Abstract
Epigenetic inactivation of tumor suppressor genes is a hallmark of cancer development. RASSF1A (Ras Association Domain Family 1 isoform A) tumor suppressor gene is one of the most frequently epigenetically inactivated genes in a wide range of adult and children's cancers and could be a useful molecular marker for cancer diagnosis and prognosis. RASSF1A has been shown to play a role in several biological pathways, including cell cycle control, apoptosis and microtubule dynamics. RASSF2, RASSF4, RASSF5 and RASSF6 are also epigenetically inactivated in cancer but have not been analyzed in as wide a range of malignancies as RASSF1A. Recently four new members of the RASSF family were identified these are termed N-Terminal RASSF genes (RASSF7–RASSF10). Molecular and biological analysis of these newer members has just begun. This review highlights what we currently know in respects to structural, functional and molecular properties of the N-Terminal RASSFs.
Key words: N-terminal RASSF, RAS, cancer, epigenetic, tumor suppressor
Introduction: The RASSF Family
The RAS-association domain family (RASSF) of proteins consists of ten members, all characterized by the inclusion of an RA-domain (RAS-association domain)1 at either their C-terminus (RASSF1–6) or N-terminus (RASSF7–10). This domain enables RAS binding of most of the classical RASSF family members albeit with a varying degree of affinity and specificity for different RAS members. However, the importance of the RA-domain in many of the classical RASSF protein functions remains to be clarified. To date, the RAS-binding status of N-terminal RASSF family members remains largely unknown. The classical RASSF members also possess a C-terminal SARAH domain (SALVADOR-RASSF-HIPPO). In Drosophila melanogaster these proteins can homo- or heterodimerize resulting in modifications to the Salvador/Warts/Hippo (SWH) signaling pathway. The SWH pathway was originally identified in the fly to control organ size. The core components of the pathway (Hippo, Salvador, Warts/Lats, Mats, Yorkie) are evolutionally conserved in mammals (MST1&2, SAV1, LATS1&2, MOB1, YAP). Recent studies have demonstrated a key role for the SWH pathway in regulating cell contact inhibition, organ size control and development of cancer in mammals.2 The SARAH domain is not present in N-terminal members, which do, however, contain predicted Coiled-coil regions with potential ability for dimerization. Lastly, RASSF1A and RASSF5/Nore1A contain a C1 domain (protein kinase C conserved region) consisting of diacylglycerol/phorbol ester-binding elements.3 This region is also absent in RASSF7–10. The structural differences between the classical and N-terminal RASSF subgroups have led to the suggestion that they are two distinct families.4 RASSF1A is one of the most frequently methylated genes in a wide range of human cancers and it is also the best characterized in terms of biological functions including a role in the highly conserved hippo tumor suppressor network (see refs. 5–7 for extensive reviews on RASSF1A gene methylation and protein function). This review highlights on what we currently know about the N-terminal RASSF genes focusing on structural, functional and epigenetic aspects.
RASSF7
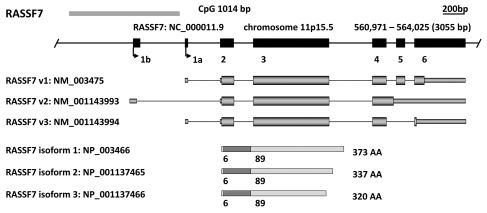
RASSF7 was first identified during the characterization of a 55-kb region of DNA surrounding HRAS1.8 The sequence 29 kb upstream of HRAS1 was originally designated HRC1 (HRAS1-related cluster protein 1) but has now been renamed as RASSF7. The RASSF7 gene is located on chromosome 11p15.5 and from its six exons generates three transcript variants due to alternative splicing at the 3′ region. These encode three main RASSF7 isoforms of 320, 337 and 373 amino acids in length that differ in their C-terminal sequences (Fig. 1). RASSF7 has orthologs in both vertebrates; Mus musculus (NM_025886.3), Danio rerio (BC044177.1), Xenopus laevis (BJ034347.1) and invertebrates; Drosophila melanogaster (CG5053) and Caenorhabditis elegans (K05B2.2) (Table 1). Xenopus RASSF7 is expressed in developing embryonic tissues including epidermis, the neural tube, eye, ear, branchial arches and kidney.4 In murine embryo and adult tissues, RASSF7 exhibits ubiquitous expression but is highly expressed in lung and brain. RASSF7 protein has also been detected in several human cell lines.9
Figure 1.
In silico characterization of the N-terminal RASSF family (RASSF7–10). For each member a genomic map is displayed with the chromosomal location, genomic position, relative position and size of each exon and CpG island as determined by the UCSC Genome Browser (genome.ucsc.edu). Immediately below are the major transcript variants (UCSC) followed by a schematic of protein isoforms as determined by the National Center for Biotechnology Information (NCBI: www.ncbi.nlm.nih.gov/). The location of the RAS-association (RA) domains were predicted by Prosite (expasy.org/prosite).
Table 1.
