Abstract
Target prediction for animal microRNAs has been hindered by the small number of verified targets available for evaluating the accuracy of predicted microRNA:target interactions. Recently, a dataset of 3404 microRNA-associated mRNA transcripts was identified by immuno-precipitation (IP) of the RNA-induced silencing complex (RISC) components, AIN-1 and AIN-2. Analysis of this dataset reveals enrichment for defining characteristics of functional microRNA target interactions, including structural accessibility of target sequences, the total free energy of microRNA:target hybridization, and the topology of base-pairing to the 5’ seed region of the microRNA. These enriched characteristics form the basis for a quantitative microRNA target prediction method, mirWIP (microRNA targets by Weighting IP dataset parameters), that optimizes sensitivity to verified microRNA:target interactions and specificity to the AIN-IP dataset. The mirWIP method can capture all of the known conserved microRNA:mRNA target relationships in C. elegans at a lower false positive rate than the current standard methods.
Introduction
The discovery of microRNAs1 and their roles in post-transcriptional gene regulation has added a new dimension to the study of animal development and disease2. microRNAs, bound to their mRNA targets, can repress gene expression through translational inhibition or by mRNA destabilization3. Under some conditions, microRNAs may also promote protein production from a target mRNA4. Animal microRNAs play a role in regulating many developmental processes and have been implicated in human disease pathways5. For these reasons, it is critical to efficiently identify the functionally important mRNA targets of microRNAs through the computational prediction of microRNA:target interactions and experimental tests of these predicted interactions.
Target prediction for microRNAs in plants is straightforward, since plant microRNAs bind with near perfect complementarity to target mRNAs. In animals, microRNAs interact with their targets predominantly by partial base-pairing, and the rules that govern the formation and functional efficacy of microRNA:mRNA interactions are not fully understood. Depending on the computational algorithm applied, the number of predicted targets for a given microRNA can range from dozens to hundreds and even thousands of genes6,7. The thorough experimental testing of such vast numbers of predicted targets has been impractical using labor-intensive transgenic reporter assays. There remains the need both for more accurate computational methods to distinguish functional from non-functional microRNA∷target interactions and also more efficient methods for the experimental testing and validation of microRNA∷target interactions in vivo.
Many computational methods have been developed to predict microRNA targets (reviewed in7). The criteria for target prediction vary widely, but often include: (A) strong, Watson-Crick base-pairing of the 5’ seed of the microRNA (nucleotide positions 2–8 of the microRNA) to a complementary site in the 3’ UTR of the mRNA transcript, (B) conservation of the microRNA binding site, (C) favorable minimum free energy (MFE) for the local microRNA∷mRNA interaction, and/or (D) structural accessibility of the surrounding mRNA sequence. Experimental support exists for each of these binding site features, but the relative importance of each feature (A–D), and how they interact to contribute to function, remains uncertain. Moreover, other important parameters for functional microRNA target interactions likely remain to be identified.
The principle of 5’ seed primacy in microRNA:target binding is well supported by experimental data. Many genetically validated microRNA:target interactions involve uninterrupted Watson/Crick base pairing in the 5’ region of the microRNA (microRNA positions 2–8, termed the “5’ seed” region2,3). Experiments show that G:U wobble pairs and bulges within the seed region can significantly disrupt repression of reporter constructs8 and that perfectly-matched seed regions are significantly enriched in the 3’ UTRs of transcripts whose levels decrease in response to microRNA over-expression 9. However, other experimental data suggests that perfectly matched microRNA seeds are neither necessary nor sufficient for all functional microRNA:target interactions. For instance, three of the genetically verified let-7 targets in C. elegans, lin-41, pha-4 and let-60/RAS, contain only imperfect binding sites with G:U wobble pairs or bulges in the seed region10–12. Two recent studies using immunoprecipitation of miRNP components indicate that only 30–45% of miRNP-associated mRNAs contain perfectly matched conserved seed elements in their 3’UTRs13,14. Therefore, target prediction algorithms need to be developed that accurately incorporate modified 5’ seed rules.
The conservation of sequences among multiple genomes has been invaluable in identifying functional regulatory elements in genomes. Most computational methods for predicting microRNA targets include an evolutionary conservation filter, often requiring strict alignment of seed-complementary sequences across multiple genomes7. However, many microRNA binding sites that do not fit the above strict definition of conservation could still be functionally important. For example, 40% of the verified microRNA targets in C. elegans reside within 3’ UTRs that align poorly between C. elegans and C. briggsae (e.g., the let-7 target sites in die-111, lss-411, pha-411, let-6012, nhr-2315, and nhr-2515). If the requirement for strict alignments is ignored in these cases, conserved sites for let-7 can be found in the orthologous 3’ UTRs, indicating evolutionary selection for a functional microRNA:target interaction. Indeed, in the case of the regulatory relationship between let-7 and let-60/RAS12, the presence of let-7 sites is conserved between worms and humans, although the sequence context of the sites is too divergent for strict alignment.
Many microRNA target prediction methods have incorporated minimum free energy (MFE) calculations into their prediction methods to identify energetically stable base pairing between a microRNA and target sequence16–20. Some methods also include estimates of the structural accessibility of microRNA binding sites in the mRNA targets18–20, and more recent methods join the two features into a single calculation19,20. Importantly, the incorporation of target structure into calculations of the free energy of microRNA:target interactions can distinguish between a set of targets that tested positive for microRNA-mediated repression and a set that were refractory to microRNA-mediated repression19. However, current prediction methods vary widely in how energy and accessibility estimates are incorporated into their calculations. Two studies18 consider accessibility of the binding sites, but differ in the amount of mRNA sequence used to calculate that parameter. Two more recent studies19,20 combine energy and accessibility calculations into a single prediction parameter, but vary in the length of sequence and in the method used to calculate accessibility. Further algorithm development is required to determine the optimal involvement of accessibility and binding energy in microRNA:target interactions.
