Abstract
There is a steadily increasing demand for speed, cost efficiency and process understanding within biopharmaceutical process development. To match this, a high-throughput method for screening of cleaning-in-place (CIP) conditions for chromatography resins has been developed. The methodology includes fouling of MabSelect SuRe chromatography resin in 96-well filter plates, cleaning of the fouled resin by incubation in different CIP agents, and finally, analysis of the residual impurities on the resin after cleaning. This article describes the improvements that transformed the method from low throughput and significant manual interference to a totally automated method with high throughput and good reproducibility.
Key words: bioprocess, cleaning-in-place, chromatography, high-throughput, monoclonal antibody, process development, protein A, screening
Introduction
Cleaning-in-place (CIP) is crucial for efficient use of a chromatography column. In order to maximize the number of cycles that a column can be reused, a cleaning procedure that efficiently removes impurities without being harmful to the chromatography resin is required. Sodium hydroxide (NaOH) efficiently removes precipitated proteins, hydrophobic proteins, nucleic acids, endotoxins and viruses and has become the gold standard for cleaning and sanitization of chromatography resins.1 The Protein A ligand on an affinity chromatography resin for capture of monoclonal antibodies (mAbs), is sensitive to harsh cleaning conditions, such as high NaOH concentrations. Therefore, this type of resin benefits in particular from optimizing the CIP protocol for cleaning efficiency and resin compatibility.
There is a steadily increasing pressure on biopharmaceutical producers to reduce process development time and cost. Regulatory authorities also require state of the art process understanding and characterization. Traditional process development approaches are time- and material-intensive. Therefore, the use of scaled-down, high-throughput methods for screening of operating conditions throughout the biopharmaceutical process has emerged over the past decade.2 PreDictor™ plates are 96-well filter plates pre-filled with chromatography resins for antibody affinity, ion exchange or multimodal chromatography. PreDictor plates are intended for screening of chromatographic conditions in all parts of the chromatographic cycle.3
Here, the development of a high-throughput method for screening of cleaning conditions for chromatography resins using PreDictor MabSelect SuRe™ plates is described. MabSelect SuRe contains an alkali-stabilized Protein A ligand that provides greater stability than conventional Protein A-based resins in the alkaline conditions used in CIP protocols.1 MabSelect SuRe is based on agarose, which is stable in alkaline conditions whereas silica-based resins are not. The method focuses on screening the efficiency of cleaning agents in removing protein impurities from a fouled chromatography resin. It is a generic method that could be used for screening of cleaning conditions for other combinations of chromatography resins and feeds than described here. The method was developed to evaluate removal of protein impurities, but could be further extended to evaluate cleaning efficiency for other impurities. The PreDictor plate format can also be used for evaluation of resin compatibility with various cleaning agents.3
The use of prefabricated plates (PreDictor) is convenient but is not a prerequisite for this type of study. Therefore, PreDictor is henceforth referred to simply as a 96-well filter plate.
In brief, the method for screening of cleaning efficiency involves three parts; (1) fouling of MabSelect SuRe resin in 96-well filter plates using harvested cell culture fluid (HCCF) with mAb; (2) cleaning of the resin by incubation of fouled resin in CIP agents and (3) analysis of residual impurities on the resin after cleaning. The method has been gradually improved for higher throughput and more automated working procedures and has been adjusted to better reflect real life column usage procedures. The original method and improvements made to it are described here.
Results
Artificial fouling of chromatography resin in 96-well filter plates.
In order to mimic the fouling that takes place in a column reused for repeated cycles, artificial fouling of the resin was initially performed in the 96-well filter plates.3–5
During the artificial fouling procedure, the MabSelect SuRe resin in the wells of the filter plate, was equilibrated using phosphate buffered saline (PBS) and then incubated in clarified E. coli lysate spiked with polyclonal IgG. Loading of E. coli lysate spiked with IgG was repeated four times. After the final incubation, the resin was washed with PBS. A precipitation solution, fouling agent, composed of salt and acid (2.9 M ammonium sulfate, 0.6 M phosphoric acid, pH 2.5), was added to the resin followed by incubation overnight. After a subsequent wash and elution of bound IgG, the resin was re-equilibrated with PBS and washed with ultrapure water (H2O) (Fig. 1). The fouled resin was then used for screening of CIP conditions. After cleaning, the residual protein impurities on the resin were analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE).
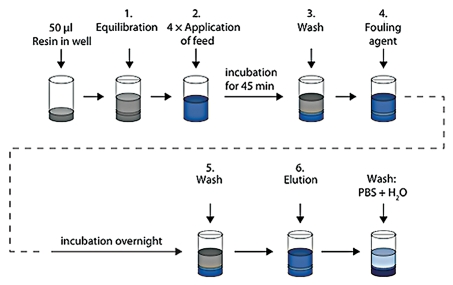
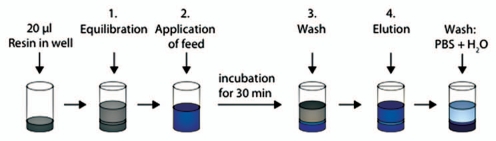
Figure 1.
Schematic description of artificial fouling of MabSelect SuRe resin in PreDictor plates. Steps 1, 3 and 5 show equilibration or wash with PBS (20 mM phosphate, 0.15 M NaCl, pH 7.4). In step 2, which was repeated four times, E. coli lysate spiked with polyclonal IgG was added to the resin and incubation was done for 45 min. In step 4, a fouling agent composed of 2.9 M ammonium sulfate, 0.6 M phosphoric acid, pH 2.5 was added, followed by incubation overnight in order to accelerate the fouling of the resin. After a subsequent wash, the bound IgG was eluted with 0.1 M sodium citrate pH 3.0. The resin in the wells was mixed at every step and liquid was removed by centrifugation between each step. H2O, ultrapure water.
Evaluation of cleaning conditions.
To evaluate cleaning conditions, the fouled resin was incubated with different cleaning chemicals, during mixing on a microtiter plate shaker at 1,100 revolutions per minute (rpm). An incubation time of 15 min was used, which corresponds to the CIP contact time in a column. Prior to the screening, deep-well plates with CIP solutions according to the experimental set-up in the 96-well filter plate were prepared. During the experiment, the CIP-solutions were transferred from the deep-well plate to the 96-well filter plate using an 8-channel pipette. It was possible to screen single cleaning steps, as well as sequences of multiple cleaning steps. Up to four cleaning steps in sequences have been evaluated. Between each cleaning step in a sequence, the resin was washed with 300 µL PBS followed by wash with 300 µL ultrapure water. After the last cleaning step the resin was washed three times with 300 µL PBS and three times with 300 µL ultrapure water (Fig. 2).
Figure 2.
