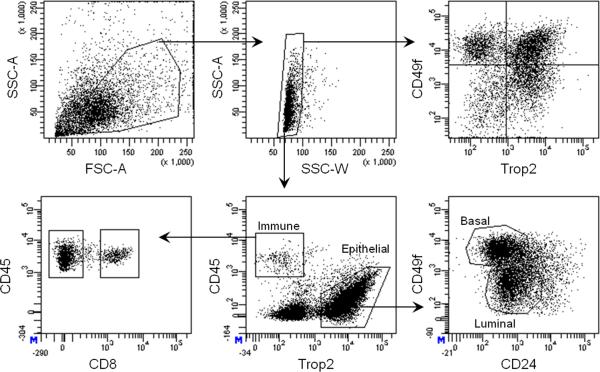
Run individual antibody controls and unstained cells on FACSAria II to set the correct compensation and voltage for sorting. Set FACS gates as indicated in Figure 2, and collect up to four distinct cell populations simultaneously. If greater than four populations are desired, switching of collection tubes between samples is required. Immune cells are marked by expression of the pan-leukocyte marker CD45 and can be subdivided into CD45+CD8+ lymphocytes and the remaining CD45+CD8- immune cells. Trop2+CD45- epithelial cells can be further subdivided into basal (Trop2+CD49fhiCD24lo/+) and luminal (Trop2+CD49floCD24+) subsets (Fig 2). Collect cells in 1 ml of 50% PrEGM/50% FBS. ! CAUTION Sorting can create an aerosol. All sorting of human cells should be done with a vacuum.
-
30
Spin down cells at 1800 rpm (754 xg) for 5 min at 4 °C. Aspirate supernatant and resuspend in 1 ml PrEGM. ■ PAUSE POINT Sorted cells can be stored on ice prior to counting and infection. For other types of analysis including protein, RNA or DNA, sorted cell pellets or lysates can be stored at -80 °C long-term.
-
31
Mix 10 μl cells/PrEGM with 10 μl 1:4 Trypan Blue Mix, and count the sorted prostate epithelial cells by hemacytometer. Viable healthy cells exclude the blue dye. This assay can be used to determine the degree of viability in a sample. Set aside a subset of sorted cells as the control sample for the transduction efficiency assay described in Box 5.
Figure 2. Gating strategy to purify basal, luminal and immune cell subsets.
FACS plots demonstrate the gating strategy used to isolate total cells based on forward scatter (FSC) and side scatter (SSC), and to exclude doublets using SSC width vs. area. Total cells can be analyzed by expression of CD49f and Trop2, as has been previously reported. A more complex sorting strategy includes separation of immune cells (CD45+) and epithelial cells (Trop2+). The CD45+ fraction is comprised of both CD8+ lymphocytes and CD8- immune cells, while the Trop2+ epithelium can be subdivided into basal (Trop2+CD49fhiCD24lo/+) and luminal (Trop2+CD49floCD24+) fractions.
Lentivirus infection ●TIMING 1 h
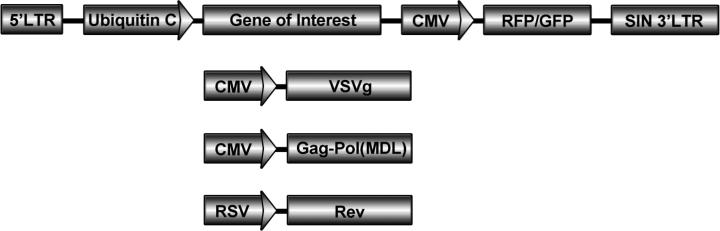
! CAUTION Lentivirus is highly infectious and the use of third generation lentiviral vectors is highly recommended (Fig. 3). All lentivirus is pseudotyped with VSV-g for efficient transduction using either mouse or human cells.
-
32
Pipet the appropriate number of cells (5 × 104- 5 × 105 per condition) into polystyrene FACS tubes depending on the number of grafts or unique genes you want to over-express. Usually a target cell number of 5 × 104 - 1 × 105 cells per graft are desirable. For multiple grafts, combining up to 5 × 105 cells in a single tube is sufficient. Total volume should be between 100 – 500 μl per tube.
-
33
Add Polybrene to each tube at a final concentration of 8 μg/ml (include the amount needed for lentivirus into the calculated total volume).
