Abstract
Changes in DNA methylation may represent an intermediate step between the environment and human diseases. Little is known on whether behavioral risk factors may modify gene expression through DNA methylation. To assess whether DNA methylation is associated with different levels of physical activity, we measured global genomic DNA methylation using bisulfite-converted DNA and real-time PCR (MethyLight) for LINE-1 in peripheral blood of 161 participants aged 45–75 years enrolled in the North Texas Healthy Heart Study and levels of physical activity using an accelerometer (Actigraph GT1M Monitor). We found that individuals with physical activity 26–30 min/day had a significantly higher level of global genomic DNA methylation compared to those with physical activity ≤10 min/day (β = 2.52, 95% CI: 0.70, 4.35). However, the association was attenuated and became statistically insignificant after multivariate adjustment (β = 1.24, 95% CI: −0.93, 3.40). There were some suggestions of a positive association between physical activity and global genomic DNA methylation in non-Hispanics (β = 1.50, 95% CI: −0.08, 3.08) that warrants further investigation.
Key words: DNA methylation, physical activity, peripheral blood
Introduction
Animal studies provide compelling evidence that patterns of DNA methylation can be affected by environmental exposures to nutritional factors,1 leading to the hypothesis that changes in DNA methylation may be an intermediate step between the environment and human diseases. In humans, previous studies have found that dietary folate restriction or folic acid supplementation can change DNA methylation.2–4 Cross-sectional studies in humans also reported that exposure to airborne benzene, polycyclic aromatic hydrocarbons (PAHs) and arsenic were significantly associated with levels of global methylation.5–7 Studies among monozygotic twins revealed that the difference in DNA methylation was more apparent in older twins than in younger ones, and the individual differences in methylation are not stable over time.8,9 These observations support further study of the potential impact of the external environment or behaviors on DNA methylation in humans.
It is estimated that about half of the human genome contains repetitive elements that are often intensively methylated. A reduced level of methylation in repetitive elements such as long interspersed repeat sequences (LINE-1) has been associated with genomic instability and chromosomal aberrations,10 leading to an increased risk of cancer. In fact, hypomethylation in repetitive elements has been reported in many cancer cells and appears to parallel overall genomic hypomethylation.10 More recently, global DNA methylation has been examined in surrogate tissues such as leukocyte DNA. A lower level of leukocyte DNA methylation was associated with an increased risk of head and neck squamous cell carcinoma,11 bladder cancer,12 breast cancer,13 gastric cancer14 and colorectal adenoma and cancer15,16 after adjusting for known risk factors. These results suggest that leukocyte DNA may serve as a surrogate biomarker for systemic genomic methylation and leukocyte DNA hypomethylation may be an independent risk factor for cancer development.
Little is known about the environmental impact on DNA methylation in humans that are amenable to intervention. In the presents study, we investigated the relationship between one of the modifiable lifestyle factors, physical activity and global genomic DNA methylation measured in the peripheral blood of a cancer-free population.
Results
Among 161 subjects who completed the baseline survey and were given an accelerometer to wear for three weekdays and one weekend, three subjects did not return the accelerometer to the study coordinator, and three subjects did not have any recorded data in the returned accelerometers. Among 155 subjects with recorded accelerometer data, 24 subjects had less than two days of valid accelerometer wear (i.e., ≥10 h wear time for a given day). This resulted in 131 subjects who had two or more days of valid accelerometer wear for the final analysis. The subjects with and without two valid days of accelerometer wear were comparable in age, gender, race/ethnicity (non-Hispanic white, non-Hispanic black, Hispanic), body mass index, smoking status (non-smokers, former smokers, current smokers), alcohol drinking (nondrinkers, monthly or less, ≥2–4 times/month) and dietary folate intake (<median vs. ≥median).