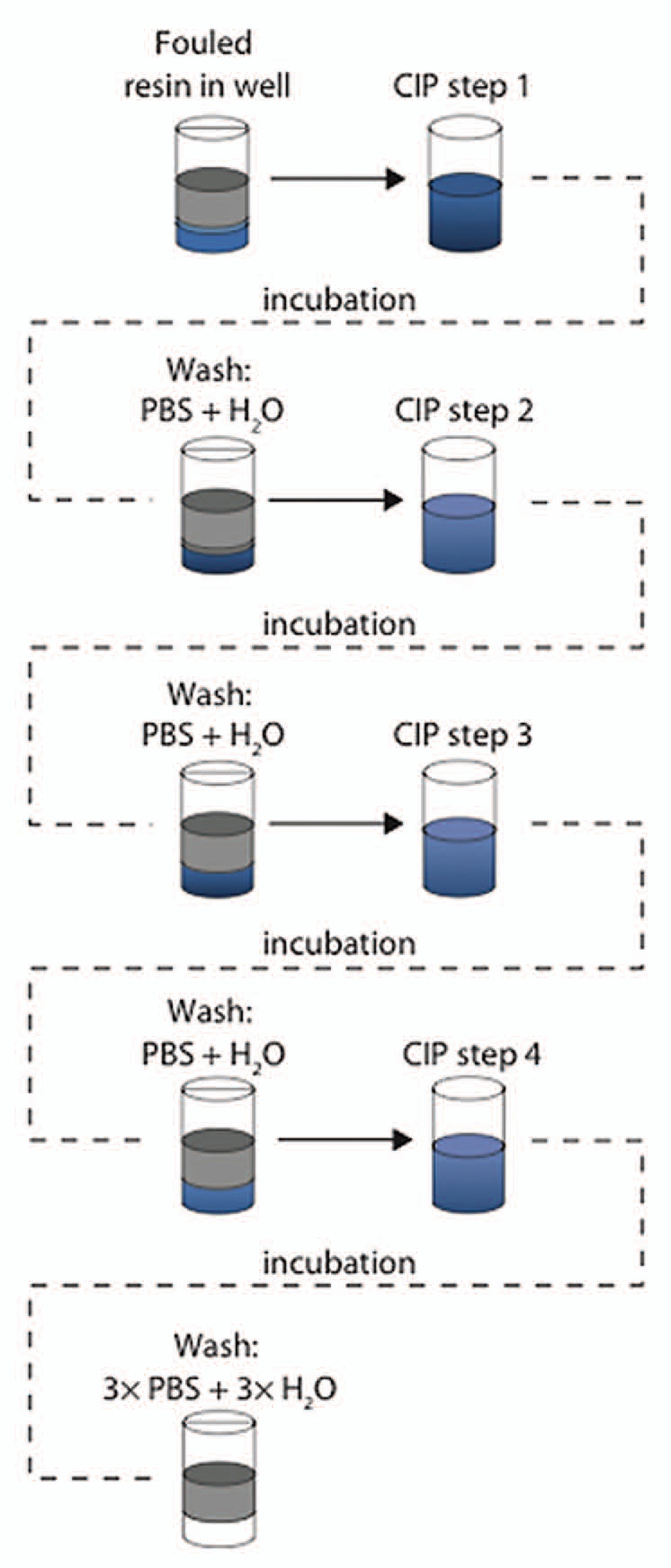
Evaluation of cleaning conditions. Different cleaning conditions were evaluated by incubation of the fouled MabSelect SuRe resin in cleaning chemicals and sequences of cleaning steps. An incubation time of 15 min was used, which corresponds to the CIP contact time in a column. Between each cleaning step in a sequence, the resin was washed with 300 µL PBS followed by wash with 300 µL H2O. In each step, mixing was done and centrifugation was used for liquid removal between each step.
SDS-PAGE analysis of residual protein impurities on the resin after cleaning of artificially fouled MabSelect SuRe.
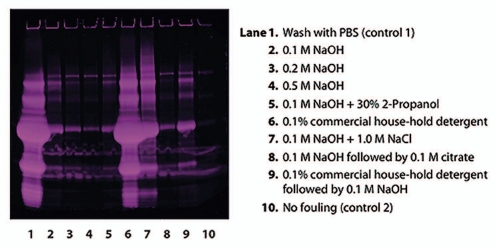
Subsequent to the artificial fouling, eight different cleaning protocols, with one or two CIP steps, were applied to the fouled resin. The fouled and cleaned resin samples were then heat treated with an SDS/Dithiothreitol (DTT) containing sample buffer for extraction of protein impurities from the resin. SDS-PAGE analysis of the remaining protein impurities on the resin after cleaning showed that increasing NaOH concentration from 0.1 to 0.5 M resulted in a decreasing amount of residual protein impurities (Fig. 3). A mixture of 0.1 M NaOH and 30% 2-propanol resulted in slightly increased cleaning efficiency compared to NaOH alone as did a two-step sequence with 0.1 M NaOH followed by 0.1 M citrate. However, the differences were small and it was not possible to tell if this was significant as the data were qualitative, and from single samples. 0.1 M NaOH + 1.0 M NaCl resulted in a significant decrease in cleaning efficiency compared to 0.1 M NaOH without salt. A commercial house-hold detergent (Yes, Procter & Gamble) was also evaluated for cleaning efficiency, but this did not contribute to cleaning of the fouled resin. The control sample, which was washed with PBS, showed that there was a large amount of proteins on the fouled resin.
Figure 3.
Reduced SDS-PAGE (Deep Purple™ stained) for analysis of proteins remaining on the artificially fouled MabSelect SuRe resin after cleaning with different agents. The control sample, which was washed with PBS (lane 1), showed that there was a large amount of proteins on the artificially fouled resin. NaOH proved to be very effective in cleaning, and increasing NaOH-concentration resulted in increasing cleaning efficiency (lanes 2–4). 0.1 M NaOH + 1.0 M NaCl (lane 7) resulted in a significant decrease in cleaning efficiency compared to 0.1 M NaOH without salt (lane 2).
Comparison of sample preparation in 96-well filter plate and collection plate.
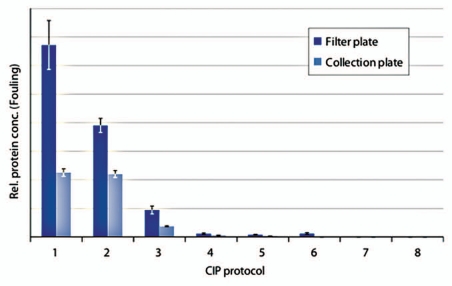
Two different modes of sample preparation prior to analysis using electrophoresis were compared. One approach was to prepare the samples in the 96-well filter plate where the resin had been artificially fouled and cleaned. The other approach was to transfer the drained resin to a collection plate before sample preparation. The residual protein impurities on the resin were analyzed using chip electrophoresis. In terms of ranking of the cleaning efficiency of the different CIP protocols, the two methods for sample preparation gave approximately the same results (Fig. 4). However, the samples prepared directly in the 96-well filter plate contained significantly more impurities compared to the samples prepared in the collection plate. The control (wash with PBS) contained a lot of impurities as did cleaning using a proprietary CIP solution. Phosphoric acid at pH 1.6 removed some of the precipitated proteins but even more efficient was 0.1 M NaOH + 1.0 M NaCl and 6.0 M Guanidine hydrochloride. Cleaning with 0.1 M NaOH resulted in a complete clearance of precipitated proteins when the sample was prepared in the collection plate, whereas after sample preparation in the 96-well filter plate, this sample contained a substantial amount of proteins. After cleaning with 0.5 M NaOH, no impurities could be detected on the resin, irrespective of mode of sample preparation. In this experiment, the samples were extensively diluted prior to analysis. In the improved method, the samples were prepared after transfer to a collection plate and dilution of the samples was limited.