-
34
Add the appropriate high-titer lentivirus to the tube in a typical multiplicity of infection (MOI) in the range of 25-75, as determined by infection on 293T cells18. ! CAUTION All experiments involving lentivirus must conform to institutional regulations and appropriate PPE must be worn at all times.
-
35
Place the tubes at 37 °C in a sterile tissue culture incubator, designated for lentivirus only, for 1 hour making sure to mix the tubes every 15 minutes by flicking.
Figure 3. Schematic of third generation lentiviral and packaging vectors.
The lentiviral vector (top) expresses your gene of interest along with a fluorescent color marker such as GFP or RFP for titering and expression in vivo. The packaging vectors (pVSVg, pMDL, pRev) are necessary for proper assembly of the lentivirus production. Please see Tiscornia, et al. (2006) for more detailed information on this system and how to efficiently produce high titer lentivirus18.
Lentivirus spinfection, washing and preparation of grafts ● TIMING 1.5 h
-
36
After 1 hour, centrifuge the tubes at 1800 rpm (754 xg) for 1 hour at 25 °C. Wash the tubes 3 times with PBS to remove any unbound lentivirus. Set aside a subset of transduced cells as the test sample for the transduction efficiency assay described in Box 5.
-
37
Combine cells with equal numbers of cultured Urogenital Sinus Mesenchyme (UGSM) cells. See Box 1 and ref. 10 for UGSM isolation, expansion and preparation. Spin down cells at 1800 rpm (754 xg) for 5 min at 4 °C. ▲CRITICAL STEP UGSM cells are essential for supporting the regenerative capacity of primary epithelium.
-
38
Aspirate supernatant, place cell pellet on ice. To prepare Matrigel grafts, follow option A. Prepare collagen grafts according to option B. ■ PAUSE POINT Grafts can be kept in a standard tissue culture incubator at 37 °C overnight prior to surgery the following day. After 24 hours, UGSM cells start to migrate out of grafts and collagen grafts can come apart.
Option A : Preparing Matrigel grafts
-
i
Resuspend cell pellet in approximately 30 μl per graft, transfer to an eppendorf tube and keep on ice.
▲CRITICAL STEP Thaw Matrigel on ice (may take 1-2 hours) and ensure that this step is carried out on ice, as Matrigel can solidify quickly at warmer temperatures. Pipet carefully and avoid bubbles when resuspending cell pellets.
▲CRITICAL STEP To prepare multiple Matrigel grafts using cells from the same condition, aliquot 30 μl of Matrigel/cells per replicate into individual eppendorf tubes.
Option B: Preparing collagen grafts
-
i
Resuspend cell pellet in approximately 15 μl per graft.
▲CRITICAL STEP Ensure that this step is carried out on ice, as collagen can solidify quickly at warmer temperatures. Pipet carefully and avoid bubbles when resuspending cell pellets.
-
ii
Pipet 15 μl of cells/collagen mixture onto tissue culture dish and incubate 20 min at 37 °C to solidify.
-
iii
Add UGSM media to the dish to cover grafts prior to implantation into mice.
▲CRITICAL STEP If multiple collagen grafts are desired, prepare a master mix of cells/collagen and pipet 15 μl per graft onto separate wells of a 6-well tissue culture dish and place the grafts at 37 °C for 20-30 minutes to allow them to solidify. Multiple grafts of the same type can be placed into a single well.
-
39
In vivo transplantation options are described in Box 3 for subcutaneous injection and Box 4 for subrenal (kidney capsule) regeneration. Harvest grafts after 8-16 weeks in vivo and fix tissue using standard techniques for fixation. ! CAUTION All experiments involving live rodents must conform to national and institutional regulations.
? TROUBLESHOOTING
Steps 1-6, Weighing and preparing tissue for overnight enzymatic digestion: 1 h
Step 7, Overnight enzymatic digestion of tissue: 8-12 h
Steps 8-17, Preparation of single cells: 1-2 h
Steps 18-28, Staining cells for FACS: 1 h
Steps 29-31, Isolation of distinct epithelial and non-epithelial cell subsets by FACS: 2-4 h
Steps 32-35, Lentivirus infection: 1 h
Steps 36-39, Lentivirus spinfection, washing and preparation of grafts: 1.5 h
TROUBLESHOOTING
Troubleshooting advice is provided in Table 2.
Table 2.
