Abstract
Fibroblast growth factors (Fgfs) encode small signaling proteins that help regulate embryo patterning. Fgfs fall into seven families, including FgfD. Non-vertebrate chordates have a single FgfD gene; mammals have three (Fgf8, Fgf17, and Fgf18); and teleosts have six (fgf8a, fgf8b, fgf17, fgf18a, fgf18b, and fgf24). What are the evolutionary processes that led to the structural duplication and functional diversification of FgfD genes during vertebrate phylogeny? To study this question, we investigated conserved syntenies, patterns of gene expression, and the distribution of conserved non-coding elements in FgfD genes of stickleback and zebrafish, and compared them to data from cephalochordates, urochordates, and mammals. Genomic analysis suggests that Fgf8, Fgf17, Fgf18 and Fgf24 arose in two rounds of whole genome duplication at the base of the vertebrate radiation; that fgf8 and fgf18 duplications occurred at the base of the teleost radiation, and that Fgf24 is an ohnolog that was lost in the mammalian lineage. Expression analysis suggests that ancestral subfunctions partitioned between gene duplicates and points to the evolution of novel expression domains. Analysis of conserved non-coding elements (CNEs), at least some of which are candidate regulatory elements, suggests that ancestral CNEs partitioned between gene duplicates. These results help explain the evolutionary pathways by which the developmentally important family of FgfD molecules arose and the deduced principles that guided FgfD evolution are likely applicable to the evolution of developmental regulation in many vertebrate multigene families.
Keywords: Fgf8, fgf8a, fgf8b, Fgf17, Fgf18, fgf18a, fgf18b, fgf18l, genome duplication, ohnolog, subfunctionalization, gene family evolution
INTRODUCTION
The development of body form and the differentiation of cells depend on a variety of processes, including cell adhesion accomplished by cell adhesion proteins (e.g. cadherins, integrins, selectins), cell communication performed by extracellular signaling proteins (e.g. Notch, Hedgehog, Wnt, Tgf-β, Fgf), and differential gene expression controlled by transcription factors (e.g. homeobox, bHLH, C2H2-zinc finger, bZIP). Families of structurally related genes encode these proteins and constitute an ancestral toolkit for the construction of animal embryos. Recent genomic investigations reveal a surprisingly complete gene set already present in the last common ancestor of all metazoans (Technau et al., ‘05, Guder et al., ‘06, Nichols et al., ‘06, Kasbauer et al., ‘07, Matus et al., ‘07), but this ancestral toolkit diversified by gene duplication and gene deletion in the great lineages of animal life.
Stem chordates appear to have inherited a subset of toolkit genes present in stem metazoans, and these genes amplified into several members of each subfamily, many in two rounds of whole genome duplication (R1 and R2) that appear to have occurred at the base of the vertebrate radiation before the divergence of chondrichthyans and osteichthyans about 530 million years ago (Kumar and Hedges, ‘98, Kortschak et al., ‘01, Ornitz and Itoh, ‘01, Lundin et al., ‘03, Dehal and Boore, ‘05, Garcia-Fernandez, ‘05, Bourlat et al., ‘06, Delsuc et al., ‘06, Guder et al., ‘06, Jacob and Lum, ‘07, Kitisin et al., ‘07, Sundstrom et al., ‘08, Yu et al., ‘08, Lynch and Wagner, ‘09). Genes derived from a whole genome duplication event (WGD) are called ‘ohnologs’ to emphasize their special characters with respect to genes duplicated by other mechanisms, such as tandem gene duplication, unequal crossing-over, or retrotransposition (Wolfe, ‘00, Postlethwait, ‘07). An example is the family of Hox clusters, which initially amplified by tandem duplication into a gene cluster that then replicated in vertebrates into four clusters that appear to be related to each other in the relationship ((A,B)(C,D)), as would be expected by the 2R model (Meulemans and Bronner-Fraser, ‘07, Amemiya et al., ‘08).
The widespread occurrence of multigene families and subfamilies raises two major questions regarding the evolution of developmental mechanisms: What are the processes by which individual members of gene families evolve their specialized roles in development? And, to what extent does the origin of individual family members by gene duplication contribute to morphological innovation and lineage divergence?
A classic multigene family essential for animal development, the fibroblast growth factor (Fgf) gene family, encodes small signaling proteins deployed in many aspects of embryo patterning (Ornitz and Itoh, ‘01). Human and mouse genomes have 22 Fgf genes grouped into seven subfamilies, each containing two to four members (FgfA (1/2), FgfB (3/7/10/22), FgfC (4/5/6), FgfD (8/17/18), FgfE (9/16/20), FgfF (11/12/13/14) and FgfG (19/21/23)) (Itoh and Ornitz, ‘04, Popovici et al., ‘05). The Fgf gene complement of the urochordate Ciona intestinalis, whose lineage diverged from the vertebrate lineage before the R2 events (Oda et al., ‘02, Delsuc et al., ‘06, Vienne and Pontarotti, ‘06, Wada et al., ‘06, Delsuc et al., ‘08), helps elucidate the pattern of diversification of mammalian Fgf subfamilies (Satou et al., ‘02). Based on strong evidence from sequence similarities, two of the six C. intestinalis Fgf genes are orthologs of FgfD and FgfF subfamilies, respectively, and moderate support, based on conserved protein signature motifs and conserved genomic syntenies, suggests that three other genes are orthologs of the FgfB, FgfC, and FgfE subfamilies (Satou, Imai and Satoh, ‘02, Popovici et al., ‘05). It is not clear whether Ci-FgfL is orthologous to the FgfA subfamily or whether it is unique to C. intestinalis.
Consider the origin of the FgfB, FgfC, and FgfG subfamilies. A representative of FgfG appears to be missing from Ciona and thus either arose by gene duplication in vertebrates or was secondarily lost in the urochordate lineage. Because FGF3, FGF4, and FGF19 of the FgfB, C, and G subfamilies are contiguous in a cluster in human chromosome 11q13.3, these three genes likely arose by tandem duplication. Those tandem duplications must have occurred before R1 because FGF6 and FGF23 (FgfC and FgfG subfamilies) are nearest neighbors in chromosome 12p13.32 (the other FgfB member was apparently lost from this chromosome), and FGF21 and FGF22 (FgfB and FgfG) are linked on human chromosome 19 (Hsa19) (the FgfC member was apparently deleted from this chromosome). This genomic pattern suggests the model that an ancient FgfB/C/G gene tandemly duplicated to FgfB and FgfC/G before the divergence of urochordates and vertebrates and that subsequently, FgfC/G duplicated to FgfC and FgfG before R1; after R2, chromosome rearrangements and gene loss led to the current human genomic arrangement.
Unexpectedly, the cnidarian Nematostella vectensis has at least 13 Fgf genes, twice as many as Ciona and five or six times more than Drosophila melanogaster and Caenorhabditis elegans (Matus et al., ‘07). Phylogenetic analysis suggests that nine Nv-Fgfs may be species-specific with no clear relationship to vertebrate Fgfs, while four Nv-Fgfs fall within the FgfD subfamily (Matus et al., ‘07).
In mammals, the FgfD subfamily includes Fgf8, Fgf17 and Fgf18 (Ornitz and Itoh, ‘01) and in zebrafish, fgf24 (Itoh and Ornitz, ‘04, Popovici et al., ‘05). The vertebrate FgfD phylogeny suggests either the hypothesis that Fgf24 was lost in the tetrapod lineage (Jovelin et al., ‘07), or the hypothesis that fgf24 was an innovation in teleosts. FgfD members play critical roles in mesoderm and neuronal induction across large phylogenetic distances. For instance Fgf8, Fgf17 and Fgf18 are expressed at the midbrain-hindbrain border (MHB) (Sato et al., ‘04). FGF18 is required for skeletal development (Ohuchi et al., ‘00, Liu et al., ‘02, Ohbayashi et al., ‘02), and Fgf8 plays an important role in MHB and limb bud development (Lewandoski et al., ‘00, Moon and Capecchi, ‘00, Sun et al., ‘02, Boulet et al., ‘04). Remarkably, FGF signaling in cnidarians may be important during gastrulation and neural induction as well (Matus et al., ‘07).
Until recently, the comparison of Fgf gene content between mammals and zebrafish did not reflect the R3 hypothesis, a whole-genome duplication occurring in the teleost lineage after the teleosts diverged from other ray-fin fish (Sidow, ‘96, Amores et al., ‘98, Postlethwait et al., ‘98, Wittbrodt et al., ‘98, Meyer and Schartl, ‘99, Christoffels et al., ‘04, Hoegg et al., ‘04, Naruse et al., ‘04, Meyer and Van de Peer, ‘05, Crow et al., ‘06). Our previous phylogenetic analysis of the vertebrate FgfD subfamily showed that fgf8 and fgf18 were duplicated early in the teleost lineage and provided evidence for a diversification compatible with the ray-fin fish genome duplication (Jovelin et al., ‘07). Duplicates of human fgf6, fgf10 and fgf20 genes have also recently been identified in zebrafish (Itoh and Konishi, ‘07).
Because of the central role of FGF signaling in development, mutations in Fgf genes and their receptors cause hereditary human diseases (Itoh, ‘07). An understanding of the precise evolutionary relationships among Fgf genes and insights into the mechanisms of their functional diversification are required to fully connect teleost research to human biology. For instance, in mouse, Fgf8 is required for heart development (Abu-Issa et al., ‘02, Frank et al., ‘02, Ilagan et al., ‘06). In zebrafish, fgf8a is required for the expression of cardiac genes (Reifers et al., ‘00b), but fgf8b is not expressed in the heart; conversely, in stickleback, fgf8b but not fgf8a is expressed in the heart (Jovelin et al., ‘07). Following the ray-fin fish genome duplication, independent evolution of regulatory elements in lineages leading to zebrafish and stickleback led to the differential partitioning of fgf8 subfunctions in heart development between paralogs so that orthologous genes in the two species have come to express different functions (Jovelin et al., ‘07).
In this study, we investigated how the developmental roles of FgfD subfamily members evolved as these genes originated from R1 to R3. First, we analyzed the conservation of syntenies among chordate FgfD genes to discern evolutionary relationships ambiguous in phylogenies (Satou, Imai and Satoh, ‘02, Itoh and Ornitz, ‘04, Popovici et al., ‘05, Jovelin et al., ‘07). Second, we used in situ hybridization in stickleback and zebrafish, whose lineages diverged early in teleost phylogeny, about 300 million years ago (Hedges, ‘02), to examine how functions of duplicate genes evolve in separate lineages. Third, we analyzed the conservation of non-coding regions in the vicinity of FgfD genes in the context of gene phylogeny and gene expression to learn how divergence in regulatory elements in the face of differential gene loss could affect functional divergence of surviving ohnologs.
Results confirmed the presence of an Fgf24 gene at the base of gnathostome vertebrates, the partitioning of functions between Fgf24 and Fgf8, and revealed the origin of teleost fgfD paralogs. Expression analyses identified shared ancestral functions and newly evolved functions in various members of the FgfD group, and exploration of conserved non-coding elements gave strong support for a pattern of functional divergence through subfunctionalization. The general pattern of gene duplication and functional divergence that this study reveals is likely to apply to many other vertebrate multiple gene families.
Materials and Methods
Conserved synteny
Genome sequences were investigated using the Ensembl databases (http://www.ensembl.org/index.html) for zebrafish Zv7, human NCBI 35, mouse NCBI m36, and Ciona intestinalis version 2.0. Orthologies were determined by best reciprocal BLAST hit (RBH) analysis (Wall et al., ‘03) automated in a genome-wide scale in our Synteny Database genomic analysis software (Catchen et al., ‘09). From the elephant shark genomic database we isolated the following FgfD gene fragments: Fgf8: exon2 AAVX01442442; Fgf17: no fragments identified; Fgf18: exon2 AAVX01015792 and AAVX01460739, exon3 AAVX01088747, and exon4 AAVX01419960; Fgf24: exon2 AAVX01560519 and AAVX01583584, exon3 AAVX01466056 and AAVX01521786, and exon4 AAVX01108229. The absence of Fgf17 is not significant due to the low (1.4x) coverage of the shark genome (Venkatesh et al., ‘07).
Gene expression
In situ hybridization probes were made by in vitro transcription of linearized clones and labeled with digoxigenin-UTP. Stickleback probes were synthesized from TOPO/Not-1 linearized clones using T3 RNA polymerase. Zebrafish fgf8a, fgf8b, fgf17, and fgf24 and stickleback fgf8a and fgf8b probes were as described (Reifers et al., ‘00a, Draper et al., ‘03, Cao et al., ‘04, Jovelin et al., ‘07). Additional probes were: zebrafish fgf18, Fgf18+73 GTGTTTGGGGTGGACGGTGTGAAT and Fgf18-578 TTTTTGCTCCGCTTGCTGACTGTAG (NM_001013264) covering the coding region; zebrafish fgf18l, Fgf18L+1 ATGCGGTCCGTCCTGTGGTCT and Fgf18L-620 ATGGCAGTGGCGGAGGAGAGG (NM_001012379) covering the coding region; stickleback fgf18, Gacfgf18+95 TCAGCGTGCACGTGGAGAACC and Gacfgf18-585 GCTTGCCCCGCTTGCTGACG (ENSGACT00000023719) covering most of the coding region but missing some of the 3′ portion of the gene; stickleback fgf17ex3F CAACGGCCGGAGCAGGGATT and fgf17-3′R TTCTTTACTCTACTTCAGT (ENSGACG00000003432) covering 442bp of the last exon and 3′ UTR; stickleback fgf24-ex1F TTACATCGAGAACCACAC and fgf24-3′R GCTGTCCCGACGTTCCGT (ENSGACG00000016697) covering 552bp of coding sequence. Embryos were fixed in 4% paraformaldehyde at 4°C for at least 2 days an then dechorionated by hand. In situ hybridizations were performed as described (Yan et al., ‘02). Stickleback embryos were staged relative to the zebrafish staging series (Kimmel et al., ‘95), which showed that at 20°C, stickleback develop about 2.5 times slower than zebrafish does at 28.5°C (Cresko et al., ‘03). The University of Oregon IACUC approved protocols for this study.
Analysis of Conserved Non-coding Elements
Genomic sequences including the immediate neighbors of each Fgf gene were obtained from Ensembl release 47 (Hubbard et al., ‘07) for human and teleosts (zebrafish, and the four percomorph fish: stickleback, medaka, fugu, and green-spotted pufferfish). Some neighbors flanking medaka fgf24 and fugu fgf17 were unavailable due to short scaffolds, and a flanking neighbor of zebrafish fgf18 was too distant to be included in the analysis. Sequences were aligned using the program LAGAN (Brudno et al., ‘03) and conserved non-coding elements (CNEs) were identified using the VISTA server (Mayor et al., ‘00, Frazer et al., ‘04). Repeated elements in the reference sequence were masked using human and fugu masks.
RESULTS
Vertebrate FgfD genes originated in two rounds of duplication
To learn the origins of vertebrate FgfD genes, we investigated conserved syntenies among chordate genomes, beginning with non-vertebrate chordates.