The mean minutes per day of moderate physical activity in the 131 subjects with two or more days of valid accelerometer wear were 17.5, whereas the mean minutes per day for vigorous physical activity was 0.6. Thus, the combined physical activity, i.e., moderate plus vigorous, was 18.1 min per day. Because the level of vigorous physical activity was low, the level of combined physical activity assessed in this population represented primarily the level of moderate physical activity. Women had significantly lower levels of physical activity than men (11.6 vs. 29.6 min/day for combined physical activity, p < 0.0001). The levels of physical activity were also different among different racial/ethnic groups. Among non-Hispanic whites, the mean min/day for combined physical activity were 28.3, which was significantly higher than that in non-Hispanic blacks and Hispanics (15.3 and 15.7, respectively) (p = 0.01).
Table 1 presents the mean minutes per day for moderate, vigorous and combined physical activity by different population characteristics among men and women separately. Among women, the level of combined physical activity declined significantly with age and body mass index (p = 0.04 and 0.004, respectively). Non-Hispanic whites had a higher level of physical activity than non-Hispanic blacks (21.0 vs. 11.0 min/day, p = 0.09) and Hispanics (21.0 vs. 9.5 min/day, p = 0.04). Among men, the level of physical activity also declined with age. Non-Hispanic whites and Hispanics had similar level of physical activity (33.6 and 31.1 min/day, respectively), both higher than that in non-Hispanic blacks (23.2 min/day). Overweight (BMI: 25–30 kg/m2) men appeared to have the highest level of physical activity (32.3 min/day) whereas normal weight (BMI <25 min/day) men had the lowest level of physical activity (19.0 min/day). However, none of the differences in men reached statistical significance. In both men and women, the level of physical activity did not differ significantly by education, alcohol drinking, cigarette smoking and dietary folate intake.
Table 1.
Duration of moderate and vigorous activity (minutes/day) by characteristics of the study population in the North Texas Healthy Heart study, 2006–2008
| Women | Men | ||||||||
| N | Moderate | Vigorous | Combined | N | Moderate | Vigorous | Combined | ||
| Age (yrs) | |||||||||
| 40–49 | 38 | 14.8 | 0.9 | 15.7a | 20 | 34.3 | 0.8 | 35.1 | |
| 50–59 | 22 | 8.3 | 0.1 | 8.4a | 19 | 24.0 | 1.6 | 25.7 | |
| 60+ | 22 | 7.7 | 0.0 | 7.7a | 10 | 23.0 | 0.2 | 23.3 | |
| Race/ethnicity | |||||||||
| Non-Hispanic white | 10 | 19.1 | 1.9 | 21.0a | 14 | 32.5 | 1.0 | 33.6 | |
| Non-Hispanic black | 37 | 10.6 | 0.4 | 11.0a | 20 | 21.5 | 1.7 | 23.2 | |
| Hispanic | 35 | 9.5 | 0.0 | 9.5a | 14 | 31.1 | 0.0 | 31.1 | |
| Education | |||||||||
| High school or less | 34 | 9.3 | 0.0 | 9.3 | 14 | 30.1 | 0.0 | 30.2 | |
| Some college/college graduate | 48 | 12.4 | 0.7 | 13.2 | 15 | 27.2 | 1.4 | 28.6 | |
| Body mass index (kg/m2) | |||||||||
| <25 | 11 | 20.3a | 1.5 | 21.8a | 4 | 18.5 | 0.6 | 19.0 | |
| 25–29 | 25 | 14.3a | 0.1 | 14.4a | 29 | 31.9 | 0.4 | 32.3 | |
| ≥30 | 46 | 7.4a | 0.3 | 7.6a | 16 | 23.5 | 2.2 | 25.6 | |
| Smoking status | |||||||||
| Non-Smoker | 57 | 11.2 | 0.4 | 11.6 | 27 | 27.4 | 0.7 | 28.1 | |
| Former Smoker | 16 | 11.9 | 0.8 | 12.6 | 17 | 26.5 | 1.8 | 28.3 | |
| Current Smoker | 8 | 10.0 | 0.0 | 10.0 | 5 | 36.5 | 0.0 | 36.5 | |
| Alcohol drinking | |||||||||
| Non-drinker | 33 | 11.2 | 0.5 | 11.7 | 20 | 26.0 | 1.7 | 27.7 | |
| Monthly or Less | 28 | 8.1 | 0.1 | 8.2 | 9 | 32.2 | 0.2 | 32.4 | |
| ≥2–4 times/month | 21 | 15.2 | 0.7 | 15.9 | 20 | 28.2 | 0.6 | 28.8 | |
| Dietary folate intake (µg/1,000 kcal) | |||||||||
| <median | 36 | 12.3 | 0.4 | 12.6 | 20 | 28.9 | 2.0 | 30.9 | |
| ≥median | 36 | 11.1 | 0.6 | 11.8 | 25 | 28.2 | 0.4 | 28.6 | |
| LINE-1 methylation (%) | |||||||||
| <median | 50 | 10.3 | 0.3 | 10.6 | 16 | 35.0 | 0 | 35.0 | |
| ≥median | 32 | 12.5 | 0.6 | 13.1 | 33 | 24.7 | 1.5 | 26.1 | |
p < 0.05.