Optimizing algorithms based on sequence features alone has been complicated by the lack of a large dataset of verified microRNA:target relationships. The number of targets that have been tested by rigorous genetic or reporter assays in various organisms has increased, but the assays vary in terms of how closely they model the endogenous characteristics of the interaction being tested7. Genome-scale datasets linking specific microRNAs to specific mRNA targets have emerged from microarray hybridization experiments that assay mRNA transcript levels after introduction of a particular microRNA by transfection9,21. Although these datasets have provided important insights into parameters associated with functional interactions, this approach is limited to the detection of microRNA:target interactions that result in transcript destabilization and does not identify stable, translationally-repressed target mRNAs. Recently, immuno-precipitation (IP) of the RNA-induced silencing complex (RISC) has been employed to identify mRNAs that stably associate with the endogenous RISC13,14,22. This approach provides a means of directly identifying endogenous stable complexes between microRNA RISC (miRISC) and target mRNAs, providing large datasets of high-confidence microRNA:target interactions that can, in principle be applied to derive target prediction algorithms of increased accuracy. One study in C. elegans22 recovered 3404 mRNA transcripts that specifically co-precipitate with the miRISC proteins AIN-1 and AIN-2. This “AIN-IP” set of mRNA transcripts forms the first biologically derived estimate for the number of genes that are targeted by microRNAs genome-wide -- in this case, at least one sixth of C. elegans genes.
We found several contextual features of microRNA binding sites that were enriched in sites in the AIN-IP set of transcripts: structural accessibility of target sequences, the total free energy of microRNA:target hybridization, and the topology of base-pairing to the 5’ seed region of the microRNA. These features were employed to develop a microRNA target prediction algorithm, mirWIP, that scores microRNA target sites based on weighting site characteristics in proportion to their enrichment. MirWIP exhibits improved overall performance compared to previous algorithms, in terms of the recovery of the AIN-IP transcripts, and in the correct identification of genetically-verified microRNA:target relationships without a requirement for alignment of target sequences. The mirWIP genome-wide predictions for C. elegans are available through a searchable web interface at www.ambroslab.org. Application of the mirWIP scoring method to any microRNA and target combination in any genome can be accessed individually through the Sfold web interface at http://sfold.wadsworth.org/starmir.pl.
Results
Initial Target Prediction
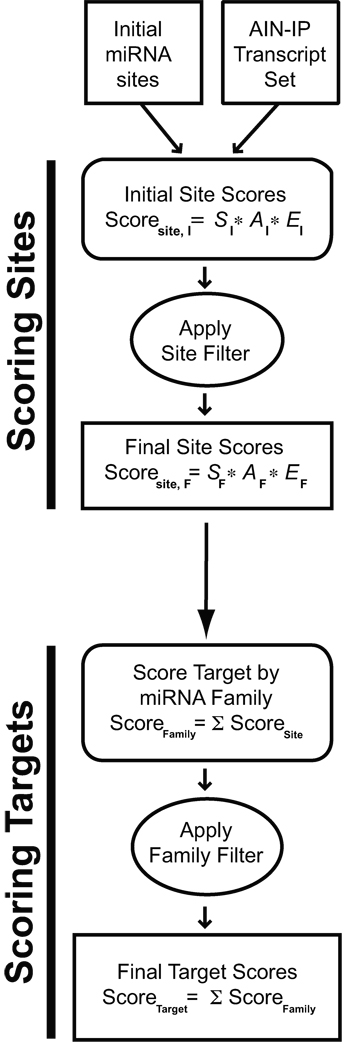
We employed RNAhybrid17 with modifications (see Supplemental Methods) to generate a list of all microRNA:target matches in C. elegans and C. briggsae. This initial set of raw microRNA target matches was filtered on the basis of minimal free energy, phylogenetic conservation, and seed pairing configuration (see Supplemental Methods and Supplementary Fig. 1) to produce an initial list of conserved C. elegans microRNA binding sites (“Initial microRNA sites” in Fig. 1). This set of sites was analyzed as described below to identify contextual features enriched in AIN-IP transcripts. Based on these analyses, an algorithm was developed that scores microRNA binding sites and mRNA targets based on characteristics enriched in the AIN-IP set of transcripts (Fig. 1). The set of 14 experimentally verified C. elegans microRNA:target interactions were omitted from this analysis to retain their independence as a test of the method (Supplemental materials).
Figure 1. Flow chart for the mirWIP target prediction method.
Analysis of predicted microRNA binding sites in the 3’ UTR sequences of AIN-IP transcripts reveals enriched contextual features. An initial set of predicted microRNA sites was obtained and analyzed for enriched features, and these enriched features were used to score individual predicted binding sites (see Methods and Supplemental Methods). Binding site scores were then combined into total microRNA family scores for each target, which estimates the likelihood that a given transcript is regulated by a particular microRNA family. Finally, the microRNA family scores were combined into a total target score for each transcript, estimating the likelihood that a given transcript is regulated by microRNAs (see Results, Methods and Supplemental Methods sections).