N-terminal RASSF family characteristics
| Gene | Synonym (www.genecard.org) | Orthologs | CpG Island methylation status | Tumor association[ref] | Putative functions | Putative interactions | Key references |
| RASSF7 | HRC1, C11orf13, HRAS1, HRAS1-related cluster protein 1, MGC126069, MGC126070 | Mus musculus—NM_025886.3 Danio rerio—BC044177.1 Xenopus laevis—BJ034347.1 Drosophila melanogaster—CG5053 Caenorhabditis elegans—K05B2.2 | none found | pancreatic ductal carcinoma,10–12 islet cell tumor,13 endometrial cancer,14 ovarian clear cell carcinoma.15 | mitotic microtubule dynamics, Aurora B activation, necroptotic cell death | DISC1, CHMP1B, TSC1, Aurora B | 4, 8, 9, 48 |
| RASSF8 | Carcinoma-associated protein HOJ-1, C12orf2, FLJ11542, DKFZp434O0227 | Mus musculus—NM_027760 Danio rerio—BC053201 Xenopus laevis—NM_001105273 Drosophila melanogaster—CG5053 | infrequent in leukemia (ALL) | lung carcinoma,31 adult, male germ cell tumor,32 leukemia(ALL),28 lymphoma.33 | cell growth, cell-cell adhesion, cytoskeletal regulation, Wnt signaling, NFκB signaling, necroptotic cell death | ASPP1 & 2, E-cadherin, 14-3-3γ, FRMD6, PSMD4 | 24, 25, 27, 35 |
| RASSF9 | P-CIP1, PAMCI, Peptidylglycine α-amidating mono-oxygenase COOH-terminal interactor | Mus musculus—NM_146240.3 Danio rerio—XM_001922320.1 Xenopus laevis—B1A192 Drosophila melanogaster—CG13875 | no CpG island | none–unstudied | endosomal recycling | N- and K-Ras | 4, 41 |
| RASSF10 | Peptidylglycine α-amidating mono-oxygenase COOH-terminal interactor-like, LOC644943 | Mus musculus—NM_175279 Xenopus laevis—NM_001115020 Drosophila melanogaster—CG32150 | leukemia, (ALL) thyroid gliomas | leukemia (ALL),28 medullary thyroid carcinoma,44 glioma.45 | mitotic progression, hedgehog signaling | No reports thus far | 28, 44, 45 |
This table provides a summary of current knowledge regarding RASSF family methylation status, tumor associations, putative functions and confirmed/putative protein interactions. See text and references therein for full details.
RASSF7 methylation.
RASSF7 has a CpG island comprising 115 CpGs across 1,015 bp (UCSC genome browser) situated upstream of and inclusive of the transcription start site. A region of this CpG island has been analyzed by CoBRA in a large number of cell lines (n = 63) covering a wide range of cancers (lung, breast, colorectal, kidney, glioma, neuroblastoma and leukemia) however no methylation has been identified in these samples.9 As an alternative form of inactivation, mutation screening has also been assessed in a panel of epithelial tumor cell lines however no mutations were found.9
RASSF7 upregulation in tumors.
RASSF7 is upregulated in pancreatic ductal carcinoma,10–12 islet cell tumors,13 endometrial cancer14 and ovarian clear cell carcinoma.15 Evidence for upregulation of RASSF7 by hypoxia-induced gene expression comes from an investigation on hypoxia-induced MicroRNA expression in MCF-7 breast tumor cells.16 As well as detecting hypoxia-mediated hsa-miR-210 overexpression, RASSF7 was also found to be upregulated in a HIF-1a-dependent fashion. Similarly, RASSF7 is upregulated in hypoxic umbilical endothelial cells17 and more recently increased RASSF7 protein was detected in human cell lines exposed to hypoxic conditions.9 Thus RASSF7 is one of a number of proteins that exhibit elevated levels in the hypoxic tissue of epithelial tumors. Additionally, RASSF7 expression is increased in cancer cells that have lost BRCA1 expression or tumor suppressor function.18
RASSF7 function.
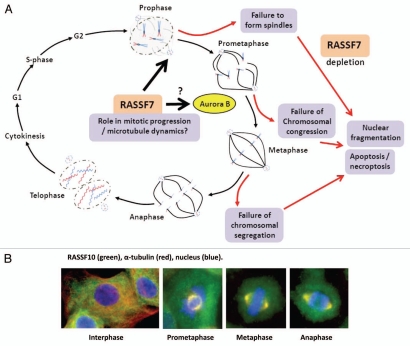
RASSF7 has putative roles in the regulation of cell growth and apoptosis. In a genome-wide RNAi screen for regulators of necroptosis (passive cell death caused by overwhelming stress, the term necroptosis is a particular type of programmed necrosis that depends on RIP1 kinase activity in apoptosis deficient conditions), Hitomi et al. found four Ras-related proteins including RASSF7 and RASSF8.19 Evidence of a role for RASSF7 in cell proliferation came from the discovery of mitotic abnormalities (e.g., reduction in mitotic index) resulting from RNAi-mediated depletion of the RASSF7 Drosophila homolog.20 RASSF7 knockdown studies on developing Xenopus embryos has revealed phenotypic defects including aberrations to body axis development, reduced eye pigmentation and severe defects to the neuroepithelial cell layer resulting in degradation of normal tissue architecture.4 Neuroepithelial cells exhibit nuclear fragmentation representing apoptotic and non-apoptotic morphologies, the latter consistent with mitotic catastrophe. RASSF7 knockdown in neural tube cells have been found to display mitotic arrest and failure to form spindles. This presumably leads to the characteristic nuclear fragmentation and subsequently, cell death. These observations clearly indicated a role for RASSF7 in mitotic progression. Recently, Recino et al. have further characterized the functions of human RASSF7 with respect to regulation of the microtubule cytoskeleton and spindle formation during mitosis.9 This report corroborates investigations on RASSF7 in Xenopus. RASSF7 was required for anchorage-independent growth while RASSF7 knockdown in a human cell line resulted in mitotic abnormalities in metaphase resulting in failure of chromosomal congression. Depletion of RASSF7 caused spindle defects (e.g., decreased polarization, increased numbers of multi-polar spindles). As with Xenopus, human RASSF7 localizes to centrosomes. These abnormalities are also concurrent with a failure in Aurora B activity thus are suggestive of a mechanistic basis for chromosomal alignment defects upon RASSF7 depletion. RASSF7 appears to be required for microtubule regrowth after nocodazole treatment therefore loss of RASSF7 and generation of associated mitotic defects indicates a RASSF7 regulatory role on microtubule dynamics during this phase of the cell cycle (Fig. 2A and Table 1).
Figure 2.