The human genome has four segments corresponding to the FgfD region of non-vertebrate chordates
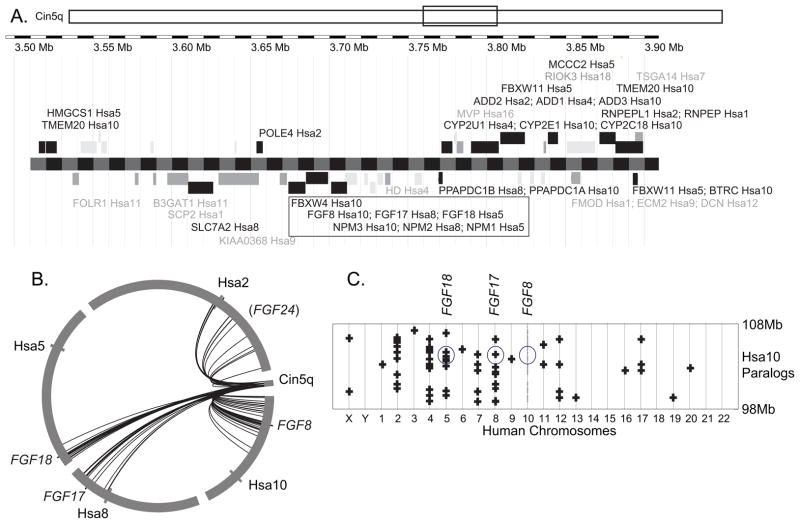
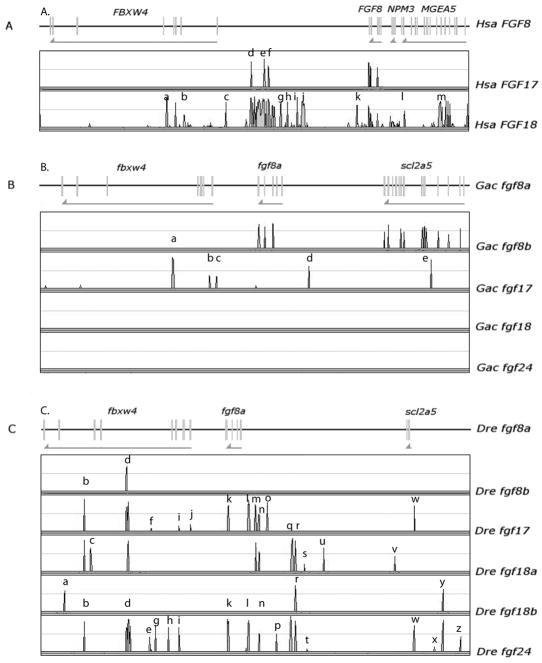
Sequence similarities between the C. intestinalis FgfD gene Ci-Fgf8/17/18 (NP_001027648, ENSCING00000009302) and vertebrate FgfD members suggest that the FgfD subfamily originated from a single gene already present in stem olfactores, the last common ancestor of urochordates and vertebrates (Satou, Imai and Satoh, ‘02, Popovici et al., ‘05). The R2 hypothesis predicts that four co-orthologs of the prototypic FgfD gene should be present in four paralogous chromosome segments in vertebrate genomes in the absence of chromosomal rearrangements or gene losses. To test this prediction, we examined the conservation of syntenies to compare the genomic neighborhood of the single C. intestinalis Fgf8/17/18 gene with genomic regions surrounding human FgfD genes (Fig. 1). Results showed that many genes flanking Ci-Fgf8/17/18 have human orthologs near FGF8, FGF17, or FGF18. Immediately to the left of Fgf8/17/18 lies ENSCINT00000018903, the ortholog of FBXW4, and immediately to the right of Fgf8/17/18 is XP_002122436, which is the ortholog of human NPM1/2/3, and all three genes are transcribed in the same orientation (Fig. 1A). Human orthologs of these three genes are also contiguous and transcribed in the same orientation on Hsa10 (FBXW4, FGF8, NPM3) (Fig 1A). This situation represents a fully conserved chromosome segment since the divergence of urochordate and vertebrate lineages, estimated to be 800 million years ago from molecular clock data (Peterson et al., ‘04, Blair and Hedges, ‘05). The orientation and nearest-neighbor relationship of FGF18/NPM1 and FGF17/NPM2 have also been preserved, although the FBXW4 paralog is missing from these paralogons (Fig. 1A). Of 24 loci with clear human orthologs in the portion of C. intestinalis chromosome 5q shown in Fig. 1A, 15 have orthologs in Hsa2, Hsa5, Hsa8, or Hsa10, suggesting that these may be paralogons from the R1 and R2 genome duplication events.
Figure 1.
The chromosome region containing Fgf8/17/18 in the urochordate Ciona intestinalis shares conserved synteny with FGFD-containing regions of the human genome. A. A portion of C. intestinalis chromosome 5q (rectangle) is enlarged to show the positions of predicted C. intestinalis genes listed with the symbols for their human orthologs, often several, and their locations in the human genome. Black font represents Ciona genes with human orthologs near FGFD genes and gray font represents other locations in the human genome. The black rectangle marks a trio of genes preserved in order and orientation for 800 million years since the divergence of urochordate and vertebrate lineages. B. A circle plot of the Fgf8/17/18 region of the C. intestinalis genome with arcs linking to human orthologs on chromosomes around the circumference of the circle demonstrating the limited position of the orthologs on each human chromosome. C. A dot plot of paralogs for genes in a 10Mb window surrounding FGF8 on Hsa10, with the location of FGF8, FGF17, and FGF18 circled. Paralogs to Hsa10 genes are shown on their chromosome directly above the location of the gene on Hsa8. Paralogs lie mainly on Hsa2, 4, 5, 8, and 10.
To determine whether the human orthologs of genes neighboring C. intestinalis Fgf8/17/18 were clustered on human chromosomes or widely distributed, we used circle plots. Fig. 1B shows that human orthologs of the neighbors of the C. intestinalis Fgf8/17/18 gene cluster around the human Fgf8, Fgf17, and Fgf18 genes rather than being splattered over the full extent of each human chromosome. We conclude that syntenies have tended to be conserved in this region of the genome, which would be expected if local inversions were more frequent than translocations during chordate evolution. Furthermore, this result is as predicted from the hypothesis that stem olfactores (urochordates + vertebrates) had a chromosome segment that duplicated twice, giving rise to paralogons on Hsa5, 8, and 10 containing FGF18, FGF17, and FGF8, and another paralogon containing parts of Hsa4 or Hsa2p that today lacks an FgfD gene.
The human genome has four FgfD-related paralogons
The hypothesis that FGFD genes arose in two rounds of chromosome duplication predicts that a dot plot that localizes paralogs of genes surrounding FGF8 on Hsa10 should identify four paralogons, including parts of Hsa5 and Hsa8, and one other chromosome segment. To test this prediction, we investigated the chromosome locations of the paralogs of all genes in the 10Mb interval surrounding FGF8, from 98 to 108Mb on Hsa10. Figure 1C shows dots for genes from the region of Hsa10 surrounding FGF8 and their paralogs on other chromosomes displayed by red pluses in a column above or below the Hsa10 gene. Results showed that chromosomes 2, 4, 5, and 8 have 13, 13, 8, and 9 paralogs of the genes indicated by gray circles on Hsa10, but all other chromosomes have six or fewer paralogs, with an average of 1.5 paralogs per chromosome. These results support the conclusion from the analysis of conserved syntenies shared between C. intestinalis and human genomes that parts of Hsa2p and/or Hsa4 are members of the FgfD-containing paralogon.
Teleost FgfD genes reflect R3 and an ohnolog gone missing
Having identified paralogous chromosome segments in the human genome that have properties expected for ‘ohnologons’ arising from the R1 and R2 whole genome duplication events, we turned to understanding the relationship of the six teleost FgfD genes to the one Ciona and three human FgfD genes.
Is fgf24 an ohnolog gone missing from the human genome?
Recognizing that the primary fate of duplicate genes is nonfunctionalization (pseudogenation) of one of the two paralogs (Bershtein and Tawfik, ‘08), we explored the hypothesis that the predicted fourth R2-derived co-ortholog of the prototypical FgfD gene is Fgf24 and that this gene was preserved in the teleost lineage but was lost in the human lineage, as suggested by phylogenetic analysis (Jovelin et al., ‘07). An alternative hypothesis to be ruled out is that fgf24 did not exist in the last common ancestor of teleosts and humans but was a teleost innovation.
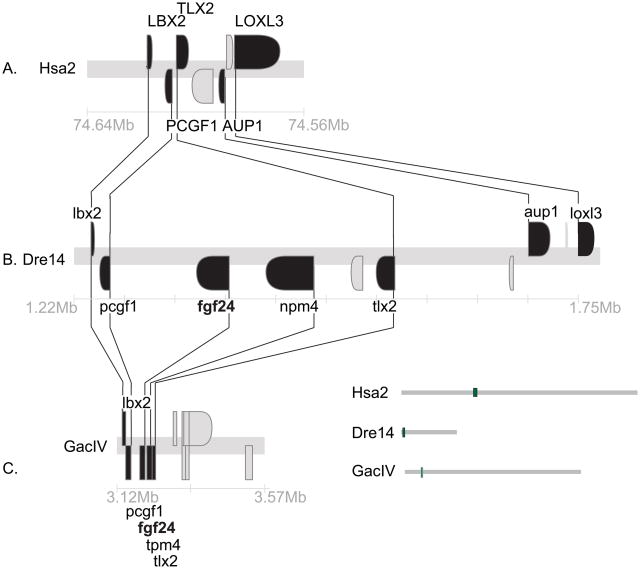
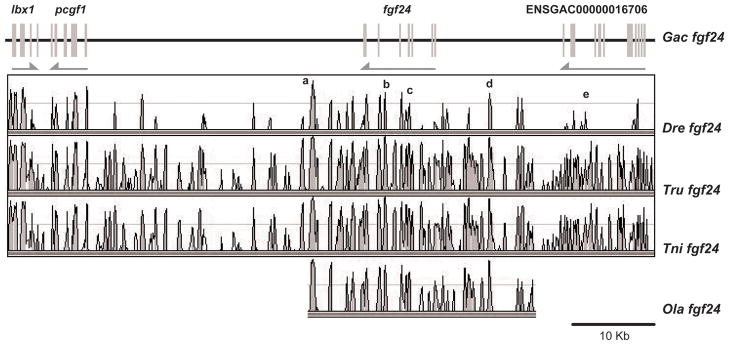
The R2 hypothesis makes three predictions. First, the teleost fgf24 gene should be embedded in a chromosome segment that is conserved with the human genome; second, the conserved region should have paralogs near other human FGFD genes, and third, this segment should show conserved synteny with the Ciona Fgf8/17/18 gene neighborhood shown in Figure 1. Here we test these predictions by examining conserved syntenies around zebrafish fgf24, starting at the first gene to its right (Fig. 2).
Figure 2.
Conserved syntenies for fgf24. A. A portion of Hsa2p with orthologs to neighbors of teleost fgf24 genes. B. A portion of zebrafish (Danio rerio) linkage group 14 (Dre14) containing fgf24. C. A portion of stickleback (Gasterosteus aculeatus) linkage group IV (GacIV) containing fgf24. This evidence is as expected if the human orthologs of fgf24 and npm4 were deleted from the human genome. Lines connect orthologs. Positions on chromosomes given in megabases (Mb).
The gene lying immediately to the right of fgf24 in the zebrafish and stickleback genomes (Q566X7 and ENSGACT00000022099, respectively) encode proteins that form a well-supported sister clade to tetrapod NPM1 and the zebrafish gene npm1; we will call this gene npm4 for now (Fig. 2B and Suppl. Fig. S1). Thus, npm4 is either an npm1 duplicate (e.g., npm1b), which would make fgf24 a duplicate of fgf18, or npm4 is a gene, like its neighbor fgf24, without a human ortholog. The human genome has three NPM genes: NPM1 (5q35), which is adjacent to FGF18 (8p21.3); NPM2, which is the nearest neighbor of FGF17, and NPM3, which is the nearest neighbor of FGF8 (10q24) (Figs. 3B, 4C, 5A). Thus, an FgfD gene and an Npm gene are neighbors for all extant human FgfD genes, as would be expected if this was the ancestral condition before R1, and neither npm4 nor its neighbor fgf24 have unique human orthologs. The next full gene to the right of fgf24 (ENSDARG00000011273) is annotated as tlx3a but is actually an ortholog of TLX2 (Wotton et al., ‘08). Human and zebrafish genomes both have three TLX family genes (Andermann and Weinberg, ‘01, Langenau et al., ‘02): TLX1 is five genes distant from FGF8 and tlx1 is three genes distant from fgf8a. TLX3 is the second gene from FGF18, while tlx3b is 2.8Mb from fgf18l. TLX2 is in Hsa2p13.1-p12 without a nearby FGFD-family gene, but tlx2 (NP_705937) is the second gene from fgf24, which favors the model that fgf24 is an ohnolog gone missing from the human genome.
Figure 3.
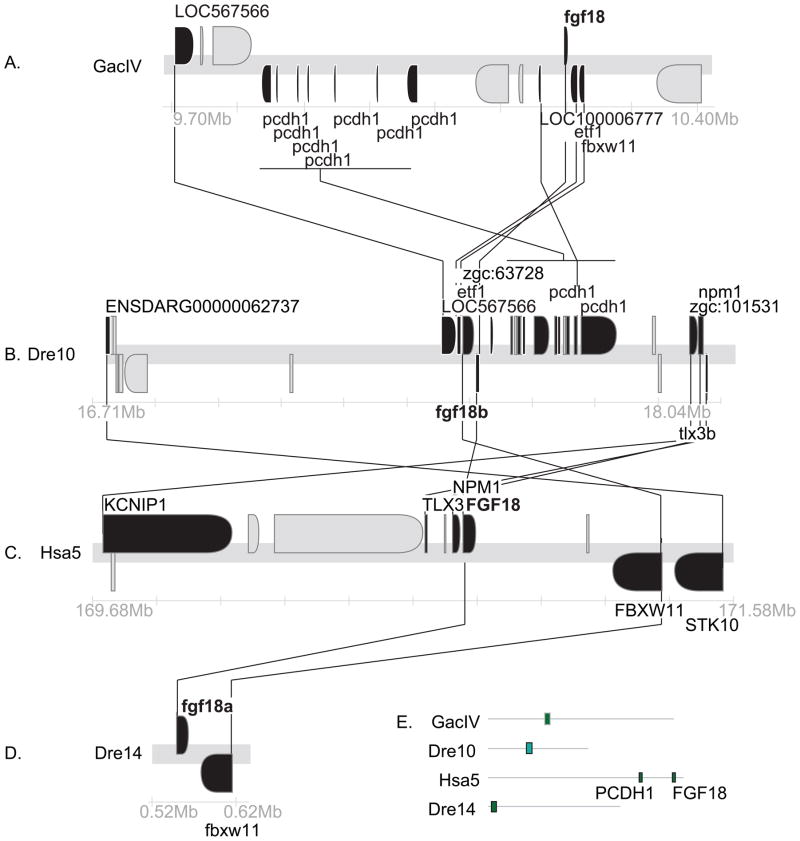
Conserved syntenies for Fgf18. A. A portion of stickleback chromosome GacIV containing fgf18. B. A portion of zebrafish chromosome Dre10 containing fgf18b. C. A portion of human chromosome Hsa5 containing FGF18. D. A portion of Dre14 containing fgf18a. E. Locations of each region shown related to the whole chromosome. Black genes have orthologs in these sections, gray genes do not.
Figure 4.