5’ Seed Match Features Are Enriched in AIN-IP Transcripts
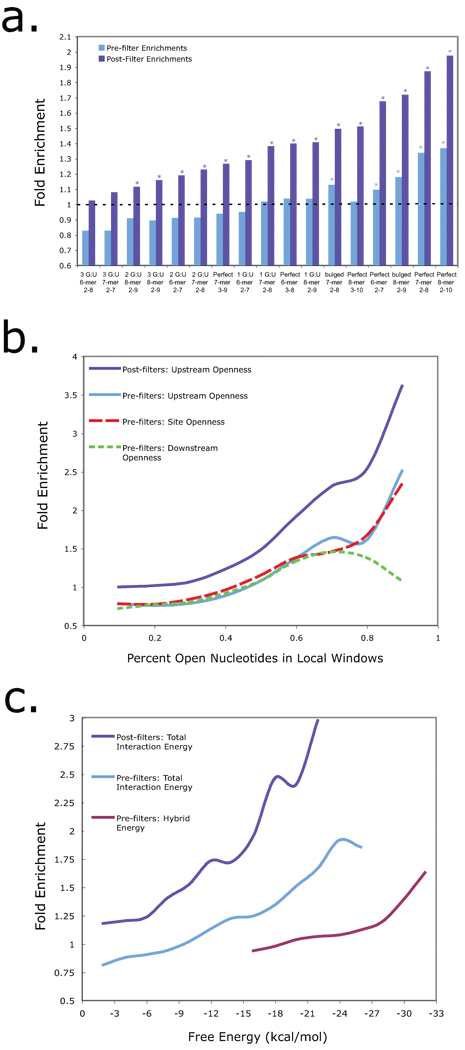
As shown in Figure 2a, the criterion of perfectly conserved seed matches to 8-mer blocks significantly enriches for AIN-IP targets over all other transcripts assayed. While extensive 5’ seed pairing shows the best enrichment in the AIN-IP list, these perfect 8-mers are relatively rare, residing within only 10% of the AIN-IP target transcripts. This is consistent with the occurrence of G:U wobble base pairs and bulges in validated microRNA:target relationships, and reinforces the conclusion that extensive 5’ seed pairing is neither necessary nor sufficient for reliable microRNA target prediction. It appears that perfectly matched seeds could be the only seed configurations enriched in the AIN-IP data (Fig. 2a, light blue bars) for the initial list of binding sites. However, note that AIN-IP transcripts are outnumbered 3:1 by all other transcripts in this list, and moreover, imperfectly paired seeds are more common than perfect matches, so these bins are more affected by the noise of false positives. With these cautions in mind, we explored the influence of other contextual features that could maximize capture of AIN-IP transcripts, and the 14 verified interactions, while striving to minimize the total number of targets predicted.
Figure 2. Characteristics of microRNA targets sites in AIN-IP Transcripts.
(a) AIN-IP Transcripts are enriched for binding sites with extensive 5’ seed pairing. The horizontal axis is ordered according to final enrichment for increasing stringency in 5’ seed matches with an indicated number of G:U wobble pairs or a single bulge on the mRNA side of the duplex. The vertical axis shows the enrichment for seed matches at the indicated stringency in AIN-IP versus all other transcripts both before (light blue) and after (dark blue) implementation of the “site filter” (see Figure 1 and Supplemental Methods). Asterisks designate significant enrichments with P < 0.05. (b) AIN-IP Transcripts are enriched for binding sites that lie within structurally accessible regions. The horizontal axis shows the calculated accessibility of local sequence windows, either: across the entire binding site (red, dashed line) or within a 25 nucleotide window upstream of the binding site (light and dark blue, solid line) or downstream (green, dotted line). After applying the site filter, enrichment was calculated for upstream windows only (dark blue, solid line). c) AIN-IP transcripts are enriched for binding sites with favorable free energies. The set of conserved binding sites in AIN-IP transcripts are more likely to have favorable (i.e., negatively-valued) total hybridization energies than their counterparts in non-AIN-IP transcripts (light and dark blue lines). Also shown is hybrid energy (purple line), which reflects the stability of the final microRNA:target duplex, and corresponds to the minimal free energy (MFE). Enrichment for ∆Gtotal significantly increases after applying the site filter (dark blue line).
AIN-IP Binding Sites Are Structurally Accessible
We used the Sfold method23 to fold whole 3’ UTR sequences plus 300 nucleotides of adjacent coding sequence for all predicted C. elegans transcripts (see Methods).The Sfold output returns the probability that each nucleotide in the 3’ UTR is predicted to be single-stranded, i.e., accessible. We used this output to calculate the average accessibility over 25 nucleotide windows around and including each potential microRNA binding site. As shown in Figure 2b, the average structural accessibility in upstream sequence windows shows the best enrichment for AIN-IP transcripts.
AIN-IP Binding Sites Are More Energetically Favorable
Hybridization between a microRNA and a structured mRNA target involves two major components: ΔGhybrid, the stability (hybrid free energy) of the microRNA:target duplex, and DGdisruption, the cost of altering the local structure of the mRNA target19. For a successful hybridization, the net energy of the process, ΔGtotal = ΔGhybrid - DGdisruption, must be thermodynamically favorable, i.e., negatively valued. As seen in Figure 2c, the binding sites in AIN-IP structures are strongly enriched for highly favorable values of ΔGtotal. Because DGtotal is an energetic measure of the target accessibility, it is highly correlated with the average structural accessibility across the binding site, as discussed above. For this reason, the trends of enrichments are similar for these two measurements (Site Accessibility in Fig. 2b and ΔGtotal in Fig. 2c). ΔGhybrid is substantially enriched, but at a lower degree than ΔGtotal.
mirWIP: microRNA Target Prediction
The three features that showed the best enrichment for AIN-IP targets (Fig. 2) were used to develop a microRNA target prediction scheme optimized to return AIN-IP transcripts and the verified microRNA:target relationships listed in Supplementary Table 1 online. This method is named “mirWIP” for microRNA targets by Weighting AIN-IP enriched parameters. Specifically, we calculated the relative enrichments for AIN-IP targets in each of the bins for: 5’ seed matching (S), upstream structural accessibility (A), and the total energy (E) of the microRNA:target hybridization, ΔGtotal. These three parameters were used to assign to each individual binding site three initial scoring parameters, SI, AI and EI.