Figure 4.
Comparison of samples prepared in a filter plate to samples prepared in a collection plate. Artificially fouled PreDictor MabSelect SuRe was cleaned using: (1) PBS (20 mM phosphate, 0.15 M NaCl, pH 7.4) (control 1), (2) Proprietary CIP solution, (3) 0.15 M phosphoric acid, pH 1.6, (4) 6.0 M Guanidine hydrochloride, (5) 0.1 M NaOH + 1.0 M NaCl, (6) 0.1 M NaOH, (7) 0.5 M NaOH, (8) PBS, no fouling (control 2). The relative protein concentration on the resin was measured using chip electrophoreses (n = 4, mean ± SD).
Evaluation of occurrence of cross-contamination between wells during resin transfer using centrifugation.
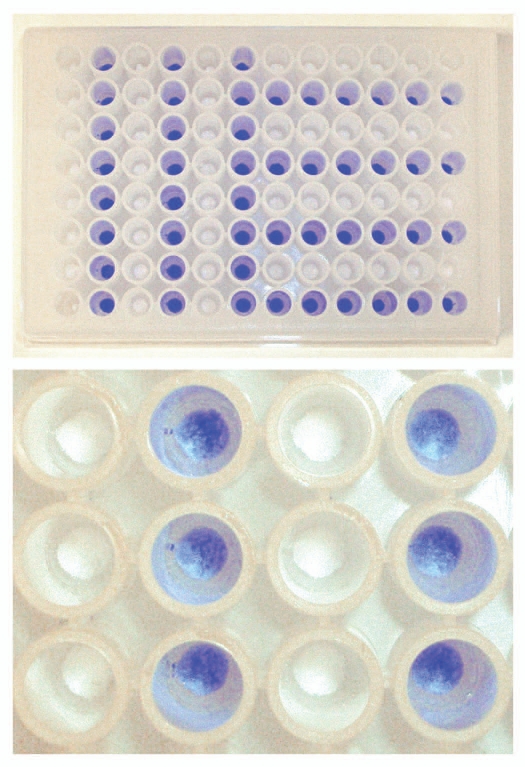
In order prepare samples for SDS-PAGE or chip electrophoresis, the resin samples were transferred from the 96-well filter plate to a collection plate. Originally, a spoon was used for manual transfer of each resin sample from the wells of the filter plate to a collection plate. In the improved method, the drained chromatography resin was rapidly transferred using centrifugation. This was achieved through precise placement of a collection plate, upside down, on the 96-well filter plate. The two plates were secured with rubber bands, inverted, and the resin in the wells was transferred from the filter plate to the collection plate using centrifugation. During method development, it was verified that no cross-contamination occurred between wells during resin transfer from a 96-well filter plate to a collection plate using centrifugation. This verification was done by coloring the MabSelect SuRe resin in every second column in the first half of the plate and in every second row in the second half of a 96-well filter plate, using bromophenol blue (Fig. 5, top). After liquid evacuation, the drained resin was transferred to a collection plate using centrifugation. By visual inspection it could be confirmed that no blue colored MabSelect SuRe particles were seen among the uncolored ones (Fig. 5, bottom). Thus, no cross-contamination occurred using centrifugation for transfer of drained resin from a 96-well filter plate to a collection plate.
Figure 5.
Evaluation of occurrence of cross-contamination between wells during resin transfer using centrifugation. MabSelect SuRe resin in every second column in the first half of a PreDictor plate and in every second row in the second half of the PreDictor plate was colored using bromophenol blue (top). After liquid evacuation the drained resin was transferred to a collection plate using centrifugation (bottom).
Evaluation of the reproducibility of resin volumes after resin transfer for sample preparation.
The reproducibility in resin volume after transfer to the collection plate was also investigated. A 96-well filter plate filled with 20 µl MabSelect SuRe was equilibrated with PBS before transfer of the drained resin to a collection plate. The transferred resin was then incubated in an excess amount of pure polyclonal IgG. The IgG in solution bound to MabSelect SuRe. Since this was a closed system, the bound IgG amount was equal to the difference between the initial IgG amount in solution and the IgG amount in the supernatant. The amount of IgG bound to the resin was assumed to be proportional to the resin volume in the wells. Thus, the standard deviation of the optical density (OD) at 280 nm in the supernatant, i.e., unbound IgG, should reflect the reproducibility in resin volume between wells. This method showed a relative standard deviation of 2% between wells. This result can be compared to the relative standard deviations for the resin volumes of PreDictor plates, which is in the order of 1–5%.3
Chip electrophoresis for analysis of residual protein impurities after cleaning of MabSelect SuRe fouled by repeated bind-elute cycles with harvested cell culture fluid.
In the improved and final version of the high-throughput method for development of CIP protocols for MabSelect SuRe, the resin in 96-well filter plates was fouled by repeated load-elution using HCCF [Chinese hamster ovary (CHO) cell supernatant] with mAb and buffers normally used in the process.6 No fouling agent was added. Equilibration, application of feed, wash and elution was repeated ten times corresponding to ten chromatography cycles (Fig. 6). No cleaning was included in this fouling procedure.
Figure 6.
Schematic description of fouling of MabSelect SuRe resin in PreDictor plates by repeated bind-elute cycles. Steps 1 and 3 show equilibration or wash with PBS (20 mM phosphate, 0.15 M NaCl, pH 7.4). In step 2, harvested cell culture fluid with mAb was added to the resin and incubation was done for 30 min. In step 4, the bound mAb was eluted with 0.1 M sodium citrate pH 3.5. Steps 1–4 were repeated ten times, corresponding to ten chromatography cycles. In each step, mixing was done and centrifugation was used for liquid removal between each step.
The fouled resin was cleaned using single cleaning steps or sequences of multiple cleaning steps (Table 1). Between each sequential cleaning step, the resin was washed with PBS followed by wash with ultrapure water (Fig. 2). The residual protein impurities on the cleaned resin were measured using chip electrophoresis (Caliper). Chip electrophoresis was selected because of the automation and high-throughput capability of the analytical method. Figure 7 shows examples of a virtual slab gel and corresponding electopherograms generated by the Caliper software. It was also possible to generate semi-quantitative data, which is convenient when evaluating hundreds of data points.
Table 1.