Emerging molecular functions of RASSF7 and RASSF10. (A) During mitosis RASSF7 localizes at centrosomes and with associated microtubules in both Xenopus and human cells. Loss of RASSF7, by RNAi-mediated knockdown, results in spindle defects and failure of chromosome alignment/segregation leading to mitotic catastrophe, DNA degradation and apoptosis. The functional mechanics of RASSF7 are unclear but recent evidence suggests a regulatory role in Aurora B kinase activation and microtubule attachment during the formation of the mitotic spindles in the early stages of mitosis.4,9 (B) The latest member of the N-terminal RASSF family, RASSF10, has been shown to display a cell cycle-dependent distribution, relocating from the cytoplasm into the nucleus at the start of the mitotic cycle where it concentrates at developing centrosomes and associated microtubules.45
RASSF7 interactions.
Thus far, yeast-two hybrid experiments have identified three interacting proteins for RASSF7. The first is a centrosome-associated protein, DISC1 (Disrupted-In-Schizophrenia 1), associated with schizophrenia.21 DISC1 interacts with cytoskeletal and centrosome proteins and co-localizes with the centrosomal complex. The second protein, CHMP1B (Charged multivesicular body protein 1b) forms part of the CHMP protein-interaction network, homologous to the yeast ESCRT III complex, a group of proteins involved in the formation of multivesicular bodies and degradation of internalized transmembrane receptor proteins.22 Lastly, RASSF7 has been reported to interact with the TSC1 (Tuberous sclerosis protein 1) gene product, hamartin, a peripheral membrane protein potentially involved in vesicular transport and docking.23
RASSF8
RASSF8 gene.
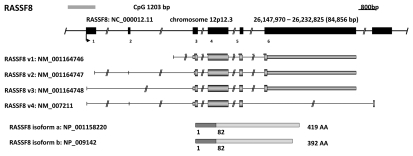
The C12orf2 (chromosome 12 open reading frame 2) gene, later renamed RASSF8 was initially characterized in a chromosomal translocation event, t(12;22)(p11.2;q13.3), with fibulin-1 (FBLN-1) and was associated with a familial form of synpolydactyly.24,25 Prior to this the genetic sequence was listed on human genome browsers as HoJ-1 carcinoma associated gene. The six exon RASSF8 gene is located on chromosome 12p12.3, is 700 kb from the KRAS2 gene26 and is predicted to generate at least four transcript variants encoding two RASSF8 isoforms of 419 and 392 amino acids. These differ at their C-terminus due to different exon usage (Fig. 3). RASSF8 has orthologs in vertebrates; Mus musculus (NM_027760), Danio rerio (BC053201), Xenopus laevis (NM_001105273) and invertebrates; Drosophila melanogaster (CG5053) (Table 1). RASSF8 is ubiquitously expressed in both murine embryos (brain, heart, kidney, liver, lung, spinal cord) and human adult tissues (liver, lung, pancreas, heart, kidney, brain, skeletal muscle, placenta, colon).27
Figure 3.
In silico characterization of the N-terminal RASSF family (RASSF7–10). For each member a genomic map is displayed with the chromosomal location, genomic position, relative position and size of each exon and CpG island as determined by the UCSC Genome Browser (genome.ucsc.edu). Immediately below are the major transcript variants (UCSC) followed by a schematic of protein isoforms as determined by the National Center for Biotechnology Information (NCBI: www.ncbi.nlm.nih.gov). The location of the RAS-association (RA) domains were predicted by Prosite (expasy.org/prosite).
RASSF8 methylation.
Although the UCSC genome browser does not recognise RASSF8 as having a CpG island, the European Bioinformatics Institute (EBI) CpG plot program detects a CpG island of 1,201 base pairs containing 182 CpGs situated upstream of and inclusive of the transcription start site. This region has been assessed for hypermethylation in a range of epithelial cancer cell lines including lung, breast, glioma, kidney and neuroblastoma but no methylation has been detected.27 Although a leukemia study identified hypermethylation in 33% of leukemia cell lines analyzed, hypermethylation was found to be infrequent among samples from leukemia patients (10% childhood T-ALL; 9% childhood B-ALL).28
RASSF8 as a tumor suppressor.
RASSF8 has been described as a potential tumor suppressor in lung carcinogenesis initially by virtue of its locality within the lung tumor-associated D12S1034 minisatellite.29 This region is homologous to a major susceptibility locus in a murine lung cancer model and contains the K-Ras2 gene.30 In 2006, Falvella et al. reported reduced RASSF8 transcript levels in lung adenocarcinoma while re-expression of RASSF8 protein in lung-tumor cell lines was found to inhibit anchorage-independent growth.31 In a reciprocal experiment, RASSF8 depletion through stable knockdown in lung tumor-derived cell lines lead to an increase in anchorage-independent growth and RASSF8-depleted cells were found to display loss of contact-dependent growth inhibition.27 In vivo, RASSF8 knockdown in Xenopus leads to enhanced cell proliferation and inoculation of RASSF8-depleted cells into SCID (severe combined immunodeficiency) mice has been found to promote the development of solid tumors.27 Taken together, these results suggest that loss of RASSF8 correlates with tumor progression and metastatic tendency in lung cancer. Common polymorphisms in RASSF8 were not found to be associated with lung adenocarcinoma risk.26 Additional studies have also indicated a tumor suppressor role for RASSF8 in other cancer cell contexts. Studies on expression levels of 12p target genes in adult, male germ cell tumors found reduced expression of C12orf2,32and in murine models of leukemia and lymphoma retroviral insertion frequently occurred in a region adjacent to the RASSF8 gene.33 One study also found an increase in extracellular RASSF8 RNA in plasma and cell-bound fractions of patients with breast cancer compared to patients with benign tumors, suggesting that RASSF8 is a potential diagnostic marker.34
RASSF8 function.