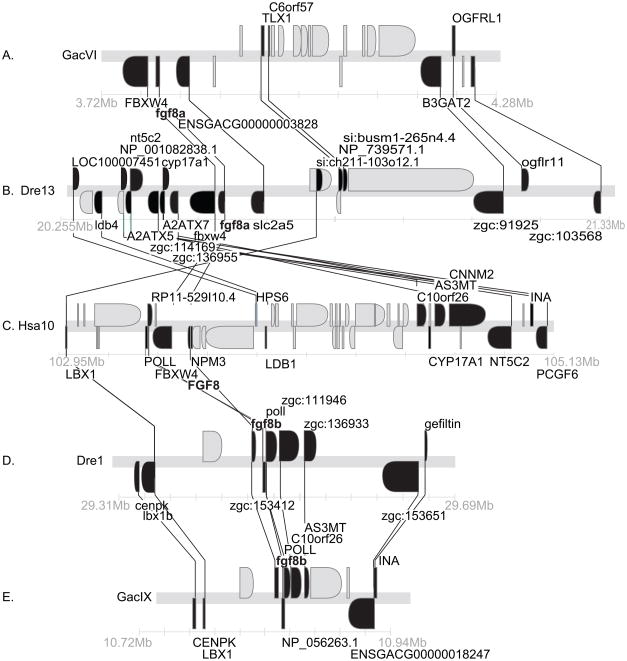
Conserved syntenies for Fgf8. A. A portion of stickleback chromosome GacVI containing fgf8a. B. A portion of zebrafish chromosome Dre13 containing fgf8b. C. A portion of human chromosome Hsa10 containing FGF8. D. A portion of Dre1 containing fgf8b. E. A portion of GacIX containing fgf8b. Black genes have orthologs in these sections, gray genes do not.
Figure 5.
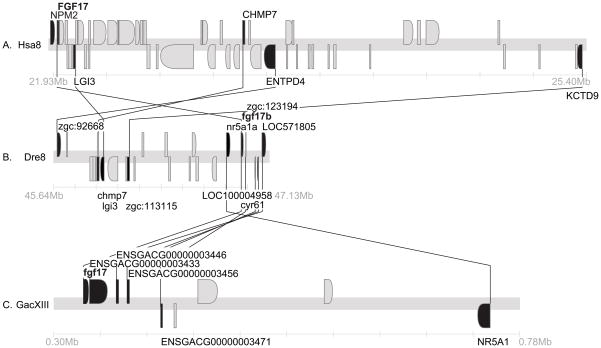
Conserved syntenies for Fgf17. A. A portion of Hsa8 containing FGF17. B. A portion of Dre8 containing fgf17. C. A portion of GacXIII containing fgf17. Black genes have orthologs in these sections, gray genes do not.
The next gene to the right, ENSDARG00000063176 from Ensembl release 45, is not annotated in Zv7 despite the continued presence of its nucleotide sequence in the position indicated in Fig. 2B. ENSDARG00000063176 has as a best human BLAST hit KAZALD1 located at 10q24.31 seven genes from FGF8, but in a phylogenetic tree, it forms, with its ortholog in the pufferfish Tetraodon nigroviridis, an outgroup to the tetrapod + zebrafish KAZALD1 clade (Suppl. Fig. S2). This phylogenetic grouping suggests that, like npm4 and fgf24, ENSDARG00000063176 is a paralog without a human ortholog and should be called kazald2 (Wotton et al., ‘08).
The two neighbors to the immediate left of zebrafish fgf24 (lbx2 and pcgf1, Fig. 2B) have orthologs adjacent on Hsa2p (Fig. 2A). In human, LBX2, PCGF1, and TLX2 are adjacent, but in the zebrafish genome fgf24 and npm4 lie between pcgf1 and tlx2 (Fig 2A, B). Moving to the right of TLX2, two of the next four genes in human, AUP1 and LOXL3, have orthologs also to the right of fgf24 in zebrafish (Fig. 2A, B). Thus, the zebrafish and human share most of the genes in this region of Hsa2p/Dre14, but orthologs of fgf24 and npm4 are specifically excised from the human genome. Stickleback linkage group IV (GacIV), which has five genes in a row orthologous to the zebrafish segment in Dre14 just discussed, and an ortholog of aup1 syntenic on GacIV, but about half a chromosome away (Fig. 2C). These results suggest that the five genes (lbx2, pcgf1, fgf24, npm4, tlx2) were neighbors before the divergence of zebrafish and stickleback lineages and that inversions in the stickleback lineage removed aup1 from its former neighbors.
The most parsimonious explanation of these data is that fgf24 and npm4 were adjacent in the FgfD paralogon in the last common ancestor of zebrafish and human, and that they were lost in the human lineage. Note that the orientation of TLX2/tlx2 relative to LBX2/lbx2 and PCGF1/pcgf1 is opposite in human and zebrafish, suggesting that a local inversion stirred the sequences, either in the human or zebrafish lineage. The human FGF18 region can serve as an outgroup to order the vector of evolutionary change: TLX3, NPM1, and FGF18 are all in the same orientation, suggesting that the deletion of the tetrapod Fgf24 and Npm4 ohnologs was associated with a small local inversion.
When did the duplication event that produced Fgf24 occur? BLAST searches of the trace files of the genome sequencing project of the elephant shark Callorhinchus milli revealed that chondrichthyes have at least an Fgf8 gene (exon 2 AAVX01442442) and both an Fgf18 gene (exon 3 AAVX01088747) and an Fgf24 gene (exon 3 AAVX01521786), as shown by exclusive amino acid positions in an alignment of exon-3 of FgfD genes from various vertebrates (Suppl. Fig. S3A). Because the shark genome, which diverged from the bony vertebrates before the divergence of teleost and tetrapod lineages, has an Fgf24 gene, this gene must have already existed in the last common ancestor of all jawed vertebrates. The inclusion of shark sequences for Fgf18 and Fgf24 in a phylogenetic tree of exon 3 results in the topology ((Fgf8,Fgf17)(Fgf18,Fgf24)) as expected from their origin in the R1 and R2 genome duplications (Suppl. Fig. S3B).
Altogether, the best interpretation of these findings is that the vertebrate FgfD subfamily originated from a single gene present in the ancestor of vertebrates and urochordates by two rounds of genome duplication. These R1 and R2 events lead to four Fgf genes in the last common ancestor of teleosts and tetrapods but the ortholog of fgf24 was subsequently lost in the tetrapod lineage.
fgf18a and fgf18b are duplicates from the teleost genome duplication
Phylogenetic analysis shows that zebrafish has two genes (fgf18a (NM_001013264) and fgf18b (also called fgf18l; NM_001012379)) in a clade with FGF18 as the most closely related human gene. Zebrafish Fgf18b is sister-taxon to a teleost Fgf18a clade (Itoh and Konishi, ‘07, Jovelin et al., ‘07, Kikuta et al., ‘07) as would be expected from gene duplication after the divergence of ray-fin and lobe-fin fish (Itoh and Konishi, ‘07, Jovelin et al., ‘07, Kikuta et al., ‘07). Four other sequenced teleost genomes have an ortholog of fgf18a but lack an annotated fgf18b and BLAST searches failed to identify an ortholog of fgf18b in genome databases for stickleback, medaka, Tetraodon, or fugu, all of which are percomorph fish. This situation could have arisen if fgf18a and fgf18b arose by the R3 duplication in stem teleosts and fgf18b was lost in the lineage of the Percomorpha but was retained in the lineage of the Ostariophysi (including zebrafish). An alternative hypothesis is that fgf18b arose by tandem duplication in the zebrafish lineage after it diverged from the percomorphs.
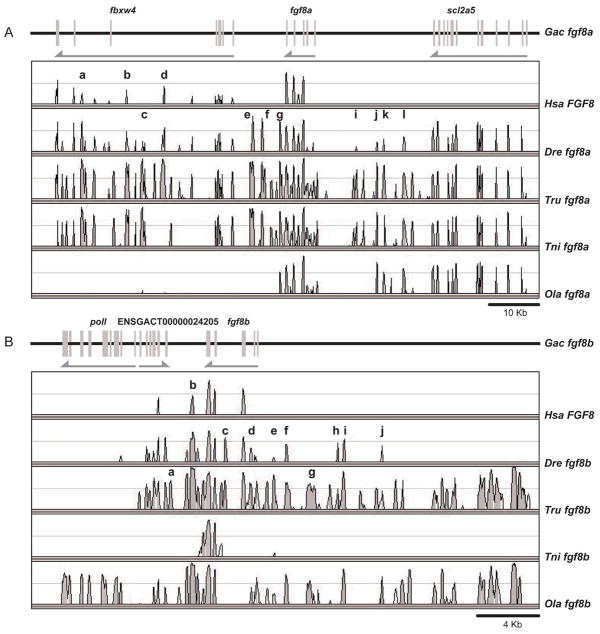
Conserved syntenies can help distinguish the R3 and tandem duplication hypotheses for the origin of fgf18 paralogs. The fgf18a gene of zebrafish is on Dre14 near fgf24 along with fbxw11, the ortholog of which is separated from FGF18 by a single hypothetical gene (Fig. 3C and D), thus supporting the orthology of zebrafish fgf18a and human FGF18. Note that the C. intestinalis genome has two orthologs of FBXW11, one located 10 genes distant and one 13 genes away from Fgf8/17/18 (NP_001027626 and ENSCINT00000018903) (Fig. 1A); this is another example of an ancient FgfD-region synteny conserved for 800 million years. A cluster of pcdh1 genes flanking fgf18 (not shown) are, as a group, co-orthologous to PCDH1 protocadherin genes in Hsa5q31, a bit distant but syntenic to FGF18 in Hsa5q35 (Fig. 3C–E). These gene clusters were apparently tandemly duplicated at least partly independently in the zebrafish and human lineages. Together, these conserved syntenies support orthology of fgf18a and FGF18.
The orthologs of several genes located near zebrafish fgf18b on zebrafish LG10 are adjacent to or near FGF18 on Hsa5q (Fig. 3B, C), consistent with the orthology of fgf18b and FGF18 as shown in the phylogeny. The nearest neighbor to fgf18b (Fig. 3B) is an ortholog of FBXW11, which is separated from FGF18 by a single small hypothetical gene (Fig. 3C). Like fgf18a and FGF18 neighborhoods, the fgf18b region of zebrafish and stickleback both have a series of tandemly duplicated pcdh genes. The region surrounding fgf18 in stickleback has at least five neighbors with orthologies to genes in the region surrounding fgf18b in zebrafish, a result expected if the two regions are orthologous (Fig. 3A, B). This is in conflict with the phylogeny, which shows that Fgf18 in stickleback (ENSGACT00000023719) is more closely related to Fgf18a than it is to Fgf18b (Jovelin et al., ‘07). If the conserved syntenies correctly suggest orthologies, then the zebrafish Fgf18b sequence is evolving more rapidly than the Fgf18a sequence. Alternatively, gene losses may account for the conflict. If orthologs neighboring stickleback fgf18 were lost near fgf18a but not near fgf18b, then the reciprocal best blast method for identifying orthologies could find only genes near fgf18b and call them as orthologs given the loss of these genes from the neighborhood of fgf18a. Both explanations, however, are consistent with the origin of fgf18a and fgf18b in the R3 event in teleost evolution.
fgf8 duplicates arose in R3
Zebrafish has one uncontested ortholog of FGF8 (Brand et al., ‘96, Reifers et al., ‘98), and a second gene (fgf17a) that was initially assigned as a co-ortholog of tetrapod Fgf17 (Reifers et al., ‘00a, Cao et al., ‘04), but that was later shown to be a fgf8 duplicate and is now called fgf8b (Itoh and Konishi, ‘07, Jovelin et al., ‘07, Kikuta et al., ‘07). A detailed analysis of conserved syntenies in human, zebrafish and stickleback supports this conclusion. Orthologs of six genes surrounding human FGF8 in a 2.16 Mb region on chromosome Hsa10q (Fig. 4C) are located in a 0.4 Mb region on zebrafish chromosome Dre1 containing fgf8b (Fig 4B). The orthologous portion of stickleback linkage group GacIX contains fgf8b and eight genes in the corresponding region of the human genome (Fig. 4A). The orthologs of a somewhat different set of human genes flanking FGF8 lie near fgf8a in zebrafish Dre13 and stickleback LG VI (Fig. 4D,E). We conclude that zebrafish fgf8 (NM_131281) and the so-called fgf17a (NM_182856) are co-orthologs of human FGF8 and support the conclusion that they should be named fgf8a and fgf8b (Itoh and Konishi, ‘07, Jovelin et al., ‘07, Kikuta et al., ‘07). Figure 4 shows that in addition to FGF8 co-orthologs, zebrafish and stickleback have co-orthologs of LBX1 (Q5CZV7 and si:busm1-265n4.1) and C10orf26 (zgc:111946 and Q1ECU4) in these genomic regions, indicating that that these duplicated loci also originated in the R3 genome duplication.
Teleosts have a single copy of fgf17
Sequenced teleost genomes have a single ortholog of FGF17. Several genes in the immediate neighborhood the gene originally called fgf17b on zebrafish Dre8 (Cao et al., ‘04) are orthologs of genes that are near FGF17 in Hsa8p21 (Fig. 5A, B). These two segments are largely conserved between zebrafish and stickleback (Fig. 5B, C). We conclude that fgf17b should be called simply fgf17 (NM_182856) and it is the zebrafish ortholog of FGF17.
Expression patterns of FgfD family genes
With this understanding of the evolutionary relationships of teleost and tetrapod FgfD genes, we wondered how paralog functions evolved as judged by gene expression patterns.
Mid-segmentation stages
fgf8 paralogs
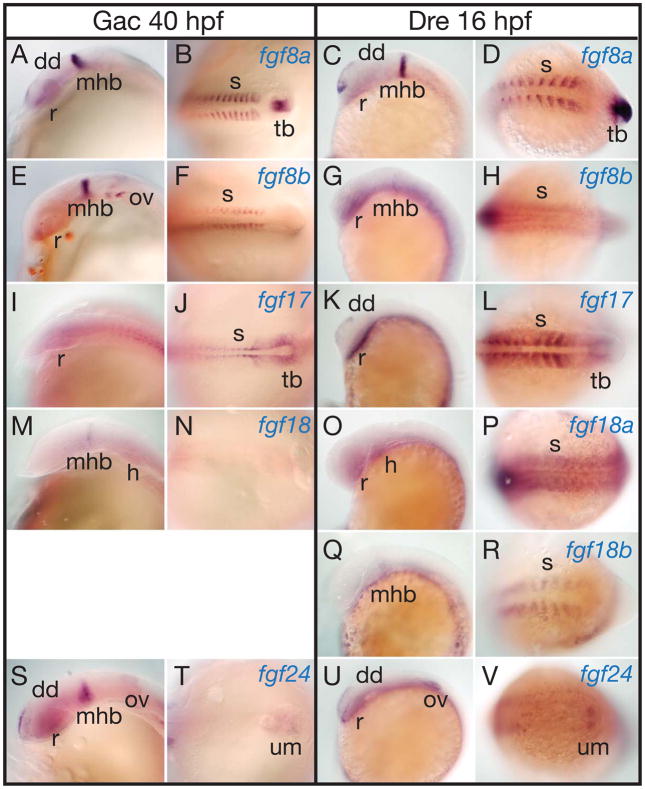
At mid-segmentation stages (40 hpf stickleback and 16 hpf zebrafish) (Kimmel et al., ‘95), the expression of fgf8a and fgf8b are similar in zebrafish and stickleback (Reifers et al., ‘98, Reifers et al., ‘00a, Draper et al., ‘03, Jovelin et al., ‘07), with strong expression of both genes in both species in the midbrain-hindbrain border (MBH) and somites (Fig. 6A–H). In addition, fgf8a is expressed in the dorsal diencephalon and the tailbud in both species. In the heart, stickleback and zebrafish express different fgf8 paralogs (Jovelin et al., ‘07).