Thirty-two different cleaning protocols including one control (wash with PBS) were evaluated for cleaning efficiency of fouled MabSelect SuRe resin
| CIP protocol | CIP step 1 | CIP step 2 | CIP step 3 | CIP step 4 |
| 1 | PBS, pH 7.4 (Control) | |||
| 2 | 0.1 M citric acid pH 3.0 | |||
| 3 | 0.1 M citric acid pH 2.5 | |||
| 4 | 6 M Gua-HCl | |||
| 5 | 100 mM 1-thioglycerol, pH 8.5 | |||
| 6 | 0.1 M citric acid pH 3.0 | 6 M Gua-HCl | ||
| 7 | 0.1 M citric acid pH 3.0 | 100 mM 1-thioglycerol, pH 8.5 | 6 M Gua-HCl | |
| 8 | 0.1 M citric acid pH 3.0 | 100 mM 1-thioglycerol, pH 8.5 | 6 M Gua-HCl | 0.1 M citric acid pH 3.0 |
| 9 | 8 M urea, 0.1 M citric acid, pH 3.0 | |||
| 10 | 8 M urea, 1 M NaCl, 0.1 M citric acid, pH 3.0 | |||
| 11 | 100 mM NaOH | |||
| 12 | 0.1 M citric acid pH 3.0 | 100 mM NaOH | ||
| 13 | 0.1 M citric acid pH 2.5 | 100 mM NaOH | ||
| 14 | 100 mM NaOH | 0.1 M citric acid pH 3.0 | ||
| 15 | 0.1 M citric acid pH 3.0 | 100 mM NaOH | 0.1 M citric acid pH 3.0 | |
| 16 | 8 M urea, 0.1 M citric acid, pH 3.0 | 100 mM NaOH | ||
| 17 | 8 M urea, 1 M NaCl, 0.1 M citric acid, pH 3.0 | 100 mM NaOH | ||
| 18 | 0.1 M ascorbic acid | 100 mM NaOH | ||
| 19 | 0.1 M citric acid pH 3.0 | 70% ethanol | 100 mM NaOH | |
| 20 | 0.1 M citric acid pH 3.0 | 30% isopropanol | 100 mM NaOH | |
| 21 | 0.5 M NaOH | |||
| 22 | 0.1 M citric acid pH 3.0 | 0.5 M NaOH | ||
| 23 | 0.1 M citric acid pH 3.0 | 0.3 M NaOH | ||
| 24 | 100 mM 1-thioglycerol, pH 8.5* | 100 mM NaOH | ||
| 25 | 100 mM 1-thioglycerol, pH 8.5 | 100 mM NaOH | ||
| 26 | 50 mM 1-thioglycerol, pH 8.5 | 100 mM NaOH | ||
| 27 | 100 mM DTT, pH 8.5 | 100 mM NaOH | ||
| 28 | 0.1 M citric acid pH 3.0 | 100 mM 1-thioglycerol, pH 8.5 | 100 mM NaOH | |
| 29 | 0.1 M citric acid pH 3.0 | 100 mM 1-thioglycerol, pH 8.5 | 100 mM NaOH | 0.1 M citric acid pH 3.0 |
| 30 | 100 mM 1-thioglycerol, pH 8.5 | 0.5 M NaOH | ||
| 31 | 0.1 M citric acid pH 3.0 | 100 mM 1-thioglycerol, pH 8.5 | 0.5 M NaOH | |
| 32 | 100 mM NaOH | 100 mM 1-thioglycerol, pH 8.5 |
The 32 CIP protocols contained sequences with 1–4 cleaning steps. The composition of the reducing agent containing solutions were: 50 or 100 mM 1-thioglycerol, 25 mM Tris, 0.15 M NaCl, 25 mM KCl, 1 M EDTA (*without EDTA), pH 8.5 or 100 mM DTT, 25 mM Tris, 0.15 M NaCl, 25 mM KCl. For the CIP steps where no chemical is indicated, ultrapure water was added to the wells in the 96-well filter plate. The remaining protein impurities on the fouled MabSelect SuRe resin after cleaning using these CIP protocols is presented in Figure 8. Note: US patent 6972327 (Immunex Corporation) apply to CIP protocols 7 and 8.
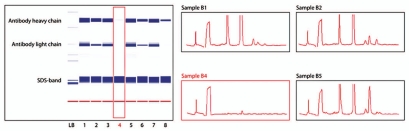
Figure 7.
Chip electrophoresis analysis of residual impurities on MabSelect SuRe after cleaning. The Caliper software generates virtual slab gels (left) and electropherograms (right). Low intensity lane in the virtual slab gel, and corresponding electropherogram, representing the most efficient CIP protocol in this figure, are highlighted. The main protein impurity on the fouled MabSelect SuRe resin was mAb; the large size molecular peak in B5 is most likely un-reduced antibody.
The relative protein impurity levels on the MabSelect SuRe resin, measured by chip electrophoresis after cleaning with 32 CIP protocols, are presented in Figure 8. Quantitative calculation of all the impurity peaks, including the antibody peaks, and baseline subtraction were automatically achieved by the software's default settings.
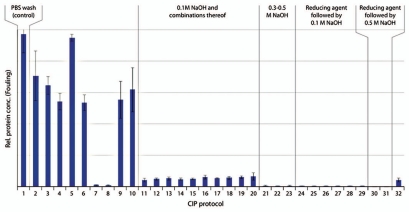
Figure 8.
Remaining impurities on fouled MabSelect SuRe resin after cleaning with 32 different protocols including one control (n = 3, mean ± SD). Detailed information about the composition of the CIP protocols is found in Table 1. Low protein concentration on the resin corresponded to efficient cleaning and vice versa. CIP protocol 11 is cleaning with 0.1 M NaOH. In CIP protocols 12–20, 0.1 M NaOH was combined with acidic steps, solvent, alcohol or chaotropic agents. CIP 21–23 corresponded to 0.3–0.5 M NaOH with/without acidic cleaning. In CIP 24–29 the resin was cleaned with reducing agent followed by 0.1 M NaOH with or without acidic cleaning and CIP 30 and 31, reducing agent was followed by 0.5 M NaOH. In CIP 32, the reversed order, i.e., 0.1 M NaOH followed by 100 mM 1-thioglycerol was used.
Efficient cleaning of MabSelect SuRe was obtained with 0.1 M NaOH (CIP protocol 11). No further improvement was seen when 0.1 M NaOH was combined with acidic steps, solvent, alcohol or chaotropic agents (CIP protocols 12–20). Even more efficient cleaning was obtained with 0.3-0.5 M NaOH (CIP protocols 21–23) or with reducing agent (50–100 mM 1-thioglycerol or 100 mM DTT) followed by NaOH (CIP protocols 24–31). No difference was observed between the samples that had been cleaned with 50 or 100 mM 1-thioglycerol. After cleaning with 100 mM reducing agent, followed by 0.5 M NaOH, no trace of protein impurities could be detected on the resin (CIP 30 and 31). The reversed order of the two steps, 0.1 M NaOH followed by 100 mM 1-thioglycerol (CIP protocol 32), did not show improved cleaning efficiency compared to 0.1 M NaOH alone. A sequence of 100 mM 1-thioglycerol followed by 6 M guanidine hydrochloride also gave very efficient cleaning (CIP protocols 7 and 8). Alone, 100 mM 1-thioglycerol did not show any clearance of protein impurities (CIP protocol 5).