Individual binding site scores were assigned in a two-step process (Fig. 1 and Supplemental Methods). After the initial scoring of all sites, we sought a mechanism to reduce noise prior to a second round of evaluating AIN-IP enrichment. Rather than cull all sites below a given initial score threshold, we chose to filter sites based on their overlap with higher-scoring sites in the same 3’ UTR. Accordingly, a window was moved along each UTR, and the best non-overlapping binding site was retained for each position in the UTR (and all overlapping binding sites were set aside). The relative enrichments were then re-calculated using this filtered site dataset (shown as dark blue bars or lines in Fig. 2). This filtering step improved the magnitude of the relative enrichments in each bin for all three features, indicating that the filtering operation improved signal to noise. These post-filter weights, SF, AF and EF (listed in Supplementary Table 2 online), were then used to re-calculate the score for the entire set of initial microRNA sites (Fig. 1), including the overlapping sites previously set aside. This calculation produced the final site scores, ScoreSite (Fig. 1).
After scoring all sites using the post-filter enrichments, we sought to again filter out the relatively low-scoring individual binding sites while calculating scores for 3’ UTR targets (Fig. 1). We found that the optimal approach was to evaluate interactions of an entire microRNA family (as defined previously24) with each target (see Supplemental Methods). We calculated the total family score for each target by adding up all non-overlapping site scores for each microRNA family member, separately. We then discarded any family:target interaction with a total mirWIP family score below “2.0” (see Supplemental Methods). Each UTR target was then given a “total target score” by adding up the contribution from each remaining microRNA family.
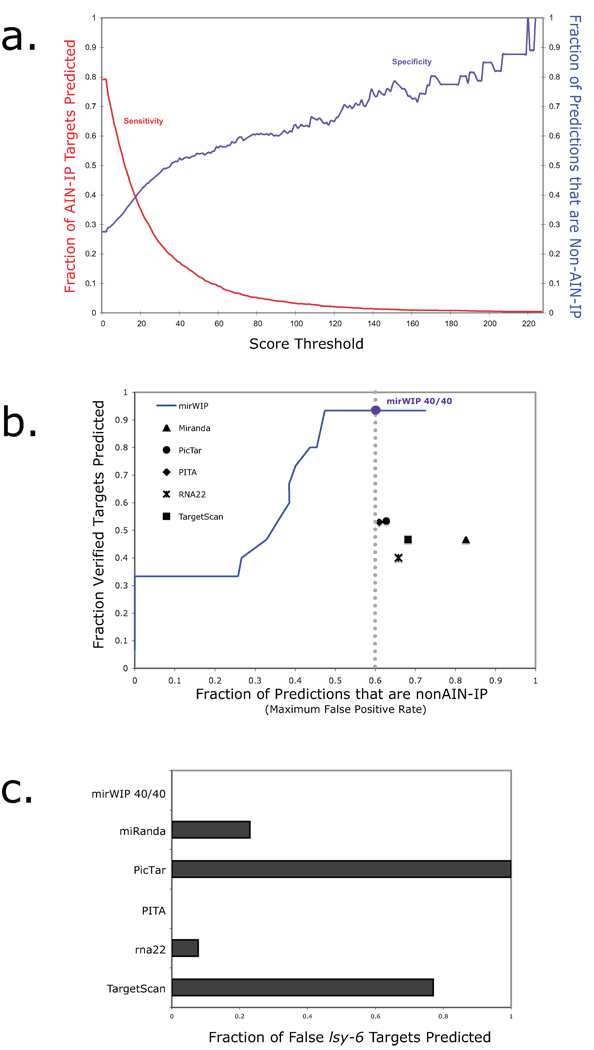
The target scores varied from approximately 2 to 400 with the highest scores going to lin-14 and hbl-1, two of the first identified microRNA targets in C. elegans. A plot of the sensitivity and specificity of our method against varying target score threshold is shown in Figure 3a. Here, sensitivity (shown in red) corresponds to the percent of AIN-IP target genes successfully recovered at each threshold. Sensitivity starts out at 79% (instead of 100%) because some targets had no strong, conserved microRNA binding sites, as will be discussed later. Specificity (shown in blue) represents the percentage of total predicted targets that are AIN-IP genes. For instance, with no threshold, the number of transcripts in the AIN-IP list is roughly ¼ of the total number of mRNAs examined, which corresponds to a specificity of ~ 27%. A compromise point can be found at a mirWIP score of 18, where the sensitivity and specificity are both ~ 40%. At this score level, 1214 AIN-IP transcripts and 1915 non-AIN-IP mRNAs are predicted as targets. This threshold easily accommodates the 14 verified C. elegans target genes, all of which exhibit a score greater than 47. Note that the experimentally validated true and false targets from Supplementary Table 1 were deliberately omitted from the enrichment analysis so that these provide an independent experimental validation set for the method.
Figure 3. Sensitivity and specificity of mirWIP.