Troubleshooting Table
| Step | Problem | Possible Reason | Solution |
|---|---|---|---|
| 8 | Incomplete pelleting of entire sample. | Residual fat tissue is present and will not spin down. | Increase spin speed and/or time to pellet cells. Addition of FBS has been shown to increase pelleting. |
| 12 | Sample will not easily be drawn through the syringe. | Cells can be clumpy and/or stringy due to incomplete digestion or dead cell debris clumping together. | For clumping, add an additional 5 minute digest in trypsin. For stringy cell debris, incubate the cells with DNase I solution for longer time points, and pipet/plunge with force against the bottom of a tissue culture dish to break up into smaller pieces. |
| 13 | Sample does not filter easily. | The amount of tissue exceeds the capacity of the filter and blocks the flow of sample through it. | Use multiple filters or a filter with a larger surface area. Alternatively, flushing the sample through the filter at an increased rate will reduce the amount of filters needed. |
| 17 | Low cell yield. | Increased number of dead cells due to lengthy digest of a small sample. | Reduce the length of collagenase/dispase digestion with smaller samples. See step 7 for more details. |
| 39 | Poor regeneration | Disparity in health of patient cells, supportive nature of UGSM due to passage, and lot to lot variation of Matrigel. | Do not use patient cells for in vivo experiments if trypan blue staining post sort is high. Keep UGSM passage low. Ensure that Matrigel can support cell growth in vitro before using in vivo. |
ANTICIPATED RESULTS
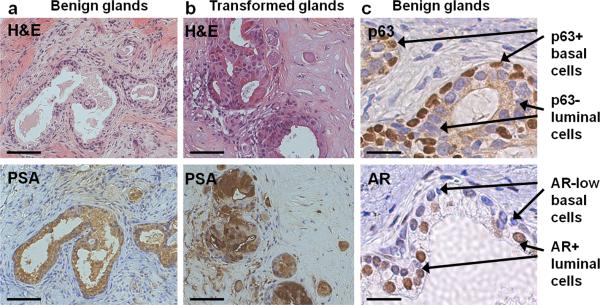
Normal prostate regeneration (Fig. 4a) or transformation (Fig. 4b) can be analyzed by staining paraffin-embedded tissue with Haematoxylin & Eosin (H&E) and human prostate luminal cell markers such as prostate-specific antigen (PSA). Normal-regenerated tubules (Fig. 4c) contain distinct layers of basal cells (p63+, AR-low) and luminal cells (p63-, AR+). Prostate tubule regeneration is a property of basal stem-like cells, purified based on the antigenic profile CD45-Trop2+CD49fhi. Total prostate cells or total epithelial cells can also be used as the source of regenerative cells, however purifying the basal fraction allows for the greatest regenerative capacity. As few as 5000 basal cells are sufficient to regenerate prostatic tubules. We have reported that introduction of two (AKT, ERG) or three (AKT, ERG, AR) oncogenes can transform cells from the basal fraction2. While basal cells are an efficient target cell for transformation, additional oncogenic influences may be capable of transforming other cell populations. We have previously demonstrated that both basal and luminal cells can be transduced with lentivirus, and that the resulting structures retain expression of both fluorescent markers and transgenes2. To ensure that target cells are infected with lentivirus, a subset of infected cells and control non-transduced cells can be left in culture for several days to look for evidence of fluorescent marker and transgene expression, as indicated in Box 5 in the transduction efficiency assay. A typical experiment will yield approximately 30-40% transduced cells.
Box 1. Preparation of tissue from robotic radical prostatectomy surgery and Pathology.
Following Institutional Review Board Approval, patients scheduled to undergo radical prostatectomy [robot-assisted laparoscopic radical prostatectomy (RALP) or open radical retropubic prostatectomy (RRP)] are consented for research participation to collect blood and prostate tissue. After an overnight fast, patients are brought to the operating room where general anesthesia is induced and a standard RALP or RRP is performed. The Pathology department is contacted just prior to removal of the specimen to facilitate immediate transportation of fresh tissue to Pathology for subsequent analyses. ▲CRITICAL STEP While every attempt is made to standardize the surgical procedure, it should be noted that differences in surgeon, surgical technique, intra-operative variables, and operative time may all affect the time at which the specimen is sent for analysis. As a result, tissue hypoxia times may vary considerably between specimens.