The median level of global genomic DNA methylation as measured by LINE-1 was 73.9%. When study participants were categorized into low and high levels of LINE-1 methylation according to the median, levels of combined physical activity were slightly higher among subjects with high levels of LINE-1 methylation (≥median) as compared to those with low levels of LINE-1 methylation (<median) (19.7 vs. 16.5) but the differences were not statistically significant (p = 0.39). In both males and females, levels of physical activity were not significantly different according to levels of LINE-1 methylation (Table 1).
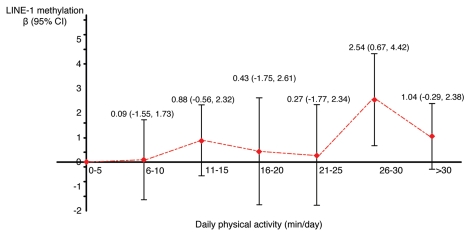
We then analyzed the association between physical activity and global genomic DNA methylation first in a linear regression model where levels of physical activity were categorized into seven groups with a 5-min/day interval (0–5, 6–10, 11–15, 16–20, 21–25, 26–30, >30 min/day). Compared to those with daily physical activity ≤5 min/day (i.e., the reference group), individuals who had physical activity 26–30 min/day had a significantly higher level of global leukocyte methylation (β = 2.54, 95% CI: 0.67, 4.42) (Fig. 1). Because the regression coefficient was close to 0 for individuals with physical activity 6–10 min/day we combined this group with the reference group. We also combined the 16–20 min/day group with the 21–25 min/day group due to the small sample size in each group for further analysis (N = 7 and 8 in each group). Table 2 presents the findings from linear regression models assessing the association between physical activity and global genomic DNA methylation. Compared to those with physical activity ≤10 min/day, individuals who had physical activity 26–30 min/day had a significantly higher level of global genomic DNA methylation in leukocytes (β = 2.52, 95% CI: 0.70, 4.35).
Figure 1.
Association between physical activity and global genomic DNA methylation (LINE-1) in the North Texas Healthy Heart Study 2006−2008. Regression coefficients (βs) and 95% confidence intervals (CIs) were generated from a simple linear regression model where LINE-1 methylation was modeled as a continuous outcome and daily physical activity was categorized into seven groups (0–5, 6–10, 11–15, 16–20, 21–25, 26–30, >30 min/day). 0–5 min/day was the reference group. Regression coefficients (βs), shown as the red dots in the figure, represent the difference in LINE-1 methylation (%) comparing each physical activity category with the reference. The vertical line represents the 95% CI with upper bar indicating the upper confidence limit and lower bar indicating the lower confidence limit.
Table 2.