(a) Choosing the optimum score threshold. The AIN-IP sensitivity of the algorithms is defined as the percent of AIN-IP targets correctly identified as a target of any microRNA (shown in red, above). The specificity of the algorithm is defined as the percentage of total predicted UTRs that are in the AIN-IP list (shown in blue, above). A compromise, for balancing the trade-off between sensitivity and specificity is defined by the point where the two curves meet: a mirWIP score threshold of 18. This corresponds to a sensitivity and specificity of approximately 40%. (b) Training mirWIP for AIN-IP sensitivity also optimizes true positive identification. A Receiver Operator Characteristic (ROC) curve is shown for mirWIP and five other microRNA prediction methods. The vertical axis shows the true positive rate (TPR), here represented by the number of verified targets correctly matched to the regulating microRNA. The horizontal axis shows the maximum false positive rate (FPR), the fraction of predicted UTRs that are not in the AIN-IP list. The performance of the mirWIP algorithm as a function of scoring threshold is shown as a blue line, and the 40% sensitivity/specificity compromise point (defined in panel a) is indicated by the large blue dot. mirWIP outperforms all five other methods by nearly doubling the TPR at a lower FPR (vertical gray line). (c) mirWIP is specific enough to reject all of the known false lsy-6 targets (Listed in Supplementary Table 1).
mirWIP Enrichements, Weights and Thresholds are Robust
To evaluate the robustness of the optimized algorithm, (in particular, to ensure that the predictions were not biased by a few high-scoring transcripts), we performed a 50% cross-validation calculation. That is, we randomly divided the data in half, derived the weights from the first half of the data, then tested how well the algorithm predicted AIN-IP vs. non-AIN-IP transcripts from the remaining half of the data set. We repeated this analysis 100 times, finding that the accuracy calculations were stable against random data shuffling, with an accuracy of 63.6% and a standard deviation of 0.6%. This calculated “accuracy” is likely to be a significant underestimate since it was measured by the ability of the algorithm to separate AIN-IP from non-AIN-IP targets, while many non-AIN-IP targets are likely to be real.
Comparison of mirWIP Performance to Other Methods
We compared our algorithm to the three most commonly used target prediction methods for C. elegans (PicTAR16, TargetScanS21, and miRanda25). We also included rna226 in our comparisons since this method does not use any of the typical prediction criteria (seed matching, conservation, energy, or structure). Lastly, we included a recent method, PITA20, which is similar to our technique in that PITA also employs seed, structure, and energy calculations to predict target transcripts, but without sequence conservation (PITA, online release 5, with the suggested threshold ΔΔG < −10 kcal/mol). These methods were selected to show the improvements gained by using our AIN-IP derived weights and our particular combination of contextual features.
We used two metrics to compare the performance of mirWIP to that of the other algorithms. First, we considered the ability of the algorithms to return the experimentally verified C. elegans microRNA:target matches listed in Supplementary Table 1. While this dataset is small, it represents the strictest test of the sensitivity of microRNA prediction methods, and a true experimental validation set for mirWIP, as these sites were not included in our enrichment analysis. We compared this to the percentage of predicted targets that are not in either the AIN-IP or verified target list – an estimated maximum false positive rate (FPR). A Receiver Operator Characteristic (ROC) plot is shown in Figure 3b, displaying these results. The blue line represents the performance of the mirWIP algorithm at varying target score thresholds, while the blue circle marks the performance of mirWIP at the 40% sensitivity cutoff defined in Figure 3a and discussed above. The mirWIP algorithm outperforms these five prediction methods by returning more verified microRNA targets at a lower FPR. The ability of mirWIP to correctly predict the weakest of the verified targets without a corresponding increase in the false positive rate is the strongest finding of this study, and highlights the utility of RISC IP assays in improving microRNA target prediction.
A second estimate of algorithm specificity is shown in Figure 3c where we compare each method’s recovery of a set of well characterized false targets of lsy-626. The mirWIP algorithm does not predict any of these genes as a target of lsy-6, similar to PITA release 5, while the other 4 methods vary in predicting 7% –100% of these interactions. This comparison may be biased against PicTAR (as compared to the other methods) since these lsy-6 targets were specifically selected from the PicTAR predictions to illustrate an instance where “conserved seed” predictions fail. However, many of the validated true targets were also selected from seed-based prediction catalogs, making the true negative comparison set as fair as the true positive set with regard to mirWIP success rates.