Figure 6.
Expression of stickleback and zebrafish FgfD genes at midsegmentation stages. A, B, E, F, I, J, M, N, S, T, 40 hpf stickleback embryos. C, D, G, H, K, L, O, P, Q, R, U, V, 16 hpf zebrafish embryos. A–D, fgf8a. E–H, fgf8b. I–L, fgf17. M–P, fgf18a. Q, R, fgf18b. S–V, fgf24. Abbreviations: dd, dorsal diencephalon; h, heart; mhb, midbrain-hindbrain boundary; ov, otic vesicle; r, retina; s, somite; tb, tail bud; um, unsegmented mesenchyme.
fgf17
At mid-segmentation in stickleback and zebrafish, fgf17 is expressed weakly in the CNS, and more strongly and specifically around the tail bud and in the segmental plate and segmented somites (Fig. 6I–L) (Cao et al., ‘04, Hamade et al., ‘06). The major difference between sfgf17 (stickleback fgf17) and zfgf17 (zebrafish fgf17) is that the zebrafish gene is expressed in the dorsal diencephalon (Fig. 6K). For both species, fgf8a and fgf8b are both expressed in the MHB, but fgf17 is not, and all three genes are expressed in the somites. In the tailbud, fgf8a in both species is expressed in the tailbud itself; fgf8b is not expressed in the tailbud region; and fgf17 is expressed in the segmental mesoderm that surrounds the tailbud in a pattern complementary to that of fgf8a. Both zfgf17 and zfgf8a are expressed in the dorsal diencephalon.
fgf18
The expression of zfgf18a and zfgf18b have not been previously described. At mid-segmentation, the single copy of fgf18 in stickleback is expressed weakly in the MHB and in the heart primordium, but not in the somites (Fig. 6M, N). In zebrafish, zfgf18b, but not zfgf18a, is expressed in the MHB like zfgf8a, and reciprocally, zfgf18a, but not zfgf18b, is expressed in the heart like zfgf8b (Fig. 6O–R). Although sfgf18 is expressed only weakly if at all in the somites (Fig. 6N), zfgf18a and zfgf18b are both expressed in the somites, but zfgf18a is expressed broadly in somites like zfgf8b (Fig. 6H, P), but zfgf18b is expressed in just a part of each somite like zfgf8a (Fig. 6D, R).
fgf24
In mid-segmentation stage stickleback embryos, sfgf24 is expressed in the dorsal diencephalon, MBH, and tailbud, but not in the somites, like sfgf18 (Fig. 6S, T). In zebrafish as in stickleback, fgf24 is expressed in the dorsal diencephalon like fgf8a (Draper et al., ‘03). In contrast to sfgf24, zfgf24 is not expressed in the MHB (Fig. 6U, V). In both stickleback and zebrafish, fgf24 is expressed in the unsegmented presomitic mesoderm.
Long pec stage
fgf8
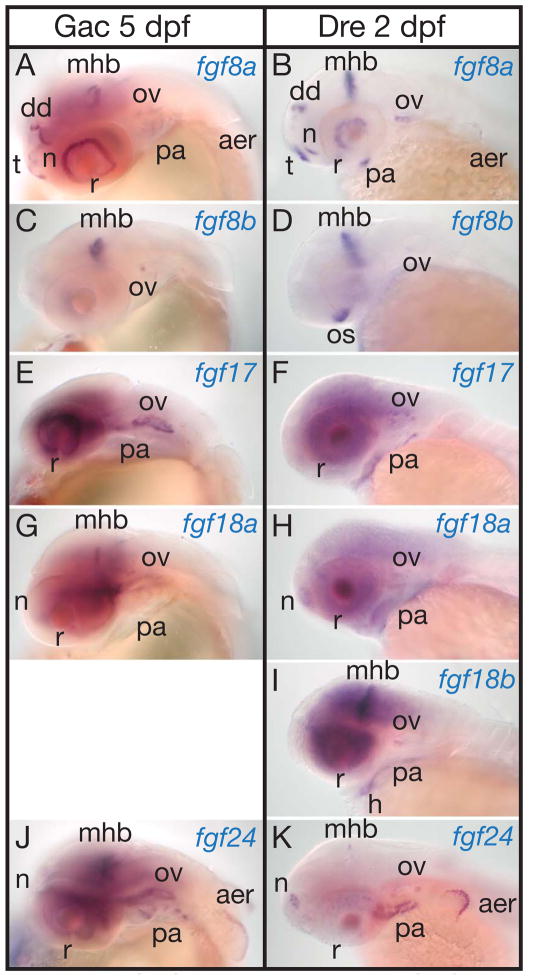
At the ‘long pec’ stage (120 hpf stickleback and 48 hpf zebrafish) (Kimmel et al., ‘95), the expression patterns of both fgf8 genes in both species occupy several of the earlier domains, but in addition, in both species, new domains appear for fgf8a (but not fgf8b) in the telencephalon, olfactory epithelium, pharyngeal arches, (Meulemans and Bronner-Fraser, ‘07)), and apical epidermal ridge (AER) of the pectoral fin bud (Fig. 7A–D).
Figure 7.
Expression of stickleback and zebrafish FgfD genes at long pec stage. A, C, E, G, J, 5 dpf stickleback embryos. B, D, F, H, I, K, 2 dpf zebrafish embryos. A, B, fgf8a. C, D, fgf8b. E, F, fgf17. G, H, fgf18a. I, fgf18b. J, K, fgf24. Abbreviations: aer, apical ectodermal ridge of pectoral fin; dd, dorsal diencephalon; h, heart; mhb, midbrain-hindbrain boundary; n, nose; ov, otic vesicle; os, optic stalk; pa, pharyngeal arches; r, retina; t, telencephalon.
fgf17
At the long-pec stage, stickleback sfgf17 is expressed in the retina (Fig. 7. E) in a pattern that overlaps in time and space expression of sfgf8a in the eye, (Fig. 7A) and is stronger than sfgf8b expression in the eye at the same stage (Fig. 7C). In addition, sfgf17 and zfgf17 are expressed in the otic vesicle (Fig. 7E, F). sfgf17 was only weakly expressed in the pharyngeal arches compared to zebrafish (Fig. 7E, F). No expression of sfgf17 was apparent in the MHB or in the pectoral fin bud. In zebrafish, zfgf17 is also expressed in the eye, but broadly in several layers. In mouse, the expression patterns of Fgf17 and Fgf8 largely overlap, but Fgf17 is expressed in a broader region of the frontal cortex and cerebellum of the brain and is required for dorsal frontal cortex and cerebellar development (Xu et al., ‘00, Cholfin and Rubenstein, ‘07, Dominguez and Rakic, ‘08). The regenerating adult zebrafish heart also expresses fgf17 (Lepilina et al., ‘06).
fgf18
To the expression domains present at mid-segmentation stages (Fig. 6M–R), at the long-pec stage, stickleback sfgf18 and zebrafish zfgf18a add expression in the retina, olfactory epithelium, otic vesicle, and pharyngeal arches 3–7. In contrast, zebrafish zfgf18b only adds expression in the retina and pharyngeal arches (Fig. 7G–I).
fgf24
At the long pec stage, stickleback sfgf24 is expressed in the MHB, olfactory epithelium, most cells of the otic vesicle, endoderm of the pharyngeal pouches, and apical epidermal ridge of the pectoral fin (Fig. 7J). Expression of sfgf24 in the eye and MHB is broader than the expression of sfgf8a and sfgf18 in these domains (Fig. 7A, G). Here we extend previous descriptions of zfgf24 expression (Draper et al., ‘03, Fischer et al., ‘03); zfgf24 is expressed in a small group of cells in the dorsal MHB, in the olfactory epithelium, endoderm of the pharyngeal pouches, and the otic vesicle. In the AER of the pectoral fin in both stickleback and zebrafish, fgf24 is expressed broader and stronger than fgf8a, with the fgf8 domain entirely within the fgf24 domain (Fig. 7A, B, J, K).
Eyes and ears
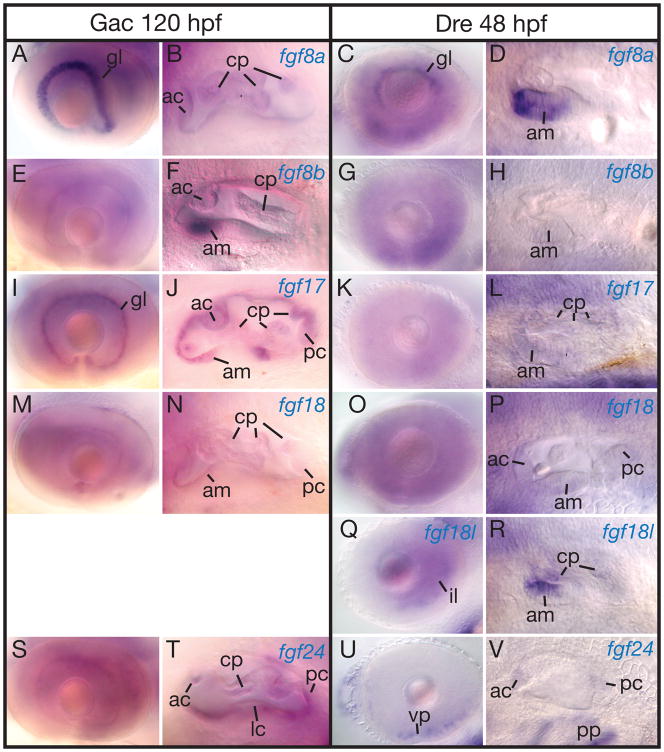
Several FgfD genes are expressed in the eyes and ears, but often in different domains. The ganglion cell layer of the eye expresses sfgf8a, zfgf8a, sfgf17 and sfgf24 (Fig. 8A, C, I, S)(Picker et al., ‘05). In the retina, zfgf24 is expressed in isolated cells scattered in various layers (Fig. 8S, U). Except for the retina and MHB, zfgf24 and sfgf24 are expressed similarly.
Figure 8.
Expression of stickleback and zebrafish FgfD genes in the ear. A, B, E, F, I, J, M, N, S, T, 5 dpf stickleback embryos. C, D, G, H, K, L, O, P, Q, R, U, V, 2 dpf zebrafish embryos. A–D, fgf8a. E–H, fgf8b. I–L, fgf17. M–P, fgf18a. Q, R, fgf18b. S–V, fgf24. Abbreviations: ac, anterior crista; am, anterior macula; cp, canal projection; fp, fusion plate; gl, gangloin layer; il, inner nuclear layer; lc, lateral crista; pc, posterior crista; pp, pharyngeal pouches; vp, ventral patch.
The developing ear expresses all FgfD genes at long-pec stages in both stickleback and zebrafish. Some genes are expressed generally throughout the ear, but others are only expressed in specific regions. The anterior sensory patch shows strong expression of sfgf8b in stickleback and its paralog zfgf8a in zebrafish, while expressing zfgf8b only weakly (Fig. 8D, F, H). In addition, the anterior sensory patch expresses zfgf18b strongly and zfgf17 and zfgf18a only weakly (Fig. 8L, P, R). The anterior and posterior cristae express zfgf24 (Fig. 8V).
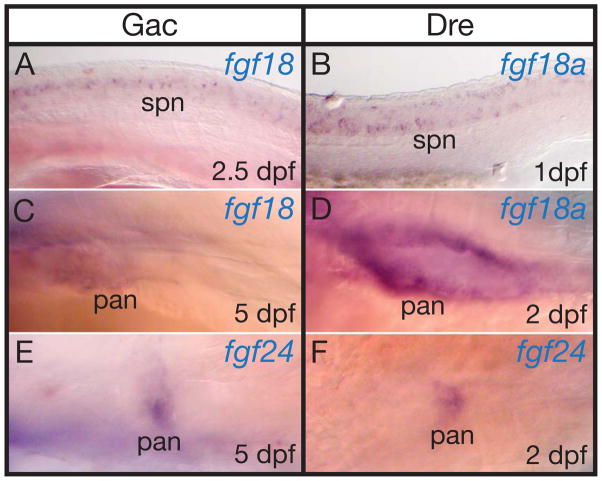
In both stickleback and zebrafish during the early pharyngula period, fgf18 and fgf18a are expressed in specific spinal cord neurons (Fig. 9A and B). None of the other FgfD genes are expressed in spinal cord neurons at this stage in zebrafish, and none of the FgfD genes in mouse are expressed in spinal cord neurons at this developmental stage, suggesting that this expression domain is either an innovation in stem teleosts, or that it was an ancestral function lost in the mammalian lineage.
Figure 9.
Expression of stickleback and zebrafish FgfD genes in the trunk. A, C, E, stickleback embryos. B, D, F, zebrafish embryos. A, B, fgf18 and fgf18a. C, D, fgf18 and fgf18a. E, F, fgf24. Abbreviations: pan, pancreas; spn, spinal cord neurons.
Of the three FgfD genes in mouse, only Fgf18 is expressed in the pancreas (Dichmann et al., ‘03). In stickleback and zebrafish long-pec stage, sfgf18, zfgf18a, sfgf24 and zfgf24 are also expressed in the pancreas (Fig. 9C–F), suggesting that a pancreas regulatory element was present before the gene duplication event that produced fgf18 and fgf24.
Analysis of Conserved Non-coding Elements
Expression patterns of the FgfD gene family just described show many examples of conservation across species and across paralogs. These conserved patterns are likely to be controlled by regulatory elements, the functions of which have been conserved for hundreds of millions of years. Because some regulatory elements conserved in function are also conserved in sequence, we searched teleost and human FgfD genomic regions for conserved non-coding elements (CNEs).
CNE sharing among FgfD paralogs within species
Human
To discover ancient conserved non-coding elements (CNEs) derived from the R1 and R2 genome duplication events, we conducted a Vista plot analysis of human FGFD genes. The analysis allowed several conclusions (Fig. 10A). First, CNEs shared between FGF8 and FGF18 were more frequent and longer than those shared by FGF8 and FGF17. Second, CNEs embedded in the genes flanking FGF8 (FBXW4 and NPM3) are conserved with those flanking FGF18, which is located adjacent to NPM1 and near FBXW11. NPM2 is adjacent to FGF17 (Fig. 5A), but CNEs embedded in NPM3 (near FGF8) and NPM1 (near Fgf18) are apparently not conserved with NPM2 (near FGF17). Third, a CNE (labeled ‘a’) shared by FGF8 and FGF18 is found within intron four of FBXW4 – this is significant because an enhancer trap element inserted in a similar position in zebrafish confers an fgf8 expression pattern (Kikuta et al., ‘07). Finally, three highly conserved CNEs located 3′ to FGF8 (labeled d, e, and f) are shared by all three human FGFD genes. We conclude that at least three non-coding regions in the genomic neighborhood of FGFD genes have been highly conserved in sequence for at least 500 million years (Kumar and Hedges, ‘98, Peterson et al., ‘04) since the R1 and R2 genome duplication events. Our results raise the hypothesis that these conserved sequences are regulatory elements that drive the expression of FGFD genes in domains common to all three genes.
Figure 10.
Distribution of CNEs among FgfD paralogs in human (A), stickleback (B) and zebrafish (C). The higher density of CNEs among human paralogs is probably the result of the R3 genome duplication in teleosts with subsequent relaxed selection and partitioning of regulatory elements (Force et al., ‘99). The reference sequence is indicated at the top. The position of exons is indicated along the toop. For each conservation plot, the top line represents 100% conservation with the reference sequence, and the bottom line indicates 50% conservation. Grey arrows show gene orientation. Lower case letters indicate peaks mentioned in the text.