Differences in global genomic DNA methylation (%) according to duration of physical activity in the North Texas Healthy Heart study, 2006–2008
| Physical activity (min/day) | N (%) | Simple linear regression model | Multivariate linear regression model 1 | Multivariate linear regression model 2 |
| β (95% CI) | β (95% CI)a | β (95% CI)b | ||
| 0–10 | 61 (47.3) | |||
| 11–15 | 20 (15.5) | 0.86 (−0.52, 2.24) | 0.69 (−0.61, 1.99) | 0.61 (−0.91, 2.13) |
| 16–25 | 15 (11.6) | 0.33 (−1.21, 1.88) | −0.30 (−1.76, 1.16) | −0.28 (−2.07, 1.51) |
| 26–30 | 10 (7.8) | 2.52 (0.70, 4.35) | 1.03 (−0.85, 2.90) | 1.24 (−0.93, 3.40) |
| >30 | 25 (17.8) | 1.20 (−0.11, 2.51) | −0.01 (−1.40, 1.40) | 0.26 (−1.34, 1.86) |
| Non-Hispanics | ||||
| 0–25 | 54 (66.7) | |||
| ≥26 | 27 (33.3) | 1.96 (0.77, 3.15) | 1.48 (0.12, 2.84) | 1.50 (−0.08, 3.08) |
| Hispanics | ||||
| 0–25 | 42 (89.4) | |||
| ≥26 | 5 (10.6) | 0.30 (−2.49, 3.09) | −0.84 (−3.48, 1.80) | −0.79 (−4.68, 3.11) |
Adjusted for age (continuous), gender, race/ethnicity (non-Hispanic white, non-Hispanic black, Hispanic).
Additionally adjusted for central obesity (yes/no), the ratio of fat mass to fat-free mass, the ratio of subcutaneous fat to visceral fat, cigarette smoking (nonsmokers, former and current smokers), alcohol drinking (never, monthly or less, and ≥2–4 times/month), dietary folate equivalent (quartiles) and education (high school or less and some college/college graduate).
To control for potential confounding effect of age, gender, race/ethnicity, social economic status and other lifestyle factors, we analyzed the association between physical activity and global genomic methylation in multivariate linear regression models (Table 2). Multivariate linear regression model 1 was adjusted for age (continuous), gender and race/ethnicity (non-Hispanic whites, non-Hispanic blacks and Hispanics). Multivariate linear regression model 2 was additionally adjusted for central obesity (yes/no), the ratio of fat mass to fat-free mass, the ratio of subcutaneous fat to visceral fat, cigarette smoking (nonsmokers, former and current smokers), alcohol drinking (never, monthly or less and ≥2–4 times/month), dietary folate equivalent (quartiles) and education (high school or less and some college/college graduate). In both multivariate models, the association between physical activity and global methylation were attenuated and became statistically insignificant.
Because gender and race/ethnicity was each significantly associated with DNA methylation in our study population, we further stratified the association by these factors to explore potential effect modification. Limited by a small sample size, we analyzed physical activity as a dichotomous variable in interaction models using 25 min/day as the cut-point. The associations between physical activity and global methylation appeared to be similar between men and women (among men, β = 0.14, 95% CI: −1.97, 2.26; among women, β = 0.52, 95% CI: −1.30, 2.34) (p for interaction = 0.86). Among non-Hispanics, levels of physical activity were positively associated with global methylation and the association was borderline significant (β = 1.50, 95% CI: −0.08, 3.08) in the fully adjusted model; there was no apparent association between physical activity and global methylation among Hispanics (β = −0.79, 95% CI: −4.68, 3.12) (p for interaction = 0.29) (Table 2).