Overlap Among microRNA Prediction Methods
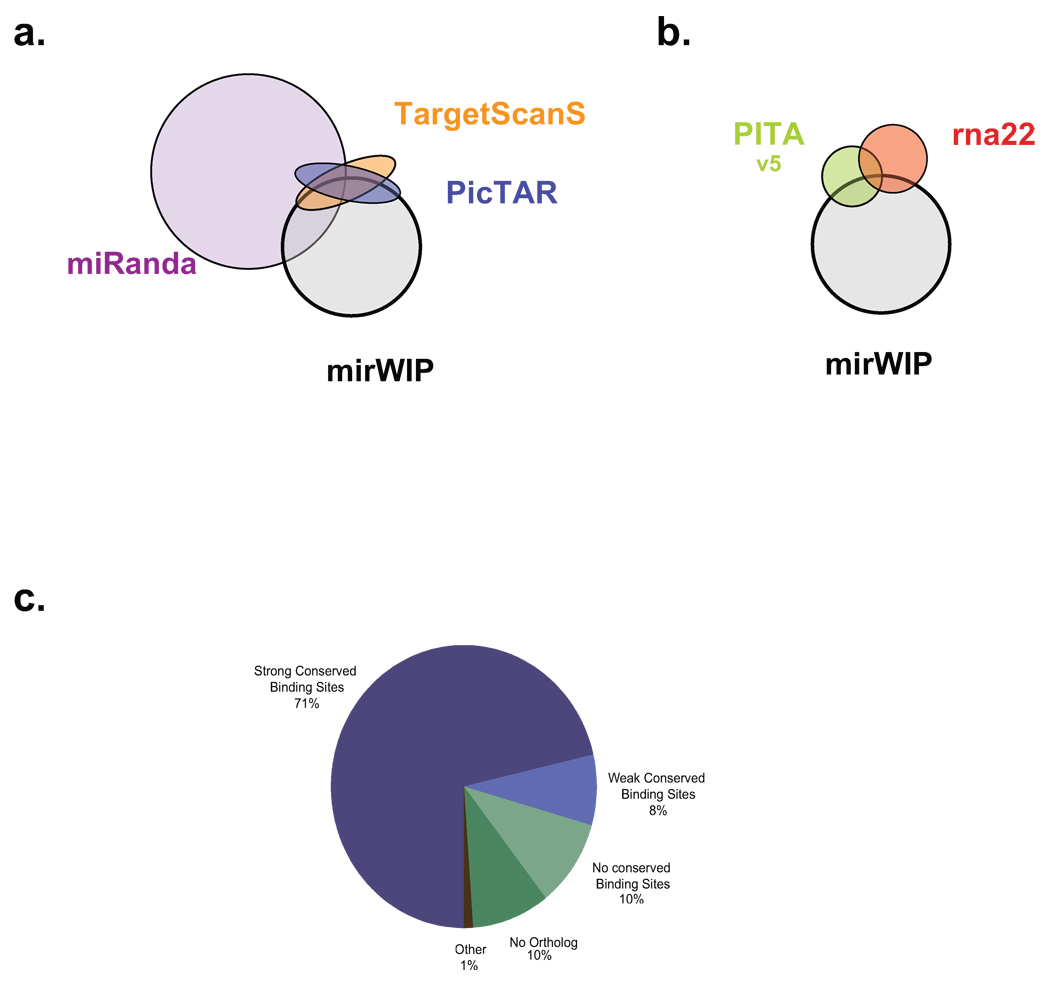
Next, we compared the overlap in predicted microRNA:target interactions for mirWIP and each of the five methods described above. The distribution of overlapping targets is illustrated as Venn diagrams (Fig. 4a–b). In Figure 4a, we compare mirWIP to those methods that consider orthologous conservation (mirWIP, Miranda, PicTAR and TargetScanS), and to two methods that do not use conservation in Figure 4b (PITA, and rna22). MiRanda predicts the largest percentage of mirWIP interactions, but it also predicts the largest number of targets overall. Interestingly, the overlap between mirWIP, PicTAR, and TargetScanS in Figure 4a shows that mirWIP tends to include predicted targets shared by PicTAR and TargetScanS, due to common predictions with strong seed signals. Most mirWIP predictions do not overlap with PicTAR and TargetScanS; these targets primarily exhibit non-canonical seeds with strong structural features or functional conservation without alignment. The lack of overlap between mirWIP and rna22 is not particularly surprising, since this method differs in all aspects from the mirWIP method. However, the lack of overlap between mirWIP and PITA is interesting given the similarity of these two methods.
Figure 4. Distribution of shared microRNA predictions & non-predictions.
(a,b) Since the various comparison methods differ in degree of similarity, Venn diagrams were split into two groups. MiRanda, PicTar, and TargetScanS all use seed-matching and conservation in their prediction method, so part (a) shows the degree of overlap between mirWIP, Miranda, PicTAR, and TargetScanS. mirWIP selects most of the targets for PicTAR and TargetScanS that these two methods share, and relatively few of the targets not shared by these two methods. PITA and rna22, neither of which uses conservation to identify microRNA targets are compared to mirWIP in panel (b). (c) Most AIN-IP transcripts can be accounted for by containing conserved binding sites for known microRNAs. However, 29% of the IP-ed genes do not have strong, conserved binding sites in their annotated 3’ UTRs. Conserved binding sites can be found for an additional 8% of the AIN-IP transcripts, but these sites fail to meet our minimum free energy threshold and have been termed “weak” sites. Lack of conservation and poor UTR annotation are the most likely reasons for the rest of these non-predictions. See Results and Supplemental Results for a discussion of the remaining AIN-IP transcripts rejected by mirWIP.
Overall there is only modest overlap amongst the six methods in the sets of microRNA:target interactions predicted. Approximately 25% of the specific microRNA:target interactions predicted by mirWIP are shared with at least one of the five other methods considered here. However, there is better agreement among these methods in terms of the mRNAs predicted to be targeted by microRNAs in general. That is, 96% of the genes in the mirWIP catalog are also predicted to be targets of microRNAs by at least one of the other methods. In other words, these prediction methods agree about many of the genes targeted by microRNAs, but disagree about which microRNA is regulating that gene. Importantly, 27% of the verified microRNA:target interactions lie in that set of predictions unique to mirWIP.
Analysis of Falsely Rejected AIN-IP Targets
The mirWIP algorithm identifies 79% of the AIN-IP transcripts on the basis of conserved binding sites in the 3’ UTR (Fig. 4c). Most of the AIN-IP transcripts that were not included by mirWIP failed to pass the initial minimum free energy (MFE) and conservation filters. By relaxing the MFE filter from −15 kcal/mol to −10 kcal/mol, we find conserved binding sites for an additional 271 AIN-IP UTRs (“weak conserved binding sites” in Fig. 4c). While there may be many true predictions in this group, relaxing the MFE filter would lead to a substantial increase in the false positive prediction rate, allowing in 940 additional non-AIN-IP target UTRs and 54% of the lsy-6 predicted sites shown to be non-functional 26. The mirWIP conservation filter rejected 10% of the AIN-IP transcripts with strong binding sites for a microRNA in C. elegans but not in C. briggsae. Finally, an additional 10% of the AIN-IP genes do not have an ortholog in which to look for conserved binding sites27. There may be many non-conserved binding sites for known microRNAs in this group as well as conserved binding sites for unknown microRNAs. Relaxing the already lenient orthology filter, however, would lead to an unacceptable false positive rate since conservation is one of the strongest filters in the algorithm.