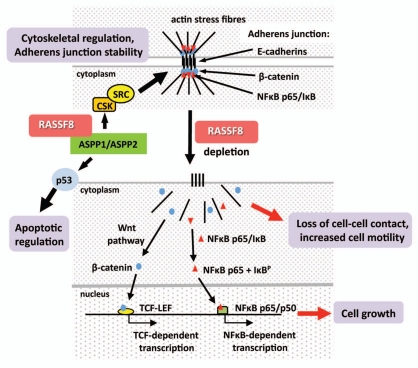
Drosophila RASSF8 (dRASFF8). RASSF8 molecular function remains unclear but studies on both Drosophila and human RASSF8 highlight a potential role in the regulation of cell-cell adhesion and cell growth. dRASSF8 is the Drosophila homolog to human RASSF8 and has been shown to bind dASPP (Drosophila apoptotic specific regulator of p53), both proteins co-localizing at adherens junctions of epithelial cells within the developing Drosophila eye.35 Langton et al.35 suggested that the dASPP-dRASSF8 complex regulates cell-cell adhesion during retinal morphogenesis since a RASSF8 mutant produced aberrant eye patterning and abnormal E-Cadherin localization. These effects are probably through the binding and modulation of dCSK activity and subsequently of Src activation, a known regulator of adherens junction stability and E-Cadherin turnover. Drosophila ASPP is a homolog to mammalian ASPP proteins which serve as stimulators of p53 pro-apoptotic function. The dRASSF8 mutant eye phenotype in Drosophila may also be partly a result of reduced apoptosis of excess interommatidial ‘pigment’ cells. dRASSF8 homozygotes also displayed altered wing morphology with an increase in wing cell number and increased body weight suggesting that dRASSF8 negatively regulates the size of developing tissues. This effect could be rescued upon expression of dRASSF8. Lastly, Langton et al.35 verified that human RASSF8 could also bind to ASPP1 and ASPP2 suggesting that RASSF8 may exhibit evolutionary conserved functions in both Drosophila and humans (Fig. 4). In zebrafish, RASSF8 has been found necessary for blood cell development36 and is also required for necroptotic cell death.19
Figure 4.
Emerging molecular functions of RASSF8. RASSF8 has been observed binding to ASPP (apoptotic specific regulator of p53) in both Drosophila and human cells.35 Drosophila ASPP acts in conjunction with dCSK (Drosophila c-SRC tyrosine kinase) and SRC to regulate adherens junction stability as well as serving as a stimulator of p53-mediated apoptosis. The role of human RASSF8 in these processes still requires further study to confirm these observations. However, loss of RASSF8, via RNAi-mediated knockdown, leads to translocation of β-catenin, a component of the canonical Wnt signalling pathway, into the nucleus and activation of the TCF-LEF promoter. NFκB p65 also relocates to the nucleus causing NFκB-dependent promoter activity. This may lead to upregulation of genes involved in cell proliferation. RASSF8 depletion is also followed by loss of intercellular contact and a concomitant increase in cell motility.27
Human RASSF8. In our studies, we have detected an interaction between human RASSF8 and the transmembrane adhesion molecule, E-cadherin.27 We observed that endogenous RASSF8 is not only found in the nucleus, but is also membrane associated at sites of cell-cell adhesion, co-localizing with the adherens junction (AJ) component β-catenin. In both stable and transient RASSF8 knockdown lung carcinoma-derived cells we observed a loss of tight cell-cell contact, enhanced cell spreading and a redistribution of both β-catenin and E-cadherin from adherens junctions into the cytoplasm and/or nucleus. β-catenin is a known mediator of the Wnt signaling pathway and we observed a marked increase in β-catenin/TCF-dependent promoter activity upon RASSF8 depletion. Similarly, RASSF8 knockdown cells displayed redistribution of NFκB subunit p65 from adherens junctions into the nucleus with a concomitant increase in NFκB promoter activity. Thus the tumor suppressor functions of RASSF8 in lung carcinogenesis may result from its ability to regulate stable formation of adherens junction complexes, which in turn suppresses β-catenin/Wnt and/or NFκB signaling (Fig. 4 and Table 1). On depletion of RASSF8, aberrant activation of these pathways could lead to the expression of target genes involved in lung carcinogenesis.37,38 Furthermore, we have observed increased cell motility and migration in RASSF8 depleted cells; thus loss of RASSF8 appears to disrupt the stable cytoskeletal architecture.27
In addition to the Drosophila dRASSF-dASPP and human RASSF8-E-Cadherin complexes there are several reports documenting additional putative RASSF8 interaction partners. In co-immunoprecipitation experiments RASSF8 bound to YWHAG, a 14-3-3γ family member and adapter protein implicated in the regulation of various vital cellular processes.39 In Yeast two-hybrid experiments RASSF8 interacted with FRMD6 (FERM domain containing protein 6), an ezrin-radixin-moesin (ERM) family protein linking actin filaments of cell surface structures to the plasma membrane and PSMD4 [proteasome (prosome, macropain) 26S subunit, non-ATPase, 4], a subunit of the 26S proteasome proteinase complex.40
RASSF9
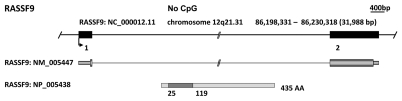
RASSF9, formerly PAM COOH-terminal interactor-1 (P-CIP1), was identified from a yeast two-hybrid screen employed to isolate proteins interacting with peptidylglycine alpha-amidating mono-oxygenase (PAM).41 PAM catalyzes the COOH-terminal a-amidation of bioactive peptides prior to their secretion from neuronal and endocrine cell vesicles. Subsequently, Chen et al. confirmed the interaction of P-CIP1 with the cytosolic domain of wild-type membrane PAM and suggested that P-CIP1 associated with vesicles in the recycling endosomal pathway.42 P-CIP1 was later renamed RASSF9 after characterization revealed its similarity to RASSF7 and RASSF8.4 Thus RASSF9 may play a role in regulating the trafficking of integral membrane PAM (Table 1). RASSF9 resides on chromosome 12q21.31 and contains two exons generating a single 435 amino acid protein (Fig. 5). RASSF9 is highly conserved between species and has orthologs including those in Mus musculus (NM_146240.3), Danio rerio (XM_001922320.1), Xenopus laevis (B1A192) and Drosophila melanogaster (CG13875) (Table 1). RASSF9 is thought to be widely expressed (GeneNote, Weizmann Institute of Science) but to date there are no reports documenting endogenous RASSF9 protein expression. Cloned RASSF9 showed preferential binding to N-Ras and K-Ras.43 Little else is known regarding RASSF9 function and it remains largely unstudied. Neither the UCSC genome browser nor the EBI CpG plot program detect RASSF9 as having a CpG island.