Stickleback
To test if CNEs near FgfD genes are also conserved in teleosts, we constructed Vista plots for stickleback. Figure 10B shows conservation for the exons of fgf8a/fgf8b and scl2a5a/scl2a5b at greater than threshold, but no CNEs shared by the two paralogs. This result is a bit surprising given the similarities of expression patterns between the two sfgf8 ohnologs. In contrast, several CNEs were apparent in the comparison of sfgf8a and sfgf17, although most exons failed to rise above threshold (Minimum Y value on the VISTA plot = 50%, Minimum conservation identity =70%, Minimum length for a CNE = 100).
Three of the stickleback CNEs (a, b, e) were in introns of fgfD neighbor genes, and two were found in intergenic region flanking the fgfD gene. Note that CNE ‘a’ in Figure 10B is in the same intron of stickleback fbxw4 as CNE ‘a’ of human FBXW4 although it is shared between the fgf8a and fgf17 regions for stickleback and FGF8 and FGF18 regions for human. Neither the exons nor any CNEs were detected that were shared between sfgf8a and either sfgf18 or sfgf24, suggesting rapid evolution, translocations, or problems with genome assembly. The sparse pattern of CNEs in stickleback fgfD genes contrasts with the rather broad CNEs found for human FGFD genes.
Zebrafish
For zebrafish (Fig. 10C), a strong CNE (labeled ‘d’ in Fig. 10B and 10C and ‘a’ in Fig. 10A) was detected in the same intron of fbxw4 as detected in stickleback and in human, and it was shared by all zebrafish fgfD regions except fgf18b. An enhancer-trap insertion nearby (CLGY667) confers reporter expression in telencephalon, optic stalk, mid-hindbrain boundary, somites, heart, olfactory pits, and tail bud in a pattern similar to that of fgf8 genes (Kikuta et al., ‘07). This CNE, conserved since before R1 and maintained by fgf8a, fgf8b, fgf17, fgf18a, and fgf24, may play a role in defining expression domains shared by vertebrate FgfD genes.
Despite their similarity in expression patterns (Fig. 8A–H, 9A–D), fgf8a shares fewer CNEs with fgf8b than it does with fgf17, fgf18a and fgf24 (Fig. 10C). Using fgf8a as base, one CNE (d) is present near 5 zebrafish fgfD genes, three (b, n, and r) are present near three fgfD genes, eight (f, i, k, l, m, q, w, and y) are near two fgfD genes, and 14 (a, c, e, g, h, j, o, p, s, t, u, v, x, and z) are shared by fgf8a and only one or another fgfD gene. fgf8a shares most CNEs (17) with fgf24 and about an equal number with fgf17 and fgf18a (11 and 10, respectively).
Enhancer trap CLG508 is located in between fgf8a and fbxw4 and confers expression in the apical ectodermal ridge of the pectoral fin bud (Kikuta et al., ‘07), but Vista detected no CNE shared by fgf8a and other fgfD genes in this intergenic region despite the expression of both fgf8 and fgf24 in the AER. Enhancer trap CLGY1030 is inserted near CNE ‘u’ in zebrafish and matches the expression pattern of fgf8a only in the tail bud (Kikuta et al., ‘07).
CNE sharing among FgfD orthologs
CNEs displayed when comparing FgfD genes within a species show elements conserved from the R2 event, while CNEs revealed by comparing a single FgfD gene among species identifies elements conserved since species divergence.
fgf17
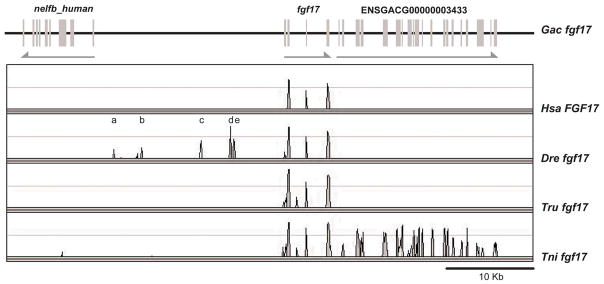
Few CNEs appeared in comparisons of the stickleback fgf17 gene to its orthologs in human and several teleosts, although exons were readily apparent (Fig. 11). At least six CNEs were apparent when comparing stickleback and zebrafish fgf17 genes, although the flanking genes were not conserved (Fig. 11). Given that stickleback and pufferfish are more closely related than stickleback and zebrafish, it was surprising to find more sfgf17 CNEs shared with zebrafish than pufferfish. The lack of CNEs near fgf17 in the two pufferfish species could be due in part to incomplete sequencing in this genomic region: fgf17 in fugu (Takifugu rubripes) lies in a small scaffold without predicted neighbors, and numerous sequencing gaps surround fgf17 in the green spotted pufferfish Tetraodon nigroviridis. Among the species examined, only stickleback and T. nigroviridis share a neighboring gene (ENSGACG00000003433) (Fig. 11). Close inspection of chromosome segments carrying fgf17 in zebrafish and stickleback shows that global synteny is conserved but gene orders are not, indicating local genomic micro-rearrangements (Fig. 5). Comparison with human reveals rearrangements over still larger genomic distances (Fig. 5). Chromosome breaks near fgf17 may have carried CNEs along with fgf17 neighbors disrupting synteny of coding and non-coding sequences in these regions.
Figure 11.
Distribution of CNEs among Fgf17 orthologs. The low number of CNEs is likely due to micro-rearrangements around the Fgf17 locus disrupting synteny of coding and non-coding sequences.
fgf18
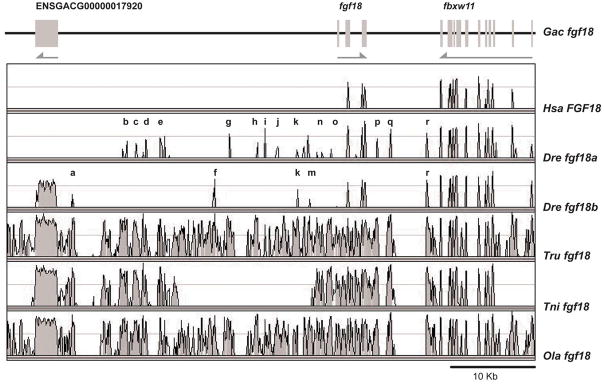
BLAST searches identified a single fgf18 gene in stickleback, medaka and both pufferfish and the topology of the fgf18 clade suggests that fgf18b may have been lost in the lineage leading to these species (Jovelin et al., ‘07). In contrast to the four percomorph species, zebrafish has two fgf18 genes. A number of sfgf18 CNEs are found associated only with zebrafish fgf18a (b, c. d, e, g, h, i, j, l, n, o, q), a few only with zfgf18b (a, f), and several with both (k, m, r) (Fig. 12). This pattern of reciprocal distribution of CNEs is what would be expected if the single stickleback fgf18 gene represented much of the ancestral distribution of CNEs and they had become distributed as predicted by subfunctionalization in the zebrafish lineage after gene duplication.
Figure 12.
Most of the CNEs conserved among stickleback, medaka and pufferfish fgf18 are partitioned between fgf18a and fgf18b in zebrafish, consistent with the DDC model (Force et al., ‘99). The high density of CNEs in teleost species having a single fgf18 gene suggests that the second fgf18 duplicate was lost shortly after the R3 duplication. The region lacking CNEs in tetraodon is due to a gap in the sequence alignment.
The high density of CNEs surrounding fgf18 suggests that fgf18b may have been lost shortly after the separation of the lineages leading to zebrafish and stickleback and that CNEs may have been maintained by selection in lineages in which only one fgf18 gene subsisted. However, in zebrafish, which retained both fgf18 duplicates, most CNEs found around fgf18 of species retaining just one fgf18 gene have been partitioned between fgf18a and fgf18b (Fig. 12).
fgf24
The high density of CNEs in proximity to fgf24 orthologs in all tested species (Fig. 13) is consistent with our inference that one fgf24 duplicate was lost before the divergence of the lineages leading to zebrafish and stickleback (Jovelin et al., ‘07). Two particularly strong CNEs (labeled a and d) lie 3′ and 5′ respectively to fgf24 and two others (b and c) are within introns of the fgf24 gene. We checked to see if Fgfd-related CNEs could be detected in the location predicted for the missing human FGF24, but found none.
Figure 13.
The high density of CNEs among teleost fgf24 orthologs suggests that the second fgf24 duplicate was lost shortly after the R3 duplication before the partitioning of fgf24 subfunctions. Medaka fgf24 is located on a short scaffold, hence the apparent lack of conservation further away from fgf24.
fgf8
In stickleback and zebrafish, fgf8 duplicates exhibit common and distinct expression patterns, and comparisons between fgf8 orthologs show that partitioning of most ancestral functions occurred before the divergence of these species (Jovelin et al., ‘07). Thus we should expect regulatory elements to be largely shared between fgf8 orthologs in stickleback and zebrafish rather than being species-specific.
For comparisons based on stickleback fgf8a, at least four CNEs were shared with the human FGF8 region (Fig. 14A). These included elements in introns of fbxw4 (CNEs a, b, and d in Fig. 14A) and an element in an intron of fgf8a itself (element h). Among the five stickleback fgfD genes, the fgf8 comparisons were the only ones that revealed any CNEs shared with human. All of the elements sfgf8a shared with human were also shared with zebrafish. In addition, zebrafish fgf8a shared with stickleback fgf8a several elements in the intergenic spaces 3′ and 5′ to fgf8a. As noted for other fgfD genes, stickleback fgf8a had more and longer CNEs shared with percomorphs than with zebrafish. With few exceptions (see element d, Fig. 14A), the percomorphs had very similar CNE patterns.
Figure 14.
Distribution of CNEs among Fgf8 orthologs. CNEs are partitioned in unique sets between teleost fgf8a (A) and fgf8b (B), reflecting the partitioning of expression domains between fgf8 duplicates (this work and (Jovelin et al., ‘07)). Fewer fgf8b CNEs are shared with human FGF8.
The comparison of the stickleback fgf8b region to that of human revealed a CNE 3′ to the stickleback fgf8b gene (element b, Fig. 14B) shared with other teleosts except T. nigroviridis. A CNE in an intron of fgf8b (element c in Fig. 16B) was shared with zebrafish and fugu. A large number of CNEs located 5′ to fgf8b (e.g., element g) were held in common by several teleosts that were not seen in the comparison with zebrafish.
Therefore, the overall pattern of conservation of non-coding sequences in the proximity of fgf8 correlates with the evolutionary history of fgf8 duplicates and their subsequent functional diversification: fgf8a and fgf8b orthologs have unique sets of CNEs, some of which are shared with human as it would be expected if subfunctions have been partitioned among paralogs.
DISCUSSION
Vertebrate FgfD genes originated as ohnologs in R1 and R2
To infer the evolutionary history of expression domains and conserved non-coding sequences, it is essential to understand the phylogenetic relationships of the four FgfD genes (Suppl. Fig. S4). The phylogenetic tree of these genes shows a history of (((Fgf8, Fgf17) Fgf18) Fgf24) (Jovelin et al., ‘07). Several other data sets, however, support the conclusion that the four FgfD genes arose in the R1 and R2 whole genome duplication events. Evidence supporting this conclusion comes from four independent sources: 1) The chromosome region surrounding the Ciona FgfD gene contains genes with human orthologs mainly on human chromosome segments containing FGF8, FGF17 and FGF18, as well as part of Hsa2p. 2) An automated search for paralogous chromosome segments around FGF8 revealed chromosome segments surrounding FGF17, FGF18, and a portion of Hsa2p. 3) The elephant shark has a copy of Fgf24, demonstrating the origin of this gene before the divergence of teleost and tetrapod genomes. 4) The immediate genomic neighborhood of the teleost fgf24 gene is preserved in the human genome with the rather precise excision of an ortholog to fgf24 and its nearest neighbor npm4. These data support the conclusion that R1 and R2 produced four ohnologs related as ((Fgf8, Fgf17)(Fgf18, Fgf24)) and that the tetropod Fgf24 gene went missing from tetrapod genomes.
Comparative analysis of expression patterns
After the R2 genome duplication event, the four FgfD genes would have been preserved either by subfunctionalization (the duplicate preservation by reciprocal partitioning of ancestral gene subfunctions) or neofunctionalization (the origin of novel domains) (Force et al., ‘99, Hughes, ‘99, Stoltzfus, ‘99, Postlethwait et al., ‘04, Chain et al., ‘08, Conant and Wolfe, ‘08), and subsequent to preservation, additional expression domains or other functions could have been lost or ‘invented’. To analyze these events, we investigated expression patterns of FgfD genes in two teleosts and compared them to published data available on their orthologs in mouse (Suppl. Fig. S4).
The single FgfD gene present in stem olfactores (urochordates + vertebrates) was an ancestor of Fgf8/17/18 in the ascidian urochordate Ciona intestinalis and its expression pattern serves as an outgroup to the vertebrate FgfD genes. In Ciona, Fgf8/17/18 is expressed in the embryonic central nervous system (CNS) coincident with En and Pax2/5/8 (Ikuta and Saiga, ‘07). Correspondingly, the vertebrate orthologs of these three Ciona genes (Fgf8, Fgf17, Fgf18, En1, En2, Pax2, Pax5, and Pax8) are co-expressed in the vertebrate CNS at the midbrain-hindbrain boundary (MHB) (Joyner, ‘96). Although urochordates may lack an MHB organizer secondarily (Canestro et al., ‘05, Ikuta and Saiga, ‘07), this gene set appears to constitute an ancient regulatory cassette in chordate CNS development. The expression of all four vertebrate FgfD ohnologs in the midbrain-hindbrain border region in at least one vertebrate species (Suppl. Fig. S4) suggests that this was an ancient expression domain existing before the divergence of urochordates and vertebrates and that this expression domain was retained by all four genes after the R1 and R2 whole genome duplication events at the base of the vertebrate radiation.
Besides its expression in the CNS, Fgf8/17/18 is also expressed in C. intestinalis in mesodermal cells lateral and anterior to the CNS expression domain, as well as cells at the tip of the tail (Imai et al., ‘02, Ikuta and Saiga, ‘07). The relationship of these two urochordate domains to similar domains in the vertebrate somitic and unsegmented mesodermal expression and tail bud domain is uncertain. In the cephalochordate amphioxus, Fgf8/17/18 is expressed transiently in the rostral central nervous system and in the pharyngeal endoderm, which are likely conserved domains with vertebrates (Muelemans and Bonner-Fraser, ‘07). A substantial number of morphological innovations or elaborations occurred after the divergence of urochordates and vertebrates but before the divergence of bony fishes into ray-fin and lobe-fin clades. These include the elaboration of neural crest and placodes, the invention of bone, and the origin of paired appendages (Gans and Northcutt, ‘83, Bassham and Postlethwait, ‘05, Jeffery, ‘06, Sauka-Spengler and Bronner-Fraser, ‘08). Also occurring at about this time period were two rounds of whole genome duplication, R1 and R2 (Kortschak et al., ‘01, Ornitz and Itoh, ‘01, Lundin et al. ‘03, Dehal and Boore, ‘05, Garcia-Fernandez, ‘05, Bourlat et al., ‘06, Delsuc et al., ‘06, Guder et al., ‘06, Jacob and Lum, ‘07, Kitisin et al., ‘07, Sundstrom et al., ‘08, Yu et al., ‘08) that we show here are highly likely to have produced the four FgfD ohnologs (Fgf8, Fgf17, Fgf18, and Fgf24). Any expression domain that appears in the orthologs of three or four of these four genes in at least one of the three species stickleback, zebrafish, and mouse, is highly likely to have been a site of expression in the last common ancestor of all bony fishes.