Discussion
The AIN-IP set of miRISC-associated mRNA transcripts represents the largest set available thus far of true microRNA targets identified from their endogenous context. This target list is not biased by selection from a particular target prediction method, allowing a fair comparison across methods. The large number of targets in the AIN-IP list allowed for a statistical analysis of both sequence and structural features associated with regulation by the miRISC complex. We found that AIN-IP transcripts are enriched for microRNA complementary sites, and that certain features of the microRNA binding sites are strongly enriched. These features include a range of 5’ seed base-pairing configurations, structural accessibility of the binding site and an upstream region, and favorable total interaction energy of the microRNA:mRNA hybridization, ΔGtotal. These findings are consistent with previous reports on the importance of both canonical and non-canonical seed matches8–11,21, target accessibility18–20, and interaction energy19,20.
The strongest enrichment values for structural accessibility and total hybridization energy, ΔGtotal, were greater than the strongest enrichment values for seed topology. We do not believe that this implies that seed matching is less predictive than the other two parameters for identifying microRNA targets. This is because all potential microRNA:target binding sites were pre-screened to meet minimal seed criteria before calculating enrichment values. Thus, it is possible that we are underestimating the contribution of seed matching relative to the two other parameters. We cannot predict to what extent the enrichment scores might reflect the relative ability of each parameter to return functional microRNA binding sites. We can say that the combination of these three parameters into a total scoring method outperforms a model where one or more of these parameters are omitted or given less weight (Supplemental Methods).
mirWIP exhibits improved target prediction in C. elegans in several respects. First, the mirWIP method returns all 14 of the conserved, verified microRNA target relationships without increasing the total false positive rate beyond that of the current standard predictions. It should be emphasized that the set of 14 validated targets were not used to train the algorithm and thus they provide an independent experimental test of the method. This list includes many non-canonical binding sites (imperfect seed matches as well as sites not conserved in aligned genomes) that cannot be identified by current target prediction methods. Secondly, mirWIP correctly rejects thirteen targets predicted by other methods, but which have been shown to be non-functional in vivo26. Lastly, we found that the miRISC association of most (79%) of the AIN-IP transcripts can be explained by the existence of conserved binding sites for known microRNAs; the remaining 21% were rejected because of a lack of conserved targeting between C. elegans and C. briggsae. These findings highlight the improvements gained by using IP-enriched features to identify the contextual features of functional microRNA binding sites. It should be emphasized that this scoring method can be applied to the output of any microRNA target prediction and secondary structure prediction method.
Among the mirWIP predicted targets, 40% were identified by the AIN-IP method while 60% of the mirWIP predicted transcripts were not stably associated with AIN proteins in the miRISC. Many of these non-AIN-IP transcripts could represent false positive predictions by mirWIP, which would imply a lower bound of 40% for our true positive predicted fraction. However, for several reasons, we believe that a significant portion of these non-AIN-IP transcripts represent bona fide microRNA targets. First, the strict cutoff implemented in defining the AIN-IP list 22 may have removed many true targets. Second, the sensitivity of the AIN-IP method is expected to be poor for interactions that involve a small fraction of the total population of the target mRNA. For example, some interactions may occur only transiently, and/or in a limited number of cells in the animal, as is the case for lsy-6 and cog-128. Third, the AIN-IP method is likely to be most effective at recovering stable microRNA:mRNA complexes, and is expected to recover unstable mRNAs much less efficiently. It is known that some microRNAs regulate their targets on the level of mRNA stability29, and hence such miRNA:mRNA complexes would be relatively short-lived and poorly detected by microarray hybridization. Finally, 4 out of the 14 genetically validated miRNA targets were not in the AIN-IP list (29%). This suggests that as many as 29% of the mirWIP predictions could be true microRNA targets that were not identified by AIN-IP. By this estimate, an upper bound on our positive prediction rate could be as high as 70%.
The sensitivity and specificity of mirWIP target predictions should be improved by analyzing additional experimental datasets. For example, the analysis of miRISC-associated RNAs from populations of developmentally-staged worms or specific cell types should help reduce the noise associated with averaging regulatory interactions over all stages and tissues. Moreover, mirWIP in its current form is supported by IP experiments that identify transcripts by their probable association with microRNAs, but without providing information directly about what particular microRNA or set of microRNAs are responsible for miRISC association. The immuno-precipitation of miRISC proteins from animals lacking a specific microRNA would allow us to match individual microRNAs to the targets they regulate. One study13 performed such an experiment with a tagged version of Argonaute in Drosophila, significantly enriching for a small number of targets for dme-miR-1. Similar experiments can be applied to C. elegans, where a comprehensive set of microRNA mutants are available 30. Finally, since the miRISC IP approach may be biased towards the identification of stable miRNA:target complexes, complementary datasets can be utilized that screen for miRNA-induced target destabilization, such as microarray assays to identify mRNA transcripts that change in response to microRNA activity.
Methods
microRNA Target Identification
We used the RNAhybrid algorithm17 to identify the raw list of possible microRNA matches in the set of orthologous 3’ UTRs of C. elegans and C. briggsae, with a few modifications to the application. These modifications, and the particular parameter choices are discussed in Supplemental Methods. Subsequent filtering and scoring of microRNA sites, and the derivation of methods for combining site scores to produce target (3’ UTR) scores, are described in Supplemental Methods and shown in Supplementary Figure 3, online.