Figure 5.
In silico characterization of the N-terminal RASSF family (RASSF7–10). For each member a genomic map is displayed with the chromosomal location, genomic position, relative position and size of each exon and CpG island as determined by the UCSC Genome Browser (genome.ucsc.edu). Immediately below are the major transcript variants (UCSC) followed by a schematic of protein isoforms as determined by the National Center for Biotechnology Information (NCBI: www.ncbi.nlm.nih.gov). The location of the RAS-association (RA) domains were predicted by Prosite (expasy.org/prosite).
RASSF10
The RASSF10 gene.
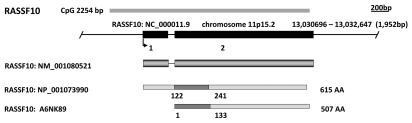
The human RASSF10 gene is located on chromosome 11p15.2 and was first described from a predicted sequence (GeneLoc: LOC644943) with homology to RASSF9/P-CIP1.4 The gene was initially listed on human genome browsers as a two-exon gene giving rise to an 1,848 base pair transcript (GenBank: NM_001080521) encoding a 615 (RefSeq: NP_001073990) amino acid protein. However, we demonstrated that a predicted alternative transcription initiation site would result in a single exon gene encoding a shorter 507 (UniProt: A6NK89) amino acid protein (Fig. 6).28 ClustalW alignment of N-terminal RASSF members in conjunction with RASSF10 expression data (see below) appear to support this notion although transcripts of the long-form have been detected in human testes.44 The N-terminal 108 amino acid sequence of RASSF10 long-form retains a high homology with several ape (Pongo abelii) and monkey (Macaca mulatta, Callithrix jacchus) RASSF10-like protein sequences suggesting the RASSF10 long-form may be a late evolutionary variant. All other RASSF10 orthologs are of the RASSF10 short-form. These include Mus musculus (NM_175279), Xenopus laevis (NM_001115020) and Drosophila melanogaster (CG32150) (Table 1).
Figure 6.
In silico characterization of the N-terminal RASSF family (RASSF7–10). For each member a genomic map is displayed with the chromosomal location, genomic position, relative position and size of each exon and CpG island as determined by the UCSC Genome Browser (genome.ucsc.edu). Immediately below are the major transcript variants (UCSC) followed by a schematic of protein isoforms as determined by the National Center for Biotechnology Information (NCBI: www.ncbi.nlm.nih.gov). The location of the RAS-association (RA) domains were predicted by Prosite (expasy.org/prosite).
RASSF10 expression.
The expression status of RASSF10 has only recently been documented. Schagdarsurengin et al. detected human RASSF10 expression (single-exon form) in thyroid, pancreas, placenta, heart, lung and kidney.44 We found expression of this transcript in human bone marrow RNA and subsequent 5′RACE verified the locality of the transcription start site.28 In on-going studies we have detected prominent expression of X. laevis RASSF10 in several brain tissues including the dorsal and ventral forebrain, the dorsal midbrain and the hindbrain as well as in other tissues including the eye, the trigeminal ganglion, dorsal fin, cement gland and branchial arches.45 In addition we have also amplified the RASSF10 gene from an IMAGE clone (4413366) derived from a human liver cDNA library. Murine RASSF10 mRNA (homologous to human RASSF10 short-form) is expressed in salivary gland, testes, kidney, lung and brain (Mouse Genome Informatics). In the fruit fly, the putative RASSF10 homolog is expressed in the developing nervous system.46 These observations suggest RASSF10 is expressed in a wide variety of tissues.
RASSF10 methylation.
RASSF10 has a CpG island of 2,254 bp with 209 CpGs (UCSC genome browser) and appears to be the most frequently methylated of the N-terminal RASSFs. We have assessed the role of RASSF10 in childhood acute lymphoblastic leukemia (ALL) by analysis of the methylation status of CpG islands associated with RASSF promoter regions in control samples and in a cohort of childhood T- and B-ALL samples.28 We found high frequency of methylation (88%) of RASSF10 in childhood T-ALL as well as in leukemia cell lines (100%). RASSF10 was unmethylated in age matched control DNA samples (from healthy individuals) from peripheral blood lymphocytes and bone marrow and displayed lower methylation frequency (16%) in childhood B-ALL. RASSF10 methylation correlated with loss of gene expression and we showed that this loss could be reversed after treatment with 5-aza-2′deoxycytidine. Thus hypermethylation of RASSF10 in childhood ALL is inversely correlated with its expression status and provides evidence to RASSF10 candidacy as a tumor suppressor gene in leukemia.