In addition to the MHB and somite/unsegmented mesoderm, at least one of the three investigated vertebrates (stickleback, zebrafish, mouse) expresses each FgfD gene in the retina, pharyngeal arches, somites, and otic vesicle (Suppl Fig. S4). We conclude that these expression domains were probably present in the four orthologous FgfD genes in the last common ancestor of ray-fin and lobe-fin fish and were differentially lost from various genes in different lineages. Because these domains were in common in all four FgfD genes, we infer that they were also in common in the unduplicated FgfD gene in the lineage leading to vertebrates before R1 and R2. An alternative hypothesis is that these shared expression domains evolved independently but convergently multiple times, a possibility that seems unlikely based on parsimony. If the hypothesis is true that expression domains shared by all four FgfD genes were present in the last pre-R1-duplication FgfD gene, then that vertebrate ancestor would have had expression domains appropriate for eyes, ears, pharyngeal arches, somites, and MHB. Available data for FgfD expression patterns in cephalochordates and urochordates do not contradict this explanation (Imai et al., ‘02, Ikuta and Saiga, ‘07); Muelemans and Bonner-Fraser, ‘07).
Expression domains in the dorsal diencephalon and the apical epidermal ridge (AER) of the pectoral fin/limb bud are shared by Fgf8 and Fgf24 (Figs. 6, 7). Furthermore, the examination of mutant phenotypes shows that Fgf8 is required for paired pectoral appendage development in tetrapods, which lack an ortholog of fgf24, and reciprocally, fgf24 is required for paired pectoral appendage development in zebrafish but fgf8a is not (Reifers et al., ‘98, Lewandoski et al., ‘00, Draper et al., ‘03) (fgf8b is not expressed in the AER of fish pectoral appendages (Jovelin et al., ‘07)). If the R1, R2 evolutionary history of the four FgfD genes is correct, then the pre-duplication FgfD gene should have had an expression domain corresponding to the dorsal diencephalon and AER domains in modern vertebrates. According to this hypothesis, the last ancestor before the vertebrate genome duplications would thus have had an expression domain for the AER, even though, paradoxically, it is unlikely that that organism had paired appendages (Coates, ‘94). Thus, the ancestral function of this expression domain would have been co-opted into appendage development after paired appendages began to evolve in early vertebrates, and it may have been used in the development of unpaired midline fins (Freitas et al., ‘06). Alternatively, one or both of these expression domains could have evolved convergently as neofunctionalizations by the two genes Fgf8 and Fgf24 well after the R1 and R2 events during the evolution of paired fins.
Some additional expression domains are displayed by just two of the four FgfD genes (Suppl Fig. 4). Expression in tailbud is shared by Fgf8 and Fgf17 and in unsegmented mesoderm and pancreas by Fgf18 and Fgf24. This distribution might be predicted by the (Fgf8, Fgf17)(Fgf18, Fgf24) lineage hypothesis if these domains had been a part of the repertoire of the Fgf8/17 and Fgf18/24 genes after the R1 whole genome duplication event. As mentioned above, the tailbud expression domain may be related to the expression domain at the tip of the tail in Ciona and inherited from stem olfactores (Imai et al., ‘02, Ikuta and Saiga, ‘07). In addition, both Fgf8 and Fgf18 share expression in the heart and Fgf17 is expressed in zebrafish heart regeneration, consistent with either inheritance from the FgfD gene of stem olfactores followed by loss in the Fgf24 clade or, less parsimoniously, independent acquisition by convergent evolution.
Several expression domains appear only on single clades in the tree. These include segmental plate only in teleost fgf17 genes, telencephalon only in Fgf8 of teleosts and tetrapods, olfactory epithelium only in teleost fgf8 genes, and spinal neurons only in fgf18a of teleosts. The most parsimonious explanation of these domains is that they represent neofunctionalization events in the corresponding clades.
Correlation of CNEs and expression domains
Because many CNEs are shared by fgf8a and other zebrafish FgfD genes (Fig. 10C), we can attempt to correlate individual CNEs with expression domains in embryonic zebrafish that are summarized in Supplementary Fig. 4. The first conclusion is that none of the CNEs identified was unique for a single tissue. CNE q is shared only by fgf8a, fgf18a, and fgf24, and only these genes express in the olfactory organ, making this a candidate regulatory element for contributing to olfactory expression, although genes possessing CNE q are expressed in tissues in addition to the olfactory epithelium. The dorsal diencephalon expresses only fgf8a, fgf17, and fgf24, and these genes share CNEs labeled b, d, i, k, l, n, and w. (Fig. 10C); of these shared elements, i, k, l, and w are absent from paralogs that do not show expression in the dorsal diencephalon, so these are candidate CNEs for positive regulatory elements contributing to expression in the dorsal diencephalon of zebrafish embryos. Elements e, g, h, p, t, x, and z are found in both fgf8a and fgf24 and in none of the other fgfD genes, and only these two genes are expressed in the aer at 2 dpf, making these elements candidates for controlling expression in the apical ectodermal ridge of the pectoral fin bud. For the retina at 16hpf, only fgf18b lacks expression, and element d is the only element possessed by all zebrafish fgfD genes except fgf18b, and so element d is a candidate for the early expression in the retina. Other factors may turn on fgf18b in the retina at pharyngula stages. Genes expressed in the heart and the spinal cord neurons have in common the same set of elements (b, c, d, m, n, q, r, s, u, and v). The unsegmented mesenchyme, segmental plate, and pancreas each have unique CNE combinations not shared with other tissues. Because vertebrates can conserve regulatory function without conserving sequence similarity (Fisher et al., ‘06), the significance of these conserved non-coding regions must be tested by experiment to draw firm conclusions.
CNEs among paralogs reflect the evolutionary history of the FgfD subfamily
Gene fates following duplication include the loss of one of the two paralogs by nonfunctionalization, or the divergence of gene function by the acquisition of new function(s) (neofunctionalization) (Ohno, ‘70) or the partitioning of ancestral functions (subfunctionalization) (Force et al., ‘99, Hughes, ‘99, He and Zhang, ‘05). Subfunction partitioning occurs by the complementary fixation of otherwise deleterious mutations in ancestral control elements of gene duplicates. This model essentially follows neutral evolution, and therefore one might expect that subfunction partitioning should increase with the time of coexistence between duplicates within a lineage. Consequently, the Duplication-Degeneration-Complementation (DDC) model predicts that more ancestral functions should have been partitioned between paralogs that have coexisted over long periods of evolutionary time than in cases where duplicates have existed for a shorter time period or where duplicates rapidly became reduced to single copy following the duplication event.
Examples of subfunction partitioning can be found for a variety of gene duplicates (e.g. Lister et al., ‘01, Cresko et al., ‘03, Liu et al., ‘05). The FgfD subfamily in teleosts provides nevertheless a rare opportunity to test predictions of the DDC model because its members share an origin through the R1, R2, and R3 genome duplication events, and because since R3, different FgfD subfamily members have spent different amounts of time as duplicate copies in various lineages. The fgf24 and fgf17 genes apparently became single copy before the divergence of zebrafish and stickleback lineages; the fgf18a and fgf18b paralogs coexisted from R3 to the present in zebrafish, but became single copy in the stickleback lineage after it diverged from the zebrafish lineage; and the fgf8a and fgf8b paralogs have co-existed from R3 to the present in several lineages. In addition, conserved non-coding elements (CNEs) identified using phylogenetic footprinting in the proximity of the Fgf8 locus have been shown to have regulatory activity in mouse (Beermann et al., ‘06) and in zebrafish (Inoue et al., ‘06). Moreover, regulatory modules of fgf8 function extend to intronic sequences of fgf8 neighbors (Kikuta et al., ‘07). It is therefore likely that CNEs identified around other Fgf genes have a functional role as well.
The large number of long CNEs detected among the three human FGFD genes contrasts with the fewer shorter CNEs found in zebrafish and stickleback. What could be responsible for this difference? It is not mere antiquity because the last common ancestor of the zebrafish fgf8 and fgf18a genes was in fact the same gene as the last common ancestor of the human FGF8 and FGF18 genes. Thus, either the human lineage preserved more broad CNEs than teleost lineages, perhaps amplifying them locally by tandem duplication, or the zebrafish lineage lost more CNEs after the divergence of human and zebrafish lineages. The pattern of just a few, short, but nevertheless well-conserved CNEs within stickleback and zebrafish genomes may relate to the additional whole-genome duplication in the teleost lineage not suffered by the tetrapod lineage (R3). Post-R3 fgfD paralogs may have experienced greater relaxation of selective constraints than post-R2 FgfD genes. After both events, we would expect the partitioning of expression domains by fixation of complementary deleterious mutations in regulatory elements (Force et al., ‘99, Lynch and Force, ‘00).
Following R3, one fgf18 duplicate was lost after the divergence of the lineages leading to zebrafish and stickleback resulting in one fgf18 gene in stickleback, medaka and pufferfish while zebrafish retained two fgf18 duplicates. Analysis of CNEs in the proximity of the stickleback fgf18 locus shows extensive conservation between stickleback and other percomorph fish (Fig. 12), suggesting that the integrity of regulatory elements in this genomic region is important for fgf18 function and has been maintained by selection in species having a single fgf18 gene. In contrast, fewer CNEs are shared between zebrafish and the percomorphs, and most CNEs have been partitioned between fgf18a and fgf18b in zebrafish. This pattern of shared and partitioned CNEs makes two predictions. First, the amount and level of conservation among CNEs surrounding fgf18 in percomorphs suggest that little subfunction partitioning may have occurred before the second fgf18 duplicate was lost in this lineage and thus fgf18 in these species may have retained most of the ancestral pre-R3 fgf18 functions. Second, the pattern of shared CNEs between fgf18a and fgf18b in zebrafish, where both duplicates have been maintained over longer evolutionary time, suggests the partitioning of ancestral pre-R3 fgf18 functions between paralogs. Supporting these predictions, stickleback fgf18 is expressed in the heart field precursor and in the MHB at mid-segmentation, while in zebrafish, these expression domains are partitioned between fgf18a and fgf18b, respectively (Fig. 6). At the long pec stage, expression of stickleback fgf18 in the olfactory epithelium and in the MHB is partitioned between zebrafish fgf18a and fgf18b (Fig. 7). Similarly, at the early pharyngula stage, expression in the spinal chord neurons partitioned to zfgf18a (Suppl. Fig. S4). Specifically, one or more of elements ‘a’, ‘f’, and ‘r’ in Figure 12 are candidates for expression of fgf18b in the MHB, and elements ‘b–e’, ‘g–j’, and ‘o–q’ are candidates for expression of fgf18a in the nose, spinal neurons, and pancreas, assuming all regulatory elements act positively, although, of course some may act to diminish expression in certain tissues.
Sequenced teleost genomes have a single fgf24 and fgf17 gene, indicating that one duplicate of each locus was lost following R3, before the divergence of the lineages leading to zebrafish and other fish species. The extensive number and length of CNEs around all fgf24 orthologs also supports this hypothesis and suggests that regulatory control elements of fgf24 have been maintained by selection (Fig. 13). By contrast, the lower number of CNEs surrounding Fgf17 genes may be mechanistically related to the multiple local chromosome rearrangements disrupting syntenies of both CNEs and Fgf17 neighboring genes between teleosts and human and within teleosts (Figs. 5, 11). CNEs detached from their target genes by chromosomal rearrangements are expected to be lost by genetic drift (Kikuta et al., ‘07). For instance, it is noteworthy that Fgf17 expression is the least conserved between mouse and teleosts among FgfD members (Suppl. Fig. S4). The expression patterns of fgf17 orthologs in stickleback and zebrafish, however, are well conserved, suggesting that important ancestral regulatory elements may have been retained in both lineages, and CNEs ‘a–e’ in Figure 11 are candidates for such elements.
Because one fgf24 duplicate was lost before the divergence of zebrafish and percomorph lineages and one fgf18 duplicate was lost after these lineages diverged, fgf24 duplicates originating from R3 have coexisted over shorter evolutionary time than fgf18 duplicates originating from the same whole-genome duplication before one copy of each gene was lost. Thus, we would expect the functions of fgf24 orthologs to be more conserved than those of fgf18 orthologs. Indeed, not only does zfgf24 share more and longer CNEs with its orthologs than zfgf18a and zfgf18b share with their orthologs (Figs. 12; 13) but zfgf24 and sfgf24 expression domains are remarkably conserved with only minor differences in the retina and MHB (Figs. 6S–V; 7J–K; 8S–V) while fgf18 orthologs show expression differences in somites, retina and ear (Figs. 6N, P, R; 8M–R).
In summary, this comparison of CNEs and expression domains verifies predictions made by the molecular mechanisms of subfunction partitioning and the DDC model (Force et al., ‘99) for all teleost fgfD members for which synteny has not been disrupted by local chromosomal rearrangements. Additionally, these results identify a clear relationship, predicted by the subfunctionalization model, between the length of time that paralogs coexist and their distribution of both CNEs and expression domains. Importantly, this work identifies a number of CNEs that are candidates for regulatory elements for specific expression domains in development. The testing of these domains for developmental functions is an ongoing project.
Supplementary Material
Supplementary Figure 1. Phylogenetic tree for NPM-family proteins. Results show that npm4 in teleosts has no apparent human ortholog. Sequences were obtained from Ensembl and NCBI and aligned by MUSCLE (Edgar, ‘04). Maximum likelihood-based phylogenetic inference analysis was conducted using PHYML (Guindon et al., ‘05). Abbreviations: Cin, Ciona intestinalis; Dre, Danio rerio zebrafish; Gac, Gasterosteus aculeatus stickleback; Hsa, Homo sapiens human. Accession numbers: npm1_Dre_NP_955460; npm1_Gac_ENSGACT00000026899; npm2a_Dre_OTTDART00000035953; npm2a_Gac_ENSGACT00000022099; npm2b_Dre_NP_001116479+AAI22198; npm3_Dre__NP_001013502; npm3_Gac_ENSGACT00000009682; npm4_Dre_XP_692942; npm_Cin_XP_002122436; NPM1_Hsa_NP_002511; NPM2_Hsa_NP_877724; NPM3_Hsa_NP_008924.
Supplementary Figure 2. Phylogenetic tree for Kazald-family proteins with IGFBP7 as outgroup shows a gene that can be called kazald2 that is missing from tetrapod genomes. Abbreviations and accession numbers: Bbe, Branchiostoma belcheri amphioxus; Dre, Danio rerio zebrafish; Gga, Gallus gallus chicken; Hsa, Homo sapiens human; Mmu, Mus musculus mouse; Tni, Tetraodon nigroviridis pufferfish; Xtr, Xenopus tropicalis frog. kazald1_Hsa_EAW49782; Kazald1_Mmu_NP_849260; MGC80370_Gga_XP_421724; Kazald1_Xtr_NP_001093733; LOC568748_Dre_NP_001098594; ENSDARG00000063176_Dre_NP_998089; Unn1_Tni_CAF94984; IGFBP7_Hsa_AAA16187; Igfbp7_Mmu_AAH47202; Unn2_Tni_CAG08989; LOC100000101_Dre_NP_001116791; PSF_Gga_XP_420577; ilgfbp_Bbe_BAB97382.