Discussion
Fouling of chromatography resin in 96-well filter plates.
When performing numerous subsequent cycles in column mode, there can be a gradual buildup of contaminants on the chromatography resin, causing fouling of the column. PreDictor plates, 96-well filter plates pre-filled with chromatography resin, are intended for single use and not for recycling. Therefore, a procedure that would accelerate the fouling of the resin in the 96-well filter plates was needed to be able to use them for evaluation of cleaning conditions. In the initial method for evaluation of cleaning conditions, an artificial fouling of the resin was performed by using E. coli lysate spiked with polyclonal IgG and a precipitation solution composed of 2.9 M ammonium sulfate, 0.6 M phosphoric acid, pH 2.5. E. coli lysate was used because it has a higher content of host cell proteins (HCP) compared to mammalian cell supernatants. The precipitation solution was used to enhance and accelerate the fouling according to previous unpublished studies (personal communication, Malin Eriksson, GE Healthcare). The artificial fouling resulted in large amount of protein impurities on the resin (Figs. 3 and 4). By using this method it was possible to distinguish between the cleaning efficiency of different CIP chemicals in removing the precipitated proteins. However, when selecting a CIP procedure for a specific process, it is desired to use the feed of interest, i.e., mammalian cell culture supernatant with mAb for a Protein A resin and buffer components that are included in the process, instead of a model system and atypical chemicals.
In the improved and final version of the high-throughput method for development of CIP protocols for chromatography resins, 96-well filter plates were fouled with CHO cell supernatant with mAb by repeated load-elute cycles. The number of load-elute cycles that were required for fouling of the resin was not fully explored. However, with the CHO cell supernatant used here, ten load-elute cycles generated a fouled resin. CIP conditions for MabSelect SuRe were described here, but the method could be used for screening of cleaning conditions for other combinations of chromatography resins and feeds. It is possible that feeds of other origins and target proteins require a different number of load-elute cycles for the chromatography resin to be sufficiently fouled for screening of CIP conditions.
Sample preparation.
After fouling and cleaning of the resin in 96-well filter plates, the residual protein impurities on the resin were analyzed using electrophoresis. Two different modes of sample preparation were compared. One approach was to prepare the samples in the 96-well filter plate, where the resin had been fouled and cleaned. The other approach was to transfer the drained resin to a collection plate before sample preparation. Even though in general the relative impurity concentration was the same for both modes of sample preparation, the samples that had been prepared in the 96-well filter plate gave higher impurity concentrations compared to the samples prepared in the collection plate. This was most significant for the least efficient CIP protocols. One explanation could be that proteins that precipitated/bound to the filter and plastic surface of the filter plate during fouling were released when the supernatant was removed through the filter after sample preparation. Instead, when transferring the resin samples from the filter plate to a collection plate before sample preparation, these proteins would remain trapped in the filter plate. Thus, the latter method would enable analysis exclusively of the impurities bound to the chromatography resin, which is preferable. The samples in this part of the study were extensively diluted prior to chip electrophoresis analysis. In the final method, 100 µL of sample buffer was added to the 50 or 20 µL resin samples. The supernatant was then taken out and analyzed without further dilution. More concentrated samples, i.e., higher impurity levels, made it easier to distinguish between the most efficient CIP protocols.
Early in method development, a small tool (spoon) was used to manually transfer the drained resin from the 96-well filter plate to a vial for sample preparation. This method was tedious, time-consuming and not very accurate. Reproducibility and high-throughput were requisite when improving the method for transfer of resin from the 96-well filter plate before sample preparation. The improved method included resin transfer from a 96-well filter plate to a collection plate by using centrifugation. This method proved to be robust and rapid with high reproducibility and no cross-contamination between wells.
CIP screening and optimization.
Cleaning conditions were screened by incubation of the fouled resin in different CIP agents. In this paper, an incubation time of 15 min was used for the Protein A-derived resin MabSelect SuRe. This incubation time corresponds to the CIP contact time in a column; for Protein A resins, typically 15 min. Other combinations of resins and feeds may require different contact times. After identifying promising CIP chemicals, the CIP procedure can be further optimized by exploring different chemical concentrations and contact times using 96-well filter plates. If a sequence is identified as a promising CIP candidate, it should be evaluated excluding the washes in between the CIP steps in order to keep the column maintenance program as short as possible.
Column verification.
The mechanism and mass transfer of fouling and cleaning will be different in a batch format compared to a column format. It is therefore important to verify the obtained results by column chromatography, with particular focus to the top material of the column, which has a different degree of fouling compared to the bottom. This is typically done in a scale-down model of the intended production scale. Several studies have proved that the correlation between the scaled-down, 96-well format and traditional columns is good.6–9 For MabSelect SuRe, a CIP using 0.1 M NaOH at a contact time of 15 min gives efficient cleaning in a mAb process, and enables use of the column for at least 150 cycles.6 More challenging feeds can benefit from a two-step CIP protocol using 100 mM reducing agent followed by 0.1 M NaOH on MabSelect SuRe.7 MabSelect, based on recombinant Protein A, which is less stable in alkaline conditions compared with MabSelect SuRe, benefits from a two-step CIP protocol using 100 mM reducing agent followed by 15 mM NaOH.8,9
Manual or automated workflow.
Fouling of resin and screening of cleaning conditions can be done in a manual workflow using multi-pipettes and manual centrifugation or vacuum filtration. The procedure has also been implemented on a fully automated robotic system such as Tecan™. Working manually, fouling of the resin using ten subsequent load-elute cycles required two working days. Using the Tecan robot, the ten cycles can be performed overnight. Screening of CIP conditions is done in half a working day and so is analysis of residual protein impurities using chip electrophoresis.
In summary, a high-throughput method for screening of CIP conditions for chromatography resins was developed to match the increasing demand for reduced process development time and cost within biopharmaceutical industry. The method includes fouling of the resin in 96-well filter plates using HCCF, cleaning by incubation of the fouled resin in CIP chemicals, and finally analysis of the residual protein impurities on the resin after cleaning.
Initially, a model feed was used and accelerated fouling of the chromatography resin was carried out by addition of a precipitation solution (acid/salt). Subsequent to a manual and tedious sample preparation, analysis was done using SDS-PAGE.
The method was gradually improved in terms of reproducibility, throughput and automation. In the up-graded method, HCCF with mAb, and buffer components included in the specific process were used. The chromatography resin was fouled by ten repeated bind-elute cycles. The throughput and reproducibility during sample preparation were also improved. Furthermore, chip electrophoresis for analysis of residual impurities on the resin after cleaning enabled quantification of protein impurities with a high level throughput and automation. It was also interesting to note that in both the E. coli lysate and the CHO cell culture system the dominant cause of fouling, as identified by SDS-PAGE or chip electrophoresis, was product related.