Methylation of the RASSF10 promoter CpG island has also been detected in both thyroid cancer cell lines and in a high frequency (66%) of primary thyroid carcinomas.44 In this study the authors detected high RASSF10 mRNA expression in a range of normal human tissues including thyroid but reduced expression in thyroid cancer cell lines. The latter was reversed by 5-aza-2′-deoxycytidine treatment and subsequent epigenetic analysis confirmed demethylation of the RASSF10 promoter correlated with increased RASSF10 transcript expression. Analysis of RASSF10 promoter methylation status showed full to partial methylation in 100% (9/9) of thyroid cancer cell lines analyzed and partial methylation in 66% (21/32) of primary thyroid carcinomas. Of these the highest methylation frequency (100%) was observed in medullary thyroid carcinoma and the lowest in undifferentiated thyroid cancer (40%). None of the normal thyroid controls showed RASSF10 methylation. Mutations within the coding region of RASSF10 have also been looked for in thyroid cancers but no mutations were found.44
More recently, we have found frequent methylation in gliomas.45 Analysis of the four WHO grades (WHO grade I, grade II and grade III astrocytomas and WHO grade IV glioblastomas) showed increasing levels of methylation from 0% (grade I) to 80% (grade III) and 60% in primary grade IV glioblastomas. Secondary grade IV glioblastomas (arising from grade II or III astrocytomas) showed a similar frequency of methylation as primary grade IV glioblastomas (arising without earlier grade lesions) (Table 1). In secondary grade IV glioblastomas RASSF10 methylation also associated significantly with worse overall survival and worse progression free survival. Analysis of the earlier stage lesions available for corresponding secondary glioblastomas suggests that RASSF10 methylation occurs at an early stage in gliomagenesis.45 In gliomas, RASSF10 methylation does not correlate with methylation of the other frequently methylated classical RASSF member, RASSF1A. In methylated glioma cell lines RASSF10 expression has been shown to be reactivated by the demethylating agent 5-aza-2′-deoxycitidine at both the RNA level and at the protein level.
Together, these studies highlight a potential tumor-suppressor role for RASSF10 through frequent epigenetic inactivation of the RASSF10 gene in at least three types of cancers.
RASSF10 function.
To date there is little published data hinting at RASSF10 function. In a genome-wide RNAi screen targeting of the Drosophila RASSF10 homolog lead to reduced hedgehog (Hh) signaling but the role in higher organism remains unstudied.47 Similar to other N-terminal RASSF members, the human RASSF10 gene lies close to the RAS gene (RRAS2) on chromosome 11p15.2 but the correlation between RAS activation and RASSF silencing has yet to be resolved.4,48 Our studies on the role of human RASSF10 in gliomagenesis has thrown up some interesting observations regarding RASSF10 localization and function.45 We raised a specific antibody to RASSF10 and used this in sub-cellular localization studies in lung and breast cancer-derived cell lines. Reminiscent of RASSF7, RASSF10 distribution appears cell cycle dependent with cytoplasmic RASSF10 becoming relocated to developing centrosomes and associated microtubules specifically during mitosis. This suggests a mitotic role for RASSF10. In biological experiments we depleted RASSF10 through RNAi-mediated knockdown in glioma-derived cell lines and observed both an increase in cell viability and cell proliferation.45 RASSF10 depleted cells also had an enhanced ability to grow in soft agar. Adversely, RASSF10 re-expression reduced the colony-forming ability of at least two glioma-derived cell lines previously found to be negative, through promoter methylation, for RASSF10 expression.45 These biological results indicate a functional role in growth-regulation and appear to correlate with the observed RASSF10 protein redistribution (Fig. 2B and Table 1). It will be exciting to discover RASSF10 protein interactions and subsequently determine more precisely the specific activities of the most recently discovered RASSF member.
Summary and Future Perspectives
The N-terminal and classical RASSF proteins are structurally different; therefore, they could be considered separate families. Yet as we have demonstrated, members of both families show frequent methylation in cancer. Furthermore they show overlapping biological characteristics, for example, acting as tumor suppressors and playing a role in mitosis. The biological properties of N-Terminal RASSFs are just beginning to be addressed and thus provide an exciting opportunity for future research.
Acknowledgements
F. Latif laboratory funded in part by Breast Cancer Campaign, Cancer Research UK and Sport Aiding Medical Research for Kids (SPARKS).
References
- 1.Ponting CP, Benjamin DR. A novel family of Ras-binding domains. Trends Biochem Sci. 1996;21:422–425. doi: 10.1016/s0968-0004(96)30038-8. [DOI] [PubMed] [Google Scholar]
- 2.Saucedo LJ, Edgar BA. Filling out the Hippo pathway. Nat Rev Mol Cell Biol. 2007;8:613–621. doi: 10.1038/nrm2221. [DOI] [PubMed] [Google Scholar]
- 3.Newton AC. Protein kinase C: Poised to signal. Am J Physiol Endocrinol Metab. 2010;298:395–402. doi: 10.1152/ajpendo.00477.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sherwood V, Manbodh R, Sheppard C, Chalmers AD. RASSF7 is a member of a new family of RAS association domain-containing proteins and is required for completing mitosis. Mol Biol Cell. 2008;19:1772–1782. doi: 10.1091/mbc.E07-07-0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agathanggelou A, Cooper WN, Latif F. Role of the Ras-association domain family 1 tumor suppressor gene in human cancers. Cancer Res. 2005;65:3497–3508. doi: 10.1158/0008-5472.CAN-04-4088. [DOI] [PubMed] [Google Scholar]