Supplementary Figure S3. Elephant shark possesses Fgf24. A. Alignment of exon 3 in vertebrate FgfD genes. Black rectangles highlight amino acid positions that characterize Fgf18 and Fgf24. Cin, Ciona intestinalis (sea squirt); Cmi, Callorhinchus mili (elephant shark); Dre, Danio rerio (zebrafish); Fru, Fugu rubripes (pufferfish); Gac, Gasterosteus aculeatus (stickleback); Hsa, Homo sapiens (human); Mmu, Mus musculus (mouse); Ola, Oryzias latipes (medakafish). B. Phylogenetic tree of exon 3 portions of FgfD genes including the shark sequences.
Supplementary Figure S4. Phylogenetic relationship among vertebrate FgfD genes and their expression patterns in mouse, zebrafish and stickleback. Abbreviations: aer, apical ectodermal ridge of pectoral fin; dd, dorsal diencephalon; h, heart; mhb, midbrain-hindbrain boundary; nc, neural crest;n, nose; oe, oral ectoderm; ov, otic vesicle; pan, pancreas; pf, pectoral fin; r, retina; s, somite; sp, segmental plate; spn, spinal cord neuron; t, telencephalon; tb, tail bud; um, unsegmented mesenchyme. stage1, segmentation; stage2, pharyngula.
Acknowledgments
We thank Dong Liu for discussions on ear morphology, Amanda Rapp, Tim Mason and the University of Oregon Zebrafish Facility for providing animals and excellent fish care, and Poh Kheng Loi and Amber Selix of the Histology Facility for sectioning. This work was supported by the National Center for Research Resources (5R01RR020833) and National Institutes of Health (P01 HD22486); the contents of this study are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH. RJ is an Associate of the IGERT Training Program for Development, Evolution, and Genomics, NSF DGE-9972830.
References
- Abu-Issa R, Smyth G, Smoak I, Yamamura K, Meyers EN. Fgf8 is required for pharyngeal arch and cardiovascular development in the mouse. Development. 2002;129:4613–4625. doi: 10.1242/dev.129.19.4613. [DOI] [PubMed] [Google Scholar]
- Amemiya CT, Prohaska SJ, Hill-Force A, Cook A, Wasserscheid J, Ferrier DE, Pascual-Anaya J, Garcia-Fernandez J, Dewar K, Stadler PF. The amphioxus Hox cluster: characterization, comparative genomics, and evolution. J Exp Zoolog B Mol Dev Evol. 2008;310:465–477. doi: 10.1002/jez.b.21213. [DOI] [PubMed] [Google Scholar]
- Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, Westerfield M, Ekker M, Postlethwait JH. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Andermann P, Weinberg ES. Expression of zTlxA, a Hox11-like gene, in early differentiating embryonic neurons and cranial sensory ganglia of the zebrafish embryo. Dev Dyn. 2001;222:595–610. doi: 10.1002/dvdy.1239. [DOI] [PubMed] [Google Scholar]
- Bassham S, Postlethwait JH. The evolutionary history of placodes: a molecular genetic investigation of the larvacean urochordate Oikopleura dioica. Development. 2005;132:4259–4272. doi: 10.1242/dev.01973. [DOI] [PubMed] [Google Scholar]
- Beermann F, Kaloulis K, Hofmann D, Murisier F, Bucher P, Trumpp A. Identification of evolutionarily conserved regulatory elements in the mouse Fgf8 locus. Genesis. 2006;44:1–6. doi: 10.1002/gene.20177. [DOI] [PubMed] [Google Scholar]
- Bershtein S, Tawfik DS. Ohno’s model revisited: measuring the frequency of potentially adaptive mutations under various mutational drifts. Mol Biol Evol. 2008;25:2311–2318. doi: 10.1093/molbev/msn174. [DOI] [PubMed] [Google Scholar]
- Blair JE, Hedges SB. Molecular phylogeny and divergence times of deuterostome animals. Mol Biol Evol. 2005;22:2275–2284. doi: 10.1093/molbev/msi225. [DOI] [PubMed] [Google Scholar]
- Boulet AM, Moon AM, Arenkiel BR, Capecchi MR. The roles of Fgf4 and Fgf8 in limb bud initiation and outgrowth. Developmental Biology. 2004;273:361–372. doi: 10.1016/j.ydbio.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Bourlat SJ, Juliusdottir T, Lowe CJ, Freeman R, Aronowicz J, Kirschner M, Lander ES, Thorndyke M, Nakano H, Kohn AB, Heyland A, Moroz LL, Copley RR, Telford MJ. Deuterostome phylogeny reveals monophyletic chordates and the new phylum Xenoturbellida. Nature. 2006;444:85–88. doi: 10.1038/nature05241. [DOI] [PubMed] [Google Scholar]
- Brand M, Heisenberg CP, Jiang YJ, Beuchle D, Lun K, Furutani-Seiki M, Granato M, Haffter P, Hammerschmidt M, Kane DA, Kelsh RN, Mullins MC, Odenthal J, van Eeden FJ, Nusslein-Volhard C. Mutations in zebrafish genes affecting the formation of the boundary between midbrain and hindbrain. Development. 1996;123:179–190. doi: 10.1242/dev.123.1.179. [DOI] [PubMed] [Google Scholar]
- Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, Green ED, Sidow A, Batzoglou S. LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 2003;13:721–731. doi: 10.1101/gr.926603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canestro C, Bassham S, Postlethwait J. Development of the central nervous system in the larvacean Oikopleura dioica and the evolution of the chordate brain. Dev Biol. 2005;285:298–315. doi: 10.1016/j.ydbio.2005.06.039. [DOI] [PubMed] [Google Scholar]
- Cao Y, Zhao J, Sun Z, Zhao Z, Postlethwait J, Meng A. fgf17b, a novel member of Fgf family, helps patterning zebrafish embryos. Developmental Biology. 2004;271:130–143. doi: 10.1016/j.ydbio.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Catchen JM, Conery JS, Postlethwait JH. Automated identification of conserved synteny after whole genome duplication. Genome Research; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chain FJ, Ilieva D, Evans BJ. Duplicate gene evolution and expression in the wake of vertebrate allopolyploidization. BMC Evol Biol. 2008;8:43. doi: 10.1186/1471-2148-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholfin JA, Rubenstein JL. Patterning of frontal cortex subdivisions by Fgf17. Proc Natl Acad Sci U S A. 2007;104:7652–7657. doi: 10.1073/pnas.0702225104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffels A, Koh EG, Chia JM, Brenner S, Aparicio S, Venkatesh B. Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. Molecular Biology and Evolution. 2004;21:1146–1151. doi: 10.1093/molbev/msh114. [DOI] [PubMed] [Google Scholar]
- Coates MI. The origin of vertebrate limbs. Dev Suppl. 1994:169–180. [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet. 2008;9:938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- Cresko WA, Yan YL, Baltrus DA, Amores A, Singer A, Rodriguez-Mari A, Postlethwait JH. Genome duplication, subfunction partitioning, and lineage divergence: Sox9 in stickleback and zebrafish. Developmental Dynamics. 2003;228:480–489. doi: 10.1002/dvdy.10424. [DOI] [PubMed] [Google Scholar]
- Crow KD, Stadler PF, Lynch VJ, Amemiya C, Wagner GP. The “fish-specific” Hox cluster duplication is coincident with the origin of teleosts. Molecular Biology and Evolution. 2006;23:121–136. doi: 10.1093/molbev/msj020. [DOI] [PubMed] [Google Scholar]
- Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Tsagkogeorga G, Lartillot N, Philippe H. Additional molecular support for the new chordate phylogeny. Genesis. 2008;46:592–604. doi: 10.1002/dvg.20450. [DOI] [PubMed] [Google Scholar]
- Dichmann DS, Miller CP, Jensen J, Scott Heller R, Serup P. Expression and misexpression of members of the FGF and TGFbeta families of growth factors in the developing mouse pancreas. Dev Dyn. 2003;226:663–674. doi: 10.1002/dvdy.10270. [DOI] [PubMed] [Google Scholar]
- Dominguez MH, Rakic P. Neuroanatomy of the FGF system. J Comp Neurol. 2008;509:141–143. doi: 10.1002/cne.21748. [DOI] [PubMed] [Google Scholar]
- Draper BW, Stock DW, Kimmel CB. Zebrafish fgf24 functions with fgf8 to promote posterior mesodermal development. Development. 2003;130:4639–4654. doi: 10.1242/dev.00671. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer S, Draper BW, Neumann CJ. The zebrafish fgf24 mutant identifies an additional level of Fgf signaling involved in vertebrate forelimb initiation. Development. 2003;130:3515–3524. doi: 10.1242/dev.00537. [DOI] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, McCallion AS. Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science. 2006;312:276–279. doi: 10.1126/science.1124070. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DU, Fotheringham LK, Brewer JA, Muglia LJ, Tristani-Firouzi M, Capecchi MR, Moon AM. An Fgf8 mouse mutant phenocopies human 22q11 deletion syndrome. Development. 2002;129:4591–4603. doi: 10.1242/dev.129.19.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas R, Zhang G, Cohn MJ. Evidence that mechanisms of fin development evolved in the midline of early vertebrates. Nature. 2006;442:1033–1037. doi: 10.1038/nature04984. [DOI] [PubMed] [Google Scholar]
- Gans C, Northcutt RG. Neural crest and the origin of vertebrates: a new head. Science. 1983;220:268–274. doi: 10.1126/science.220.4594.268. [DOI] [PubMed] [Google Scholar]
- Garcia-Fernandez J. The genesis and evolution of homeobox gene clusters. Nat Rev Genet. 2005;6:881–892. doi: 10.1038/nrg1723. [DOI] [PubMed] [Google Scholar]
- Guder C, Philipp I, Lengfeld T, Watanabe H, Hobmayer B, Holstein TW. The Wnt code: cnidarians signal the way. Oncogene. 2006;25:7450–7460. doi: 10.1038/sj.onc.1210052. [DOI] [PubMed] [Google Scholar]
- Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML Online--a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005;33:W557–559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamade A, Deries M, Begemann G, Bally-Cuif L, Genet C, Sabatier F, Bonnieu A, Cousin X. Retinoic acid activates myogenesis in vivo through Fgf8 signalling. Dev Biol. 2006;289:127–140. doi: 10.1016/j.ydbio.2005.10.019. [DOI] [PubMed] [Google Scholar]
- He X, Zhang J. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics. 2005;169:1157–1164. doi: 10.1534/genetics.104.037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB. The origin and evolution of model organisms. Nat Rev Genet. 2002;3:838–849. doi: 10.1038/nrg929. [DOI] [PubMed] [Google Scholar]
- Hoegg S, Brinkmann H, Taylor JS, Meyer A. Phylogenetic timing of the fish-specific genome duplication correlates with the diversification of teleost fish. Journal of Molecular Evolution. 2004;59:190–203. doi: 10.1007/s00239-004-2613-z. [DOI] [PubMed] [Google Scholar]
- Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, Down T, Dyer SC, Fitzgerald S, Fernandez-Banet J, Graf S, Haider S, Hammond M, Herrero J, Holland R, Howe K, Johnson N, Kahari A, Keefe D, Kokocinski F, Kulesha E, Lawson D, Longden I, Melsopp C, Megy K, Meidl P, Ouverdin B, Parker A, Prlic A, Rice S, Rios D, Schuster M, Sealy I, Severin J, Slater G, Smedley D, Spudich G, Trevanion S, Vilella A, Vogel J, White S, Wood M, Cox T, Curwen V, Durbin R, Fernandez-Suarez XM, Flicek P, Kasprzyk A, Proctor G, Searle S, Smith J, Ureta-Vidal A, Birney E. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL. Adaptive evolution of genes and genomes. New York: Oxford Univ. Press; 1999. [Google Scholar]
- Ikuta T, Saiga H. Dynamic change in the expression of developmental genes in the ascidian central nervous system: revisit to the tripartite model and the origin of the midbrain-hindbrain boundary region. Dev Biol. 2007;312:631–643. doi: 10.1016/j.ydbio.2007.10.005. [DOI] [PubMed] [Google Scholar]
- Ilagan R, Abu-Issa R, Brown D, Yang YP, Jiao K, Schwartz RJ, Klingensmith J, Meyers EN. Fgf8 is required for anterior heart field development. Development. 2006;133:2435–2445. doi: 10.1242/dev.02408. [DOI] [PubMed] [Google Scholar]
- Imai KS, Satoh N, Satou Y. Region specific gene expressions in the central nervous system of the ascidian embryo. Mech Dev. 2002;119(Suppl 1):S275–277. doi: 10.1016/s0925-4773(03)00128-x. [DOI] [PubMed] [Google Scholar]
- Inoue F, Nagayoshi S, Ota S, Islam ME, Tonou-Fujimori N, Odaira Y, Kawakami K, Yamasu K. Genomic organization, alternative splicing, and multiple regulatory regions of the zebrafish fgf8 gene. Development, Growth and Differentiation. 2006;48:447–462. doi: 10.1111/j.1440-169X.2006.00882.x. [DOI] [PubMed] [Google Scholar]
- Itoh N. The Fgf families in humans, mice, and zebrafish: their evolutional processes and roles in development, metabolism, and disease. Biol Pharm Bull. 2007;30:1819–1825. doi: 10.1248/bpb.30.1819. [DOI] [PubMed] [Google Scholar]
- Itoh N, Konishi M. The zebrafish fgf family. Zebrafish. 2007;4:179–186. doi: 10.1089/zeb.2007.0509. [DOI] [PubMed] [Google Scholar]
- Itoh N, Ornitz DM. Evolution of the Fgf and Fgfr gene families. Trends Genet. 2004;20:563–569. doi: 10.1016/j.tig.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Jacob L, Lum L. Hedgehog signaling pathway. Sci STKE. 2007;2007:cm6. doi: 10.1126/stke.4072007cm6. [DOI] [PubMed] [Google Scholar]
- Jeffery WR. Ascidian neural crest-like cells: phylogenetic distribution, relationship to larval complexity, and pigment cell fate. J Exp Zoolog B Mol Dev Evol. 2006;306:470–480. doi: 10.1002/jez.b.21109. [DOI] [PubMed] [Google Scholar]
- Jovelin R, He X, Amores A, Yan YL, Shi R, Qin B, Roe B, Cresko WA, Postlethwait JH. Duplication and divergence of fgf8 functions in teleost development and evolution. J Exp Zoolog B Mol Dev Evol. 2007;308B:730–743. doi: 10.1002/jez.b.21193. [DOI] [PubMed] [Google Scholar]
- Joyner AL. Engrailed, Wnt and Pax genes regulate midbrain--hindbrain development. Trends Genet. 1996;12:15–20. doi: 10.1016/0168-9525(96)81383-7. [DOI] [PubMed] [Google Scholar]
- Kasbauer T, Towb P, Alexandrova O, David CN, Dall’armi E, Staudigl A, Stiening B, Bottger A. The Notch signaling pathway in the cnidarian Hydra. Dev Biol. 2007;303:376–390. doi: 10.1016/j.ydbio.2006.11.022. [DOI] [PubMed] [Google Scholar]
- Kikuta H, Laplante M, Navratilova P, Komisarczuk AZ, Engstrom PG, Fredman D, Akalin A, Caccamo M, Sealy I, Howe K, Ghislain J, Pezeron G, Mourrain P, Ellingsen S, Oates AC, Thisse C, Thisse B, Foucher I, Adolf B, Geling A, Lenhard B, Becker TS. Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Res. 2007;17:545–555. doi: 10.1101/gr.6086307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Developmental Dynamics. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kitisin K, Saha T, Blake T, Golestaneh N, Deng M, Kim C, Tang Y, Shetty K, Mishra B, Mishra L. Tgf-Beta signaling in development. Sci STKE. 2007;2007:cm1. doi: 10.1126/stke.3992007cm1. [DOI] [PubMed] [Google Scholar]