The method has been used to screen numerous CIP chemicals and sequences of CIP steps for MabSelect SuRe in a mAb process. A one-step CIP procedure using 0.1 M NaOH at a contact time of 15 min was identified to efficiently clean the MabSelect SuRe resin. For more challenging feeds and for conventional Protein A resins that are less stable in alkaline conditions, two-step CIP protocols using reducing agent followed by NaOH can be an option.
Materials and Methods
Artificial fouling of the chromatography resin.
A PreDictor MabSelect SuRe, 50 µl plate (Code no. 28-9258-25, GE Healthcare, Sweden) was opened according to the manufacturer's instruction.10 The storage solution (20% ethanol) was removed and the resin was equilibrated using 200 µL PBS (20 mM phosphate, 0.15 M NaCl, pH 7.4) x3. The bottom of the plate was covered with Microseal® ‘F’ film (product number MSF1001, Bio-Rad Laboratories, USA) before adding feed/CIP solutions, to avoid liquid leakage from the plates during incubation. It was later discovered that leakage from the plates could be avoided by carefully blotting the bottom of the PreDictor plates on a soft paper tissue to remove any drops before adding new liquid, and avoiding direct contact between the PreDictor plate outlets and any surface by keeping the PreDictor plate on a collection plate throughout the experiment.3,10
E. coli [BL21 (DE3) wild type] was high pressure homogenized. The lysate was then centrifuged, and the supernatant was subsequently filtrated through a 0.45 µm pore size filter to obtain a clarified solution, and then pH adjusted to 7.3. 200 µL clarified E. coli lysate spiked with polyclonal IgG, Gammanorm (Product number 008565, Octapharma, Sweden) to a concentration of 3 mg/mL was added to the resin in the wells. To some wells used as controls, 200 µL PBS (20 mM phosphate, 0.15 M NaCl, pH 7.4) was added instead of E. coli lysate. The top of the PreDictor plate was covered with Microseal ‘F’ film and incubation was performed for 45 min on a microplate shaker (MTS 2/4 digital, IKA, Germany, 3 mm circular centripetal movement) at 1,100 rpm. After incubation, the unbound fraction was removed. Loading of 200 µL clarified E. coli lysate spiked with IgG was repeated in total four times. After the final incubation, the resin was washed with 200 µL PBS. The bottom of the PreDictor plate was covered with Microseal ‘F’ film and 200 µL precipitation solution (2.9 M ammonium sulfate, 0.6 M phosphoric acid, pH 2.5) was added to the resin. The top of the plate was covered with Microseal ‘F’ film and incubation was done at 1,100 rpm overnight (approximately 17 h). After a subsequent wash with 200 µL PBS and elution with 200 µL 0.1 M sodium citrate pH 3.0 x3, the resin was re-equilibrated with 200 µL PBS x3 and washed with 200 µL ultrapure water x3. In all steps above, an 8-channel multi-pipette was used for sample and buffer addition to the plate. Centrifugation at 500x g for 1 minute (Centrifuge 5810 R with rotor A-2-DWP, Eppendorf, Germany) was performed for liquid removal from the plates. The liquid fractions were collected into 96-well collection plates (Assay block 0.5 mL, V-bottom, product number 3957 Costar®, Corning Incorporated, USA) and discarded. The fouled resin was then used for evaluation of CIP conditions.
Fouling by repeated bind-elute cycles.
A PreDictor MabSelect SuRe, 20 µL plate (Code number 28-9258-24, GE Healthcare, Sweden) was opened according to the manufacturer's instruction.10 The storage solution (20% ethanol) was removed and the resin was equilibrated using 200 µL PBS (20 mM phosphate, 0.15 M NaCl, pH 7.4) x3. The bottom of the plate was blotted on a soft paper tissue and placed on an empty collection plate 96-well 500 µL V-shaped bottom (Code no. 28-4039-43, GE Healthcare, Sweden). 300 µL CHO cell supernatant with mAb (1.1 mg/mL) (GE Healthcare, Sweden) was applied to the wells. The top of the PreDictor plate was covered with microplate foil (96-well) (Code no. BR-1005-78, GE Healthcare, Sweden). The PreDictor plate placed on the collection plate was secured with rubber bands on a microplate shaker (MTS 2/4 digital, IKA, Germany, 3 mm circular centripetal movement) and incubation was performed for 30 min at 1,100 rpm. Following a post-load wash with 200 µL PBS, the bound mAb was eluted with 200 µL 0.1 M sodium citrate, pH 3.5 x 2. In each step above, mixing was done briefly at 1,100 rpm on a microplate shaker and centrifugation at 500x g for 1 minute (Centrifuge 5810 R with rotor A-2-DWP, Eppendorf, Germany) was performed for liquid removal from the plate. The liquid fractions were collected into collection plates (96-well, 500 µL, V-shaped bottom; Code no. 28-4039-43, GE Healthcare, Sweden) and discarded. Equilibration, application of feed, wash and elution was repeated in total ten times (Fig. 6). The procedure was done manually in two working days. After the 5th cycle, the resin in the PreDictor plate was stored overnight in 200 µL PBS at 5°C. The top of the PreDictor plate was covered with microplate foil (96-well) (Code no. BR-1005-78, GE Healthcare, Sweden) and the bottom of the plate was covered with Microseal ‘F’ film (product number MSF1001, Bio-Rad Laboratories, USA) during storage. The following day the cycles 6–10 were performed. After the last cycle, the resin in the wells was washed with 200 µL ultrapure water and stored in 200 ~−L ultrapure water until used for screening of CIP conditions. The top of the PreDictor plate was covered with microplate foil (96-well) (Code no. BR-1005-78, GE Healthcare, Sweden) and the bottom of the plate was covered with Microseal ‘F’ film (product number 132097, Bio-Rad Laboratories, USA) during storage.
Screening of CIP conditions.
The fouled resin in the PreDictor plate was washed with 200 µL ultrapure water x2. The bottom of the PreDictor plate was blotted on a soft paper tissue and the plate was placed on a collection plate 96-well 500 µL V-shaped bottom (Code no. 28-4039-43, GE Healthcare, Sweden). 300 µL CIP solutions were added to the wells and the top of the predictor plate was covered with microplate foil (96-well) (Code no. BR-1005-78, GE Healthcare, Sweden). The PreDictor plate placed on the collection plate was secured with rubber bands on a microplate shaker (MTS 2/4 digital, IKA, Germany, 3 mm circular centripetal movement) and incubation was done for 15 min at 1,100 rpm. 300 µL PBS (20 mM phosphate, 0.15 M NaCl, pH 7.4) was added followed by wash with 300 µL ultrapure water. This procedure (incubation in CIP solutions and washes) was repeated up to in total four times depending on the number of CIP steps in the sequence. After the final CIP step, the fouled and cleaned resin in the wells was washed with 300 µL PBS x3 followed by wash with 300 µL ultrapure water x3 (Fig. 2). In each step, mixing was done briefly at 1,100 rpm on a microplate shaker and centrifugation at 500x g for 1 min (Centrifuge 5810 R with rotor A-2-DWP, Eppendorf, Germany) was performed for liquid removal from the plate. The liquid fractions were collected into collection plates (96-well, 500 µL, V-shaped bottom; Code no. 28-4039-43, GE Healthcare, Sweden) and discarded.