The prostate is transported to surgical pathology at room temperature (25 °C) as soon as it is removed from the patient. A trained and licensed pathology assistant weighs and measures the prostate, inks its surface with Indian ink (to assess the status of surgical margins on histologic sections) and slices the prostate from base to apex. Under an IRB-approved protocol, slices 2 and 4 of the prostate are used for research while the rest is entirely submitted for pathologic diagnosis. Slices 2 and 4 are divided into 4 quadrants and additional ink of different colors is used for orientation indicating right vs. left, anterior vs. posterior. The procured tissue is separated into top and bottom portions with a knife, keeping enough tissue on the top for frozen section diagnosis while preserving as much fresh tissue in the bottom as possible. Tissue from the top is snap-frozen and a frozen section slide is prepared and stained with a hematoxylin and eosin (H&E)-based protocol. The pathologist examines the frozen section slides under the microscope and circles the cancer and benign areas. The fresh tissue (bottom) is then matched with the frozen section slides and the cancer and benign areas are separated manually. Care is taken to ensure that benign regions are free of cancer, however cancer regions will regularly contain surrounding benign areas.
Box 2. Urogenital Sinus Mesenchyme (UGSM) Preparation.
UGSM cells are essential for proper in vivo regeneration. For the sake of completeness and ease of implementing the procedure, the information provided here has been reproduced and adapted from a prior protocol where we described the preparation of UGSM cells10.
Set up matings for timed pregnancies. Sacrifice pregnant female mice at E16 (embryonic day 16 of pregnancy.)
Take the uterus with the embryos, and move to a 10 cm dish containing DMEM complete digestion media. Cut the uterus laterally, separate embryos from the placenta, and place in a fresh dish containing DMEM complete digestion media.
Cut embryos in half, below the liver. Place the bottom half of the embryos into a new dish containing sterile 1x PBS. Place the bottom half of the embryos in a supine position and cut the abdomen open while holding the hind legs apart with forceps.
The urogenital sinus is connected to the bladder. As in the adult, the urogenital sinus could be removed intact by gently pulling up on the bladder. Dissect the pelvic UGS, clean off the attached tubular structures, and cut off the bladder.
Place the pelvic UGS onto a concave glass slide containing 250 μl DMEM complete digestion media. When each UGS has been collected, wash all tissues 3 times with 1x PBS. Aspirate the last wash of PBS carefully, and add 1 ml 1% Trypsin. Keep the plate in 4 °C for 90 minutes to allow for digestion.
Carefully remove the Trypsin with a pipette, and add DMEM complete digestion media. Carefully pipette the media off, and add 1 ml DMEM complete digestion media containing 500U DNase I. Let sit for 5 minutes.
Wash the UGS three more times with fresh DMEM complete digestion media.
After the third wash, take 2 28-gauge needles and separate the mesenchyme away from the epithelium. The epithelium can be identified as the opaque cylinder shaped object inside the more translucent and vascular mesenchyme.
Collect all of the mesenchyme fragments into a 15 ml Falcon tube containing 10 ml Collagenase/Dispase digestion solution. Digest at 37 °C for 2 hours with rocking action on an Adams™ Nutator Mixer.
After digestion, filter the cells through a nylon mesh filter with 40 μm pore size, and wash the filter with 10 ml DMEM complete digestion media. Spin down cells at 1300 rpm (400 xg) at 25 °C for 5 minutes. Aspirate media, resuspend cells in 10 ml UGSM Media. Plate in a 10 cm tissue culture dish.
Culture for 5–7 days, and monitor cell growth. Passage and expand the UGSM cells when they get to 80% confluency. ■ PAUSE POINT UGSM cells can be frozen down and stored at −80 °C or in liquid nitrogen for up to 6 months. Do not passage UGSM cells more than 5 times, as they lose their inductive capacity in later passages.
Box 3. Subcutaneous injection.
For the sake of completeness and ease of implementing the procedure, the information provided here has been reproduced and adapted from a prior protocol where we described subcutaneous injection of primary epithelial cell grafts for in vivo regeneration10.
Anesthetize a male immune-deficient mouse with the appropriate anesthetic such as Ketamine/Xylazine or Isofluorine gas approved by national and institutional regulations. Once the animal is anesthetized, place the mouse prone and shave the back of the mouse. Sterilize the target injection site by alternating iodine and alcohol prep swabs 3 times.
Immediately before injection, gently pipet to mix the 30 μl cell mixture in eppendorf tube on ice. Draw up the mixture into an insulin syringe, making sure to avoid bubbles.