Structural Accessibility Calculations
We use the Sfold method23 to fold 3’ UTR sequences for all C. elegans transcripts, plus 300 nucleotides of coding sequence adjacent to the stop codon. Sfold returns the probability that each nucleotide in the given sequence would be single-stranded, here referred to as structural accessibility. Details for accessibility calculations and length of sequence examined are given in Supplemental Methods.
Total Interaction Energy Calculations, ΔGtotal
The calculations for ΔGtotal, are separate from the average accessibility calculations performed above but do also use the predicted accessibilities, as follows. We used the predicted structures for each binding site, calculating the energy necessary to disrupt any bound nucleotides in that region, ΔGdisruption. This disruption energy is then added to the minimal free energy, ΔGhybrid, to obtain the total interaction energy, ΔGtotal.
Statistical Analysis
We estimated the significance of the pre-filter enrichments for seed, structural accessibility measures, and total free energy shown in Figure 2 using Fisher’s Exact two-tailed contingency table. For the post-filter enrichments, which were derived from 100 random shuffles of the data, we calculated the P-values from the Z-score of a normal distribution. Individual P-values for every bin are given in Supplementary Table 2 along with a discussion of the method chosen to calculate P-values.
Genome-wide prediction of microRNA targets
C. elegans genomic microRNA target predictions generated using the mirWIP algorithm are available through a web interface at http://ambroslab.org. The mirWIP scoring method has also been implemented into the StarMir module of the Sfold package to make predictions for any microRNA:target pair from any species of interest: http://sfold.wadsworth.org/starmir.pl. Source code is available for the RNAhybrid modifications and the scoring method from Nature Methods.
Supplementary Methods
Includes: details for the intial microRNA binding site identification methods and modifications to the RNAhybrid source code, details for the calculation and statistical analysis of enrichments, alternative methods examined for scoring sites and targets, and an analysis of the robustness of the calculated accuracy of the mirWIP method.
Supplementary Material
Acknowledgments
We would like to thank C. Hammell and all members of the Ambros lab for useful discussions. The Computational Molecular Biology and Statistics Core at the Wadsworth Center is acknowledged for providing computing resources for this work. This research was supported by National Institute of Health grants GM34028 and GM 066826 to V.A., GM068726 to Y. D, and GM47869 to M. Han, as well as National Science Foundation grant DBI-0650991 to Y.D; and by the Howard Hughes Medical Institute, of which M. Han is an investigator.
Footnotes
Supplementary Material
Supplementary Figure 1 Flow chart illustrating the initial microRNA binding site identification steps.
Supplementary Figure 2 Contextual Features Not Correlated with Enrichment for AIN-IP Transcripts.
Supplementary Figure 3 ROC curves used to optimize mirWIP method
Supplementary Table 1 Verified True Positive and True Negative microRNA targets in C. elegans.
Supplementary Table 2 Lists of Enrichment Values & P-values for microRNA Binding Site Contextual Features.
Supplementary Text Supplementary Results and Methods
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 3.Jackson RJ, Standart N. How do microRNAs regulate gene expression? Sci STKE. 2007;2007(367):re1. doi: 10.1126/stke.3672007re1. [DOI] [PubMed] [Google Scholar]
- 4.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318(5858):1931. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 5.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11(4):441. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Miranda KC, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126(6):1203. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 7.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38 Suppl:S8. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 8.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3(3):e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 10.Vella MC, Reinert K, Slack FJ. Architecture of a validated microRNA∷target interaction. Chem Biol. 2004;11(12):1619. doi: 10.1016/j.chembiol.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 11.Grosshans H, et al. The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans. Dev Cell. 2005;8(3):321. doi: 10.1016/j.devcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 12.Johnson SM, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 13.Easow G, Teleman AA, Cohen SM. Isolation of microRNA targets by miRNP immunopurification. Rna. 2007;13(8):1198. doi: 10.1261/rna.563707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beitzinger M, et al. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol. 2007;4(2):76. doi: 10.4161/rna.4.2.4640. [DOI] [PubMed] [Google Scholar]
- 15.Hayes GD, Frand AR, Ruvkun G. The mir-84 and let-7 paralogous microRNA genes of Caenorhabditis elegans direct the cessation of molting via the conserved nuclear hormone receptors NHR-23 and NHR-25. Development. 2006;133(23):4631. doi: 10.1242/dev.02655. [DOI] [PubMed] [Google Scholar]
- 16.Lall S, et al. A genome-wide map of conserved microRNA targets in C. elegans. Curr Biol. 2006;16(5):460. doi: 10.1016/j.cub.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 17.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. Rna. 2004;10(10):1507. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robins H, Li Y, Padgett RW. Incorporating structure to predict microRNA targets. Proc Natl Acad Sci U S A. 2005;102(11):4006. doi: 10.1073/pnas.0500775102. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436(7048):214. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 19.Long D, et al. Potent effect of target structure on microRNA function. Nat Struct Mol Biol. 2007;14(4):287. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- 20.Kertesz M, et al. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39(10):1278. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 21.Grimson A, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27(1):91. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L, et al. Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol Cell. 2007;28(4):598. doi: 10.1016/j.molcel.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding Y, Chan CY, Lawrence CE. RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. Rna. 2005;11(8):1157. doi: 10.1261/rna.2500605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruby JG, et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127(6):1193. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 25.Enright AJ, et al. MicroRNA targets in Drosophila. Genome Biol. 2003;5(1):R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat Struct Mol Biol. 2006;13(9):849. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- 27.Stein LD, et al. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 2003;1(2):E45. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnston RJ, Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426(6968):845. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- 29.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A. 2006;103(11):4034. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bagga S, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122(4):553. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 30.Miska EA, et al. Most Caenorhabditis elegans microRNAs Are Individually Not Essential for Development or Viability. PLoS Genet. 2007;3(12):e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.