CIP protocols for cleaning of artificially fouled MabSelect SuRe.
Artificially fouled PreDictor MabSelect SuRe, 50 µL (Code no. 28-9258-25, GE Healthcare, Sweden) was cleaned using: (1) Wash with PBS (20 mM phosphate, 0.15 M NaCl, pH 7.4, control 1), (2) 0.1 M NaOH, (3) 0.2 M NaOH, (4) 0.5 M NaOH, (5) 0.1 M NaOH + 30% 2-propanol, (6) Commercial house-hold detergent (Yes, Procter & Gamble) diluted to 0.1% v/v with water, (7) 0.1 M NaOH + 1.0 M NaCl, (8) 0.1 M NaOH followed by 0.1 M citrate, (9) 0.1% commercial house-hold detergent followed by 0.1 M NaOH and (10) No fouling (control 2). Analysis of residual protein impurities after artificial fouling and cleaning was done using SDS-PAGE.
CIP protocols for comparison of sample preparation in PreDictor plate and collection plate.
Artificially fouled PreDictor MabSelect SuRe, 50 µL (Code no. 28-9258-25, GE Healthcare, Sweden) was cleaned using: (1) PBS (20 mM phosphate, 0.15 M NaCl, pH 7.4) (control 1), (2) Proprietary CIP solution, (3) 0.15 M phosphoric acid, pH 1.6, (4) 6.0 M Guanidine hydrochloride, (5) 0.1 M NaOH + 1.0 M NaCl, (6) 0.1 M NaOH, (7) 0.5 M NaOH, (8) PBS, No fouling (control 2), four replicates for each condition. Sample preparation was done directly in the PreDictor plate or after transfer to a collection plate. The samples in this study were extensively diluted prior to chip electrophoresis analysis. First 200 µL sample buffer was added to the 50 µL resin. After heat treating of the sample, a volume of 2 µL of the supernatant was then further diluted 22 times (2:44) before analysis of residual protein impurities after fouling and cleaning using chip electrophoresis.
CIP protocols for cleaning of MabSelect SuRe fouled by repeated bind-elute cycles with CHO cell supernatant with mAb.
PreDictor MabSelect SuRe, 20 µL (Code number 28-9258-24, GE Healthcare, Sweden) fouled by repeated bind-elute cycles using CHO cell supernatant with mAb was cleaned with 32 different CIP protocols including 1–4 CIP steps (Table 1). CIP solutions were prepared and dispensed from deep-well plates, Microplate 48 Deep well, 5 mL/well (Product number 360002, Seahorse Bioscience, North Billerica, MA) according to the experimental plate layout. The protocols were run in triplicates and analyzed for residual protein impurities using chip electrophoresis as described below.
Transfer of fouled and cleaned resin from the PreDictor plate to a collection plate by using centrifugation.
The liquid was removed from the PreDictor plate using centrifugation at 500x g for 1 min. A collection plate was placed very precisely upside down on the PreDictor plate. The two plates were secured with rubber bands. The plates were inverted and the drained resin in the wells was transferred from the filter plate to the collection plate using centrifugation (Centrifuge Avanti J-20 XP with rotor JS-5.3, Beckman Coulter, Brea, CA USA) at 972x g for 5 min.
Comparison of sample preparation in the PreDictor plate and sample preparation in a collection plate.
The resin in two Predictor MabSelect SuRe 50 µl plates (Code no. 28-9258-25, GE Healthcare, Sweden) was artificially fouled as described above. Some of the wells were used as controls and instead of feed, PBS buffer (20 mM phosphate, 0.15 M NaCl, pH 7.4) were added to these wells. Six model CIP solutions according to the description above were used to compare the two modes of sample preparation. Sample buffer for chip electrophoresis was prepared by dissolving 3 g Tris in 40 mL ultrapure water. The pH was adjusted to 7.5 with glacial acetic acid. The final volume was adjusted to 50 mL by adding ultrapure water. 0.5 g SDS was dissolved in 5 mL of the Tris buffer and the volume was adjusted to 50 mL by adding ultrapure water. The sample buffer was prepared by mixing 25 mL SDS buffer and 1.39 mL 1 M DTT and the volume was adjusted to 50 mL by adding ultrapure water. For one of the fouled and cleaned PreDictor plates, the drained resin samples were transferred to a collection plate by using centrifugation as described above. The liquid was drained from the other PreDictor plate using centrifugation at 500x g for 1 min. The bottom of the PreDictor plate was covered with Microseal ‘F’ film (product number MSF1001, Bio-Rad Laboratories, USA). 200 µL of SDS/DTT containing sample buffer for chip electrophoresis was added to the drained resin samples in the PreDictor plate and in the collection plate. The top of the plates were covered with Microseal ‘F’ film and the resin was incubated in sample buffer for 15 min on a microplate shaker (MTS 2/4 digital, IKA, Germany, 3 mm circular centripetal movement) at 1,100 rpm and then heated in a heating chamber at 104°C for 5 min. The microplate foil was removed from the PreDictor plate and the supernatant was collected into a collection plate using centrifugation at 500x g for 1 min. In the other sample preparation mode, the collection plate with samples heat treated in SDS/DTT was again mixed briefly at 1,100 rpm before the plate was centrifuged at 500x g for 2 min to spin down the chromatography resin. The supernatants from the PreDictor plate and the collection plate were further diluted, 2:44, before analysis by chip electrophoresis.
Investigation of cross-contamination during transfer of resin from a PreDictor plate to a collection plate using centrifugation.
A PreDictor MabSelect SuRe 50 µL plate (Code no. 28-9258-25, GE Healthcare, Sweden) was opened according to the manufacturer's instruction.10 The storage solution (20% ethanol) was removed from the PreDictor plate by using centrifugation (Centrifuge Avanti J-20 XP with rotor JS-5.3, Beckman Coulter, Brea, CA) at 500x g for 1 min. The resin in the plate was equilibrated with 200 µL PBS (20 mM phosphate, 0.15 M NaCl, pH 7.4) x3. Between each equilibration step, the liquid was removed using centrifugation at 500x g for 1 min. Two hundred microliters of bromophenol blue dissolved in water was added to the wells in every second column in the first half of the PreDictor plate and in every second row in the second half of a PreDictor plate. The plate was incubated on a microplate shaker (MTS 2/4 digital, IKA, Germany, 3 mm circular centripetal movement) at 750 rpm for 60 min. After removal of the bromophenol solution by centrifugation at 500x g for 1 min, the drained MabSelect SuRe resin was transferred from the PreDictor plate to a collection plate using centrifugation. The collection plate was inspected visually for occurrence of any colored particles among the uncolored ones.