Using a clean pair of forceps, pull up on the freshly shaved and cleaned skin of the mouse, creating a tent between the skin and musculature of the flank. Inject the mixture, and pull out the needle slowly. Hold onto the skin with the forceps for a minute to make sure the Matrigel has settled.
Implant a testosterone pellet subcutaneously in mice to ensure an environment with excess androgen. This step requires making a small incision through the outer layer of skin, implanting the pellet, and using metal clips to staple the wound in the skin.
Monitor and medicate the mice in accordance to national and institution animal care protocols. Remove staples 7 days after procedure.
Box 4. Subrenal regeneration assay.
For the sake of completeness and ease of implementing the procedure, the information provided here has been reproduced and adapted from a prior protocol where we described implanting primary epithelial cell grafts under the kidney capsule for in vivo regeneration10.
Anesthetize a male immune-deficient mouse with the appropriate anesthetic suc as Ketamine/Xylazine or Isofluorine gas approved by national and institutional regulations. Once the animal is anesthetized, place the mouse prone, and shave a 3cm × 3cm area in the mid back region. Sterilize the surgical area by alternating iodine and alcohol prep swabs 3 times.
Cut a small incision in the skin around the middle of the back, approximately 1cm lateral to the spine. Cut a second small hole in the peritoneum exposed.
Hold the peritoneum open using forceps, and search for the kidney. Once located, use the small fat pad located at the tip of the kidney to pull it to the surface, and out above the skin.
Gently grab the thin membrane on the outside of the kidney with two forceps, and tear a small hole. Lift the edge of the capsule; push the collagen graft underneath the membrane. 1 to 4 collagen grafts can be inserted per kidney capsule depending on the size of the kidney. ▲CRITICAL STEP Make sure the collagen plug is securely underneath the kidney capsule.
Grab the peritoneum with forceps, and gently allow the kidney to slip back into the body. Make sure the graft doesn't slip out. Suture the peritoneum together, careful not to puncture any organs. Insert a testosterone pellet subcutaneously. Finally, use metal clips to staple the skin closed.
Monitor and medicate the mice in accordance to national and institutional animal care protocols. Remove staples 7 days after surgery.
Box 5. Transduction efficiency assay.
Take a small aliquot of primary prostate epithelial cells that have been infected with lentivirus and washed three times (from step 36), and an aliquot of cells that have not been exposed to lentivirus as a control (from step 31). Approximately 1 × 104 - 1 × 105 cells can be evaluated in this assay. Keep transduced and control cells separate at all times. Transfer cells into an eppendorf tube in 100 μl PrEGM. Add 100 μl of pre-thawed Matrigel and mix gently by pipetting up and down.
Plate 200 μl mixture into the center of a single well in a 12-well tissue culture dish. Transfer the dish to a tissue culture incubator set at 37 °C and 5-8% CO2 to allow mixture to solidify. This step takes approximately 30-45 minutes.
When the mixture has solidified, add 1 ml PrEGM to each well.
Return dish to incubator and allow cells to grow and take up lentivirus for 72 hours until the fluorescent color marker from the lentivirus is expressed at detectable levels in transduced cells.
Remove media from each well taking care not to disturb the Matrigel. Add 1 ml Collagenase/Dispase Digestion Solution to each well and return dish to the incubator for 1 hour.
At this point, Matrigel will be disrupted and cells should be floating in each well. Collect cells into a FACS tube. Add additional PrEGM to the tube. Centrifuge the tubes at 1800 rpm (754 xg) for 5 minutes at room temperature.
Resuspend cells in 300 μl PrEGM.
Run samples on flow cytometer. Use the control tube to set the positive fluorescence gate. 0% of control cells should appear in the positive gate. Run the transduced cell samples. The percentage of cells in the positive gate represents the percentage of cells expressing detectable levels of fluorescence from the lentiviral cassette.
Figure 4. Regeneration of benign and transformed human prostate glands in vivo.
(a) Basal cells without genetic manipulation generate phenotypically benign glands with expression of prostate-specific antigen (PSA) indicating differentiation to the luminal lineage. Scale bars, 100 μm. (b) Basal cells manipulated to express selected oncogenes can generate malignant lesions, characterized by increased staining for PSA and loss of basal cells. Scale bars, 100 μm. (c) High-power images of benign tissue stained for the transcription factors p63 and AR reveal the presence of distinct layers of p63+ AR-low basal cells and p63- AR+ luminal cells. Scale bars, 25 μm.