- Kortschak RD, Tamme R, Lardelli M. Evolutionary analysis of vertebrate Notch genes. Dev Genes Evol. 2001;211:350–354. doi: 10.1007/s004270100159. [DOI] [PubMed] [Google Scholar]
- Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- Langenau DM, Palomero T, Kanki JP, Ferrando AA, Zhou Y, Zon LI, Look AT. Molecular cloning and developmental expression of Tlx (Hox11) genes in zebrafish (Danio rerio) Mech Dev. 2002;117:243–248. doi: 10.1016/s0925-4773(02)00187-9. [DOI] [PubMed] [Google Scholar]
- Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, Burns CG, Poss KD. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- Lewandoski M, Sun X, Martin GR. Fgf8 signalling from the AER is essential for normal limb development. Nature Genetics. 2000;26:460–463. doi: 10.1038/82609. [DOI] [PubMed] [Google Scholar]
- Lister JA, Close J, Raible DW. Duplicate mitf genes in zebrafish: complementary expression and conservation of melanogenic potential. Developmental Biology. 2001;237:333–344. doi: 10.1006/dbio.2001.0379. [DOI] [PubMed] [Google Scholar]
- Liu RZ, Sharma MK, Sun Q, Thisse C, Thisse B, Denovan-Wright EM, Wright JM. Retention of the duplicated cellular retinoic acid-binding protein 1 genes (crabp1a and crabp1b) in the zebrafish genome by subfunctionalization of tissue-specific expression. The FEBS Journal. 2005;272:3561–3571. doi: 10.1111/j.1742-4658.2005.04775.x. [DOI] [PubMed] [Google Scholar]
- Liu Z, Xu J, Colvin JS, Ornitz DM. Coordination of chondrogenesis and osteogenesis by fibroblast growth factor 18. Genes & Development. 2002;16:859–869. doi: 10.1101/gad.965602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin LG, Larhammar D, Hallbook F. Numerous groups of chromosomal regional paralogies strongly indicate two genome doublings at the root of the vertebrates. J Struct Funct Genomics. 2003;3:53–63. [PubMed] [Google Scholar]
- Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch VJ, Wagner GP. Multiple chromosomal rearrangements structured the ancestral vertebrate Hox-bearing protochromosomes. PLoS Genet. 2009;5:e1000349. doi: 10.1371/journal.pgen.1000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus DQ, Thomsen GH, Martindale MQ. FGF signaling in gastrulation and neural development in Nematostella vectensis, an anthozoan cnidarian. Dev Genes Evol. 2007;217:137–148. doi: 10.1007/s00427-006-0122-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- Meulemans D, Bronner-Fraser M. Insights from amphioxus into the evolution of vertebrate cartilage. PLoS ONE. 2007;2:e787. doi: 10.1371/journal.pone.0000787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A, Schartl M. Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Current Opinion in Cell Biology. 1999;11:699–704. doi: 10.1016/s0955-0674(99)00039-3. [DOI] [PubMed] [Google Scholar]
- Meyer A, Van de Peer Y. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD) Bioessays. 2005;27:937–945. doi: 10.1002/bies.20293. [DOI] [PubMed] [Google Scholar]
- Moon AM, Capecchi MR. Fgf8 is required for outgrowth and patterning of the limbs. Nature Genetics. 2000;26:455–459. doi: 10.1038/82601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naruse K, Tanaka M, Mita K, Shima A, Postlethwait J, Mitani H. A medaka gene map: the trace of ancestral vertebrate proto-chromosomes revealed by comparative gene mapping. Genome Research. 2004;14:820–828. doi: 10.1101/gr.2004004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols SA, Dirks W, Pearse JS, King N. Early evolution of animal cell signaling and adhesion genes. Proc Natl Acad Sci U S A. 2006;103:12451–12456. doi: 10.1073/pnas.0604065103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Wada H, Tagawa K, Akiyama-Oda Y, Satoh N, Humphreys T, Zhang S, Tsukita S. A novel amphioxus cadherin that localizes to epithelial adherens junctions has an unusual domain organization with implications for chordate phylogeny. Evol Dev. 2002;4:426–434. doi: 10.1046/j.1525-142x.2002.02031.x. [DOI] [PubMed] [Google Scholar]
- Ohbayashi N, Shibayama M, Kurotaki Y, Imanishi M, Fujimori T, Itoh N, Takada S. FGF18 is required for normal cell proliferation and differentiation during osteogenesis and chondrogenesis. Genes & Development. 2002;16:870–879. doi: 10.1101/gad.965702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. New-York: Springer-Verlag; 1970. [Google Scholar]
- Ohuchi H, Kimura S, Watamoto M, Itoh N. Involvement of fibroblast growth factor (FGF)18-FGF8 signaling in specification of left-right asymmetry and brain and limb development of the chick embryo. Mech Dev. 2000;95:55–66. doi: 10.1016/s0925-4773(00)00331-2. [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Itoh N. Fibroblast growth factors. Genome Biol. 2001;2:REVIEWS3005. doi: 10.1186/gb-2001-2-3-reviews3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson KJ, Lyons JB, Nowak KS, Takacs CM, Wargo MJ, McPeek MA. Estimating metazoan divergence times with a molecular clock. Proc Natl Acad Sci U S A. 2004;101:6536–6541. doi: 10.1073/pnas.0401670101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovici C, Roubin R, Coulier F, Birnbaum D. An evolutionary history of the FGF superfamily. Bioessays. 2005;27:849–857. doi: 10.1002/bies.20261. [DOI] [PubMed] [Google Scholar]
- Postlethwait J, Amores A, Cresko W, Singer A, Yan YL. Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends in Genetics. 2004;20:481–490. doi: 10.1016/j.tig.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH. The zebrafish genome in context: ohnologs gone missing. J Exp Zoolog B Mol Dev Evol. 2007;308:563–577. doi: 10.1002/jez.b.21137. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH, Yan YL, Gates MA, Horne S, Amores A, Brownlie A, Donovan A, Egan ES, Force A, Gong Z, Goutel C, Fritz A, Kelsh R, Knapik E, Liao E, Paw B, Ransom D, Singer A, Thomson M, Abduljabbar TS, Yelick P, Beier D, Joly JS, Larhammar D, Rosa F, Westerfield M, Zon LI, Johnson SL, Talbot WS. Vertebrate genome evolution and the zebrafish gene map. Nature Genetics. 1998;18:345–349. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- Reifers F, Adams J, Mason IJ, Schulte-Merker S, Brand M. Overlapping and distinct functions provided by fgf17, a new zebrafish member of the Fgf8/17/18 subgroup of Fgfs. Mechanisms of Development. 2000a;99:39–49. doi: 10.1016/s0925-4773(00)00475-5. [DOI] [PubMed] [Google Scholar]
- Reifers F, Bohli H, Walsh EC, Crossley PH, Stainier DY, Brand M. Fgf8 is mutated in zebrafish acerebellar (ace) mutants and is required for maintenance of midbrain-hindbrain boundary development and somitogenesis. Development. 1998;125:2381–2395. doi: 10.1242/dev.125.13.2381. [DOI] [PubMed] [Google Scholar]
- Reifers F, Walsh EC, Leger S, Stainier DY, Brand M. Induction and differentiation of the zebrafish heart requires fibroblast growth factor 8 (fgf8/acerebellar) Development. 2000b;127:225–235. doi: 10.1242/dev.127.2.225. [DOI] [PubMed] [Google Scholar]
- Sato T, Joyner AL, Nakamura H. How does Fgf signaling from the isthmic organizer induce midbrain and cerebellum development? Development, Growth & Differentiation. 2004;46:487–494. doi: 10.1111/j.1440-169x.2004.00769.x. [DOI] [PubMed] [Google Scholar]
- Satou Y, Imai KS, Satoh N. Fgf genes in the basal chordate Ciona intestinalis. Development Genes and Evolution. 2002;212:432–438. doi: 10.1007/s00427-002-0266-8. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. Evolution of the neural crest viewed from a gene regulatory perspective. Genesis. 2008;46:673–682. doi: 10.1002/dvg.20436. [DOI] [PubMed] [Google Scholar]
- Sidow A. Gen(om)e duplications in the evolution of early vertebrates. Curr Opin Genet Dev. 1996;6:715–722. doi: 10.1016/s0959-437x(96)80026-8. [DOI] [PubMed] [Google Scholar]
- Stoltzfus A. On the possibility of constructive neutral evolution. J Mol Evol. 1999;49:169–181. doi: 10.1007/pl00006540. [DOI] [PubMed] [Google Scholar]
- Sun X, Mariani FV, Martin GR. Functions of FGF signalling from the apical ectodermal ridge in limb development. Nature. 2002;418:501–508. doi: 10.1038/nature00902. [DOI] [PubMed] [Google Scholar]
- Sundstrom G, Larsson TA, Larhammar D. Phylogenetic and chromosomal analyses of multiple gene families syntenic with vertebrate Hox clusters. BMC Evol Biol. 2008;8:254. doi: 10.1186/1471-2148-8-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Technau U, Rudd S, Maxwell P, Gordon PM, Saina M, Grasso LC, Hayward DC, Sensen CW, Saint R, Holstein TW, Ball EE, Miller DJ. Maintenance of ancestral complexity and non-metazoan genes in two basal cnidarians. Trends Genet. 2005;21:633–639. doi: 10.1016/j.tig.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Venkatesh B, Kirkness EF, Loh YH, Halpern AL, Lee AP, Johnson J, Dandona N, Viswanathan LD, Tay A, Venter JC, Strausberg RL, Brenner S. Survey sequencing and comparative analysis of the elephant shark (Callorhinchus milii) genome. PLoS Biol. 2007;5:e101. doi: 10.1371/journal.pbio.0050101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vienne A, Pontarotti P. Metaphylogeny of 82 gene families sheds a new light on chordate evolution. Int J Biol Sci. 2006;2:32–37. doi: 10.7150/ijbs.2.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H, Okuyama M, Satoh N, Zhang S. Molecular evolution of fibrillar collagen in chordates, with implications for the evolution of vertebrate skeletons and chordate phylogeny. Evol Dev. 2006;8:370–377. doi: 10.1111/j.1525-142X.2006.00109.x. [DOI] [PubMed] [Google Scholar]
- Wall DP, Fraser HB, Hirsh AE. Detecting putative orthologs. Bioinformatics. 2003;19:1710–1711. doi: 10.1093/bioinformatics/btg213. [DOI] [PubMed] [Google Scholar]
- Wittbrodt J, Meyer A, Schartl M. More genes in fish? Bioessays. 1998;20:511–515. [Google Scholar]
- Wolfe K. Robustness -- it’s not where you think it is. Nature Genetics. 2000;25:3–4. doi: 10.1038/75560. [DOI] [PubMed] [Google Scholar]
- Wotton KR, Weierud FK, Dietrich S, Lewis KE. Comparative genomics of Lbx loci reveals conservation of identical Lbx ohnologs in bony vertebrates. BMC Evol Biol. 2008;8:171. doi: 10.1186/1471-2148-8-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Liu Z, Ornitz DM. Temporal and spatial gradients of Fgf8 and Fgf17 regulate proliferation and differentiation of midline cerebellar structures. Development. 2000;127:1833–1843. doi: 10.1242/dev.127.9.1833. [DOI] [PubMed] [Google Scholar]
- Yan YL, Miller CT, Nissen R, Singer A, Liu D, Kirn A, Draper B, Willoughby J, Morcos PA, Amsterdam A, Chung BC, Westerfield M, Haffter P, Hopkins N, Kimmel C, Postlethwait JH. A zebrafish sox9 gene required for cartilage morphogenesis. Development. 2002;129:5065–5079. doi: 10.1242/dev.129.21.5065. [DOI] [PubMed] [Google Scholar]
- Yu WP, Rajasegaran V, Yew K, Loh WL, Tay BH, Amemiya CT, Brenner S, Venkatesh B. Elephant shark sequence reveals unique insights into the evolutionary history of vertebrate genes: A comparative analysis of the protocadherin cluster. Proc Natl Acad Sci U S A. 2008;105:3819–3824. doi: 10.1073/pnas.0800398105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Phylogenetic tree for NPM-family proteins. Results show that npm4 in teleosts has no apparent human ortholog. Sequences were obtained from Ensembl and NCBI and aligned by MUSCLE (Edgar, ‘04). Maximum likelihood-based phylogenetic inference analysis was conducted using PHYML (Guindon et al., ‘05). Abbreviations: Cin, Ciona intestinalis; Dre, Danio rerio zebrafish; Gac, Gasterosteus aculeatus stickleback; Hsa, Homo sapiens human. Accession numbers: npm1_Dre_NP_955460; npm1_Gac_ENSGACT00000026899; npm2a_Dre_OTTDART00000035953; npm2a_Gac_ENSGACT00000022099; npm2b_Dre_NP_001116479+AAI22198; npm3_Dre__NP_001013502; npm3_Gac_ENSGACT00000009682; npm4_Dre_XP_692942; npm_Cin_XP_002122436; NPM1_Hsa_NP_002511; NPM2_Hsa_NP_877724; NPM3_Hsa_NP_008924.
Supplementary Figure 2. Phylogenetic tree for Kazald-family proteins with IGFBP7 as outgroup shows a gene that can be called kazald2 that is missing from tetrapod genomes. Abbreviations and accession numbers: Bbe, Branchiostoma belcheri amphioxus; Dre, Danio rerio zebrafish; Gga, Gallus gallus chicken; Hsa, Homo sapiens human; Mmu, Mus musculus mouse; Tni, Tetraodon nigroviridis pufferfish; Xtr, Xenopus tropicalis frog. kazald1_Hsa_EAW49782; Kazald1_Mmu_NP_849260; MGC80370_Gga_XP_421724; Kazald1_Xtr_NP_001093733; LOC568748_Dre_NP_001098594; ENSDARG00000063176_Dre_NP_998089; Unn1_Tni_CAF94984; IGFBP7_Hsa_AAA16187; Igfbp7_Mmu_AAH47202; Unn2_Tni_CAG08989; LOC100000101_Dre_NP_001116791; PSF_Gga_XP_420577; ilgfbp_Bbe_BAB97382.
Supplementary Figure S3. Elephant shark possesses Fgf24. A. Alignment of exon 3 in vertebrate FgfD genes. Black rectangles highlight amino acid positions that characterize Fgf18 and Fgf24. Cin, Ciona intestinalis (sea squirt); Cmi, Callorhinchus mili (elephant shark); Dre, Danio rerio (zebrafish); Fru, Fugu rubripes (pufferfish); Gac, Gasterosteus aculeatus (stickleback); Hsa, Homo sapiens (human); Mmu, Mus musculus (mouse); Ola, Oryzias latipes (medakafish). B. Phylogenetic tree of exon 3 portions of FgfD genes including the shark sequences.
Supplementary Figure S4. Phylogenetic relationship among vertebrate FgfD genes and their expression patterns in mouse, zebrafish and stickleback. Abbreviations: aer, apical ectodermal ridge of pectoral fin; dd, dorsal diencephalon; h, heart; mhb, midbrain-hindbrain boundary; nc, neural crest;n, nose; oe, oral ectoderm; ov, otic vesicle; pan, pancreas; pf, pectoral fin; r, retina; s, somite; sp, segmental plate; spn, spinal cord neuron; t, telencephalon; tb, tail bud; um, unsegmented mesenchyme. stage1, segmentation; stage2, pharyngula.