Investigation of reproducibility during transfer of resin from a PreDictor plate to a collection plate using centrifugation.
A PreDictor MabSelect SuRe 20 µL plate (Code number 28-9258-24, GE Healthcare, Sweden) was opened according to the manufacturer's instruction.10 The storage solution (20% ethanol) was removed from the PreDictor plate by using centrifugation (Centrifuge Avanti J-20 XP with rotor JS-5.3, Beckman Coulter, Brea, CA) at 500x g for 1 min. The resin in the plate was equilibrated with 200 µL PBS (20 mM phosphate, 0.15 M NaCl, pH 7.4) x3. Between each equilibration step, the liquid was removed using centrifugation at 500x g for 1 min. The resin was transferred to a collection plate using centrifugation. 250 µl of polyclonal IgG Gammanorm (Product number 008565, Octapharma, Sweden) diluted to a concentration of 3.3 mg/mL in PBS, was added to the resin in the collection plate and incubation was done for 60 min at 300 rpm on a microplate shaker (MTS 2/4 digital, IKA, Germany, 3 mm circular centripetal movement). After incubation, 100 µL of the supernatant was transferred to a UV-readable plate analyzed for OD at 280 nm. The standard deviation for the OD at 280 nm was calculated.
SDS-PAGE.
The artificially fouled and cleaned resin samples were transferred to 500 µL Eppendorf tubes by using a small spoon (Product number 149940-4, WWR, USA). Between each sample, the spoon was washed with 20% ethanol. 20 µL Novex Tris-Glycine SDS sample buffer (LC2676, Invitrogen, USA) and 2 µL 1 M DTT was added to each sample. The samples were heat treated for 3 min at 95°C. The samples (resin slurry) were mixed before 10 µL of each slurry sample (both resin and liquid) was loaded to the wells of a Novex 8–16% Tris-Glycine gel (EC6045BOX, Invitrogen) mounted in a MiniVE holder (GE Healthcare, Sweden). The electrophoresis was run using 0.25 M Tris, 0.19 M Glycine, 0.1% SDS as running buffer under following conditions: 100 V, 400 mA for 2.10 hours. The gel was stained with Deep Purple™ Total Protein stain kit (RPN0306, GE Healthcare, Sweden) according to the manufacturer's instruction and the gel was scanned using a Typhoon 9410 (GE Healthcare, Sweden) at 100–200 µm resolution.
Chip electrophoresis.
Sample buffer for chip electrophoresis was prepared by dissolving 3 g Tris in 40 mL ultrapure water. The pH was adjusted to 7.5 with glacial acetic acid. The final volume was adjusted to 50 mL by adding ultrapure water. 0.5 g SDS was dissolved in 5 mL of the Tris buffer and the volume was adjusted to 50 mL by adding ultrapure water. The sample buffer was prepared by mixing 25 mL SDS buffer and 1.39 mL 1 M DTT and the volume was adjusted to 50 mL by adding ultrapure water.
The fouled, cleaned and drained MabSelect SuRe resin in the PreDictor plate was transferred to a collection plate 96-well 500 µl V-shaped bottom (Code no. 28-4039-43, GE Healthcare, Sweden) by using centrifugation as described above. 100 µl of the SDS/DTT containing sample buffer was added to the resin in each well in the collection plate. The plate was covered with Microseal ‘F’ film (product number MSF1001, Bio-Rad Laboratories, USA) and mixed briefly at 1,100 rpm on a microplate shaker (MTS 2/4 digital, IKA, Germany, 3 mm circular centripetal movement) before heating the plate in a heating chamber at 104°C for 5 min. The collection plate was again mixed briefly at 1,100 rpm before it was centrifuged at 500x g for 2 min to spin down the chromatography resin. 40 µL of the supernatant was transferred from the collection plate to a ThermoFast 96 skirted PCR-plate (Product number AB-0800-L, Abgene Limited, UK) for analysis by chip electrophoresis.
A Caliper HT Protein Express chip (P/N 760301, Caliper LifeSciences, USA) was prepared according to the manufacturer's instruction.11 The HT Protein Express 100 high sensitivity assay was run on a Caliper LabChip™ 90 with the control software LabChip HT version 2.6.0 (Caliper Life Sciences, USA). The data was evaluated using the software DataViewer version 1.1.79.0 (Caliper Life Sciences, USA) according to default settings.
Acknowledgements
To Tomas Björkman, Enrique Carredano, Jinyu Zou, Felix Solamo and Hanna Tengliden for great contribution to the work and good discussions, and to Gunnar Malmquist, Andy Masters and Anne Barry for valuable input and critical review of the article.
References
- 1.Hagel L, Jagschies G, Sofer G. Handbook of Process Chromatography—Development, Manufacturing, Validation and Economics. Second edition. London, UK: Academic Press; 2008. Cleaning and Sanitization; pp. 147–159. [Google Scholar]
- 2.Björkman T, Blackwell J, Castillo F, Ecker D, Fulton SP, Jagschies G, et al. High Throughput Methods for Process Development. In: Levine L, Jagshies G, editors. The Development of Therapeutic Monoclonal Antibody Products—A Comprehensive Guide to CMC Activities form Clone to Clinic. Acton: MA Bioprocess Technology consultants Inc; 2010. pp. 190–197. [Google Scholar]
- 3.Handbook: High-throughput Process Development with PreDictor™ Plates, Principles and Methods 28-9403-58 AA. GE Healthcare; 12/2009. [Google Scholar]
- 4.Grönberg A, Tengliden H, Johansson HJ. Rapid development of CIP protocols for affinity media; Poster presented at the SPICA conference; Sept 29-Oct 01, 2008. [Google Scholar]
- 5.Eriksson M. Degree project in Biotechnology and Genomics Engineering. Institute of Technology, Umeå University; 2009. Development of a High Throughput Method for Screening of Cleaning-in-Place Protocols for Chromatography Media. [Google Scholar]
- 6.High-throughput process development for design of cleaning-in-place protocols. 2010 Application note 28-9845-64 AA. GE Healthcare. [Google Scholar]
- 7.Grönberg A, Ersoy M, Johansson HJ, Solamo F. Oral presentation at ACS conference. BIOT program, Washington DC: 2009. High-throughput development of cleaning-in-place protocols for chromatography resins. [Google Scholar]
- 8.Grönberg A, Johansson HJ, Eriksson KO, Carredano E. High-throughput process development technology for design of cleaning-in-place (CIP) protocols for chromatography media; Oral presentation at Miniaturisation—Micro Scale Bioprocess Development conference; UK. 2010. [Google Scholar]
- 9.Grönberg A, Johansson HJ, Eriksson KO, Carredano E. Automated HTPD Technology for design of CIP protocols for Chromatography resins; Poster presented at High-throughput Process Development meeting; Krakow Poland. 2010. [Google Scholar]
- 10. PreDictor plates instructions, 28-9258-34 AD, GE Healthcare.
- 11.HT Protein LabChip Kit User Guide, Caliper Life Sciences. 2007. [Google Scholar]