- 6.Avruch J, Xavier R, Bardeesy N, Zhang XF, Praskova M, Zhou D, et al. Rassf family of tumor suppressor polypeptides. J Biol Chem. 2009;284:11001–11005. doi: 10.1074/jbc.R800073200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richter AM, Pfeifer GP, Dammann RH. The RASSF proteins in cancer; from epigenetic silencing to functional characterization. Bba-Rev Cancer. 2009;1796:114–128. doi: 10.1016/j.bbcan.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Weitzel JN, Kasperczyk A, Mohan C, Krontiris TG. The HRAS1 gene cluster: Two upstream regions recognizing transcripts and a third encoding a gene with a leucine zipper domain. Genomics. 1992;14:309–319. doi: 10.1016/s0888-7543(05)80221-6. [DOI] [PubMed] [Google Scholar]
- 9.Recino A, Sherwood V, Flaxman A, Cooper WN, Latif F, Ward A, et al. Human RASSF7 regulates the microtubule cytoskeleton and is required for spindle formation, Aurora B activation and chromosomal congression during mitosis. Biochem J. 2010;430:207–213. doi: 10.1042/BJ20100883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brandt R, Grutzmann R, Bauer A, Jesnowski R, Ringel J, Lohr M, et al. DNA microarray analysis of pancreatic malignancies. Pancreatology. 2004;4:587–597. doi: 10.1159/000082241. [DOI] [PubMed] [Google Scholar]
- 11.Friess H, Ding J, Kleeff J, Fenkell L, Rosinski JA, Guweidhi A, et al. Microarray-based identification of differentially expressed growth- and metastasis-associated genes in pancreatic cancer. Cell Mol Life Sci. 2003;60:1180–1199. doi: 10.1007/s00018-003-3036-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Logsdon CD, Simeone DM, Binkley C, Arumugam T, Greenson JK, Giordano TJ, et al. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res. 2003;63:2649–2657. [PubMed] [Google Scholar]
- 13.Lowe AW, Olsen M, Hao Y, Lee SP, Taek Lee K, Chen X, et al. Gene expression patterns in pancreatic tumors, cells and tissues. PLoS One. 2007;2:e323. doi: 10.1371/journal.pone.0000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mutter GL, Baak JP, Fitzgerald JT, Gray R, Neuberg D, Kust GA, et al. Global expression changes of constitutive and hormonally regulated genes during endometrial neoplastic transformation. Gynecol Oncol. 2001;83:177–185. doi: 10.1006/gyno.2001.6352. [DOI] [PubMed] [Google Scholar]
- 15.Tan DS, Lambros MB, Rayter S, Natrajan R, Vatcheva R, Gao Q, et al. PPM1D is a potential therapeutic target in ovarian clear cell carcinomas. Clin Cancer Res. 2009;15:2269–2280. doi: 10.1158/1078-0432.CCR-08-2403. [DOI] [PubMed] [Google Scholar]
- 16.Camps C, Buffa FM, Colella S, Moore J, Sotiriou C, Sheldon H, et al. hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res. 2008;14:1340–1348. doi: 10.1158/1078-0432.CCR-07-1755. [DOI] [PubMed] [Google Scholar]
- 17.Liang GP, Su YY, Chen J, Yang ZC, Liu YS, Luo XD. Analysis of the early adaptive response of endothelial cells to hypoxia via a long serial analysis of gene expression. Biochem Biophys Res Commun. 2009;384:415–419. doi: 10.1016/j.bbrc.2009.04.160. [DOI] [PubMed] [Google Scholar]
- 18.Welcsh PL, Lee MK, Gonzalez-Hernandez RM, Black DJ, Mahadevappa M, Swisher EM, et al. BRCA1 transcriptionally regulates genes involved in breast tumorigenesis. Proc Natl Acad Sci USA. 2002;99:7560–7565. doi: 10.1073/pnas.062181799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hitomi J, Christofferson DE, Ng A, Yao J, Degterev A, Xavier RJ, et al. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell. 2008;135:1311–1323. doi: 10.1016/j.cell.2008.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goshima G, Wollman R, Goodwin SS, Zhang N, Scholey JM, Vale RD, et al. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morris JA, Kandpal G, Ma L, Austin CP. DISC1 (Disrupted-In-Schizophrenia 1) is a centrosome-associated protein that interacts with MAP1A, MIPT3, ATF4/5 and NUDEL: regulation and loss of interaction with mutation. Hum Mol Genet. 2003;12:1591–1608. doi: 10.1093/hmg/ddg162. [DOI] [PubMed] [Google Scholar]
- 22.Tsang HT, Connell JW, Brown SE, Thompson A, Reid E, Sanderson CM. A systematic analysis of human CHMP protein interactions: Additional MIT domain-containing proteins bind to multiple components of the human ESCRT III complex. Genomics. 2006;88:333–346. doi: 10.1016/j.ygeno.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 23.Yasui S, Tsuzaki K, Ninomiya H, Floricel F, Asano Y, Maki H, et al. The TSC1 gene product hamartin interacts with NADE. Mol Cell Neurosci. 2007;35:100–108. doi: 10.1016/j.mcn.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 24.Debeer P, Schoenmakers EF, Thoelen R, Holvoet M, Kuittinen T, Fabry G, et al. Physical map of a 1.5 mb region on 12p11.2 harbouring a synpolydactyly associated chromosomal breakpoint. Eur J Hum Genet. 2000;8:561–570. doi: 10.1038/sj.ejhg.5200497. [DOI] [PubMed] [Google Scholar]
- 25.Debeer P, Schoenmakers EF, Twal WO, Argraves WS, De Smet L, Fryns JP, et al. The fibulin-1 gene (FBLN1) is disrupted in a t(12;22) associated with a complex type of synpolydactyly. J Med Genet. 2002;39:98–104. doi: 10.1136/jmg.39.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Falvella FS, Spinola M, Manenti G, Conti B, Pastorino U, Skaug V, et al. Common polymorphisms in D12S1034 flanking genes RASSF8 and BHLHB3 are not associated with lung adenocarcinoma risk. Lung Cancer. 2007;56:1–7. doi: 10.1016/j.lungcan.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 27.Lock FE, Underhill-Day N, Dunwell T, Matallanas D, Cooper W, Hesson L, et al. The RASSF8 candidate tumor suppressor inhibits cell growth and regulates the Wnt and NFkappaB signaling pathways. Oncogene. 2010;29:4307–4316. doi: 10.1038/onc.2010.192. [DOI] [PubMed] [Google Scholar]