ACKNOWLEDGEMENTS
We thank members of the Witte lab for helpful comments and testing of the protocol. A.S.G. is supported by the Warsaw Family Research Fellowship and a Dissertation Year Fellowship from the Graduate Division at UCLA. J.M.D. is supported by the UCLA Tumor Biology Program (USHHS Ruth L. Kirschstein Institutional NRSA #T32 CA009056). D.B. is supported by a Postgraduate Scholarship from the Natural Sciences and Engineering Research Council of Canada. J.H. is supported by the American Cancer Society, the Department of Defense (DOD) Prostate Cancer Research Program, and the UCLA SPORE in Prostate Cancer (principal investigator, R. Reiter). J.H. and O.N.W. are supported by a Prostate Cancer Foundation Challenge Award. O.N.W. is an investigator of the Howard Hughes Medical Institute.
Footnotes
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
REFERENCES
- 1.Lawson DA, et al. Basal epithelial stem cells are efficient targets for prostate cancer initiation. Proc Natl Acad Sci U S A. 107:2610–5. doi: 10.1073/pnas.0913873107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldstein AS, Huang J, Guo C, Garraway IP, Witte ON. Identification of a cell of origin for human prostate cancer. Science. 2010;329:568–71. doi: 10.1126/science.1189992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mulholland DJ, et al. Lin-Sca-1+CD49fhigh stem/progenitors are tumorinitiating cells in the Pten-null prostate cancer model. Cancer Res. 2009;69:8555–62. doi: 10.1158/0008-5472.CAN-08-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker N, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–11. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 5.Vries RG, Huch M, Clevers H. Stem cells and cancer of the stomach and intestine. Mol Oncol. 2010;4:373–84. doi: 10.1016/j.molonc.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins AT, Habib FK, Maitland NJ, Neal DE. Identification and isolation of human prostate epithelial stem cells based on alpha(2)beta(1)-integrin expression. J Cell Sci. 2001;114:3865–72. doi: 10.1242/jcs.114.21.3865. [DOI] [PubMed] [Google Scholar]
- 7.Olumi AF, et al. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res. 1999;59:5002–11. doi: 10.1186/bcr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldstein AS, et al. Trop2 identifies a subpopulation of murine and human prostate basal cells with stem cell characteristics. Proc Natl Acad Sci U S A. 2008;105:20882–7. doi: 10.1073/pnas.0811411106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xin L, Ide H, Kim Y, Dubey P, Witte ON. In vivo regeneration of murine prostate from dissociated cell populations of postnatal epithelia and urogenital sinus mesenchyme. Proc Natl Acad Sci U S A. 2003;100(Suppl 1):11896–903. doi: 10.1073/pnas.1734139100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lukacs RU, Goldstein AS, Lawson DA, Cheng D, Witte ON. Isolation, cultivation and characterization of adult murine prostate stem cells. Nat Protoc. 2010;5:702–13. doi: 10.1038/nprot.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawson DA, et al. Basal epithelial stem cells are efficient targets for prostate cancer initiation. Proc Natl Acad Sci U S A. 2010;107:2610–5. doi: 10.1073/pnas.0913873107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein KA, et al. Progression of metastatic human prostate cancer to androgen independence in immunodeficient SCID mice. Nat Med. 1997;3:402–8. doi: 10.1038/nm0497-402. [DOI] [PubMed] [Google Scholar]
- 13.Wang S, et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell. 2003;4:209–21. doi: 10.1016/s1535-6108(03)00215-0. [DOI] [PubMed] [Google Scholar]
- 14.Ellwood-Yen K, et al. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 2003;4:223–38. doi: 10.1016/s1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 15.Greenberg NM, et al. Prostate cancer in a transgenic mouse. Proc Natl Acad Sci U S A. 1995;92:3439–43. doi: 10.1073/pnas.92.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Priolo C, et al. Establishment and genomic characterization of mouse xenografts of human primary prostate tumors. Am J Pathol. 2010;176:1901–13. doi: 10.2353/ajpath.2010.090873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLaughlin J, et al. Sustained suppression of Bcr-Abl-driven lymphoid leukemia by microRNA mimics. Proc Natl Acad Sci U S A. 2007;104:20501–6. doi: 10.1073/pnas.0710532105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nat Protoc. 2006;1:241–5. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]