- 28.Hesson LB, Dunwell TL, Cooper WN, Catchpoole D, Brini AT, Chiaramonte R, et al. The novel RASSF6 and RASSF10 candidate tumour suppressor genes are frequently epigenetically inactivated in childhood leukaemias. Mol Cancer. 2009;8:42. doi: 10.1186/1476-4598-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yanagitani N, Kohno T, Sunaga N, Kunitoh H, Tamura T, Tsuchiya S, et al. Localization of a human lung adenocarcinoma susceptibility locus, possibly syntenic to the mouse Pas1 locus, in the vicinity of the D12S1034 locus on chromosome 12p11.2-p12.1. Carcinogenesis. 2002;23:1177–1183. doi: 10.1093/carcin/23.7.1177. [DOI] [PubMed] [Google Scholar]
- 30.Gariboldi M, Manenti G, Canzian F, Falvella FS, Radice MT, Pierotti MA, et al. A major susceptibility locus to murine lung carcinogenesis maps on chromosome 6. Nat Genet. 1993;3:132–136. doi: 10.1038/ng0293-132. [DOI] [PubMed] [Google Scholar]
- 31.Falvella FS, Manenti G, Spinola M, Pignatiello C, Conti B, Pastorino U, et al. Identification of RASSF8 as a candidate lung tumor suppressor gene. Oncogene. 2006;25:3934–3938. doi: 10.1038/sj.onc.1209422. [DOI] [PubMed] [Google Scholar]
- 32.Korkola JE, Houldsworth J, Chadalavada RS, Olshen AB, Dobrzynski D, Reuter VE, et al. Downregulation of stem cell genes, including those in a 200 kb gene cluster at 12p13.31, is associated with in vivo differentiation of human male germ cell tumors. Cancer Res. 2006;66:820–827. doi: 10.1158/0008-5472.CAN-05-2445. [DOI] [PubMed] [Google Scholar]
- 33.Weiser KC, Liu B, Hansen GM, Skapura D, Hentges KE, Yarlagadda S, et al. Retroviral insertions in the VISION database identify molecular pathways in mouse lymphoid leukemia and lymphoma. Mamm Genome. 2007;18:709–722. doi: 10.1007/s00335-007-9060-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rykova E, Skvortsova TE, Hoffmann AL, Tamkovich SN, Starikov AV, Bryzgunova OE, et al. Breast cancer diagnostics based on extracellular DNA and RNA circulating in blood. Biomed Khim. 2008;54:94–103. [PubMed] [Google Scholar]
- 35.Langton PF, Colombani J, Chan EH, Wepf A, Gstaiger M, Tapon N. The dASPP-dRASSF8 complex regulates cell-cell adhesion during Drosophila retinal morphogenesis. Curr Biol. 2009;19:1969–1978. doi: 10.1016/j.cub.2009.10.027. [DOI] [PubMed] [Google Scholar]
- 36.Eckfeldt CE, Mendenhall EM, Flynn CM, Wang TF, Pickart MA, Grindle SM, et al. Functional analysis of human hematopoietic stem cell gene expression using zebrafish. PLoS Biol. 2005;3:e254. doi: 10.1371/journal.pbio.0030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Downward J. Cancer: A tumour gene's fatal flaws. Nature. 2009;462:44–45. doi: 10.1038/462044a. [DOI] [PubMed] [Google Scholar]
- 38.Katoh Y, Katoh M. Hedgehog target genes: mechanisms of carcinogenesis induced by aberrant hedgehog signaling activation. Curr Mol Med. 2009;9:873–886. doi: 10.2174/156652409789105570. [DOI] [PubMed] [Google Scholar]
- 39.Jin J, Smith FD, Stark C, Wells CD, Fawcett JP, Kulkarni S, et al. Proteomic, functional and domain-based analysis of in vivo 14-3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr Biol. 2004;14:1436–1450. doi: 10.1016/j.cub.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 40.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 41.Alam MR, Caldwell BD, Johnson RC, Darlington DN, Mains RE, Eipper BA. Novel proteins that interact with the COOH-terminal cytosolic routing determinants of an integral membrane peptide-processing enzyme. J Biol Chem. 1996;271:28636–28640. doi: 10.1074/jbc.271.45.28636. [DOI] [PubMed] [Google Scholar]
- 42.Chen L, Johnson RC, Milgram SL. P-CIP1, a novel protein that interacts with the cytosolic domain of peptidylglycine alpha-amidating monooxygenase, is associated with endosomes. J Biol Chem. 1998;273:33524–33532. doi: 10.1074/jbc.273.50.33524. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez-Viciana P, Sabatier C, McCormick F. Signaling specificity by Ras family GTPases is determined by the full spectrum of effectors they regulate. Mol Cell Biol. 2004;24:4943–4954. doi: 10.1128/MCB.24.11.4943-4954.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schagdarsurengin U, Richter AM, Wohler C, Dammann RH. Frequent epigenetic inactivation of RASSF10 in thyroid cancer. Epigenetics. 2009;4:571–576. doi: 10.4161/epi.4.8.10056. [DOI] [PubMed] [Google Scholar]
- 45.Hill VK, Underhill-Day N, Krex D, Robel K, Sangan CB, Summersgill HR, et al. Epigenetic inactivation of the RASSF10 candidate tumour suppressor gene is a frequent and early event in gliomagenesis. Oncogene. 2011;30:978–989. doi: 10.1038/onc.2010.471. [DOI] [PubMed] [Google Scholar]
- 46.Reeves N, Posakony JW. Genetic programs activated by proneural proteins in the developing Drosophila PNS. Dev Cell. 2005;8:413–425. doi: 10.1016/j.devcel.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 47.Nybakken K, Vokes SA, Lin TY, McMahon AP, Perrimon N. A genome-wide RNA interference screen in Drosophila melanogaster cells for new components of the Hh signaling pathway. Nat Genet. 2005;37:1323–1332. doi: 10.1038/ng1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sherwood V, Recino A, Jeffries A, Ward A, Chalmers AD. The N-terminal RASSF family: A new group of Ras-association-domain-containing proteins, with emerging links to cancer formation. Biochem J. 2009;425:303–311. doi: 10.1042/BJ20091318. [DOI] [PubMed] [Google Scholar]