Discussion
In a cancer-free adult population, we evaluated levels of moderate and vigorous physical activity using data collected from accelerometers. Our results indicated that the average level of moderate- or greater-intensity physical activity was 17–18 min per day, and the majority of the study population (i.e., over 70%) was physically inactive. Men tended to be more active than women. However, in both men and women, levels of physical activity declined with age. These findings are consistent with reports from the NHANES that also used accelerometers to access levels of physical activity in the US population.17
There is accumulating evidence suggesting that physical activity may protect against cancer. Early studies in large US populations showed that a higher level of physical activity was associated with a lower risk of cancer occurrence and mortality in general.18 The report from the Surgeon General on Physical Activity and Health and the report from the World Cancer Research Fund and the American Institute for Cancer Research reviewed studies examining the effect of physical activity on various cancer and found convincing evidence that physical activity lowers the risk of colorectal cancer in a dose-response fashion.19,20 A number of biological mechanisms have been proposed to explain the beneficial effect of physical activity including reduced insulin resistance, body fatness and inflammatory response and changes in endogenous steroid hormone metabolism.21
Exercise training has been associated with changes in gene expression and also more directly with changes in methylation of specific genes. Flynn et al. reported significantly lower mRNA expression of toll-like receptor 4 (TLR4) and CD 14 shortly after resistance exercise training in elderly women.22 Nakajima et al. found that elderly men and women following a high-intensity interval walking regimen for six months (N = 230) had a significantly higher degree of methylation compared to controls (N = 153) for ASC, a gene responsible for the secretion of pro-inflammatory cytokines IL-1β and IL-18.23 DNA methylation may therefore link physical activity and chronic inflammation, an underlying mechanism for cancer development. Nevertheless, none of the previous studies have investigated the association between physical activity and global DNA methylation.
In a cancer-free population, we identified a higher level of global DNA methylation among individuals with physical activity 26–30 min/day compared to those with physical activity ≤10 min/day. However, the association was not dose-dependent and became statistically insignificant after adjusting for age, gender, race/ethnicity and other lifestyle factors. The study may not be powered to detect a more subtle relationship between physical activity and DNA methylation. In addition, low levels of physical activity in our study population may make it difficult to identify a significant impact of physical activity on DNA methylation. However, the levels of physical activity in our study population parallel those reported in the NHANES study population, and physical activity was negatively associated with metabolic syndrome and its components on central obesity, elevated triglycerides and low HDL cholesterol. Although DNA methylation is not stable over time, environmental influences on DNA methylation may be most important during prenatal and early postnatal development.24 We studied levels of physical activity during adulthood, which may have limited impacts on DNA methylation.
Gender appeared to be a strong confounder for the association between physical activity and global methylation. After controlling for gender in the linear regression model, the regression coefficient was attenuated and a high level of physical activity (i.e., 26–30 min/day) was no longer significantly associated with global methylation (β = 1.55, 95% CI: −0.34, 3.44). The confounding effect can be due to the strong negative association between female gender and global methylation (i.e., women had significantly lower levels of global methylation than men), as well as the negative association between female gender and physical activity (i.e., women had significantly lower levels of physical activity than men). In contrast, controlling for age, race/ethnicity, education, body composition, smoking, alcohol drinking and dietary folate intake did not substantially change the regression coefficient of physical activity on global methylation.
There were some suggestions from our study that physical activity is positively associated with global methylation in non-Hispanics. We observed a lower level of leukocyte LINE-1 methylation in non-Hispanic blacks and Hispanics compared to non-Hispanic whites. The racial/ethnic difference in global methylation was also reported in a previous study measuring LINE-1 methylation in leukocytes where Caucasians had a significantly higher level of global methylation than non-Caucasians.11 This difference may reflect some genetic or environmental factors in different racial/ethnic groups that have not yet been clearly identified. Effect modification by race/ethnicity on the association between physical activity and global methylation needs to be further investigated.
A limitation of our study is that we used LINE-1 methylation by MethyLight as a single surrogate measure of global methylation. In contrast to LINE-1 pyrosequencing that provides the average level of methylation of each CpG site, MethyLight requires that all CpG sites within the promoter and probe regions be methylated to detect a signal. In addition, both assays interrogate the promoter region of LINE-1 which is only found in the subset of active elements. Thus, results may differ from that of other assays such as measurement of total 5-methylcytosine.
In summary, we did not find a significant impact of physical activity on LINE-1 methylation but there are some suggestions of an elevated risk of global hypomethylation associated with low levels of physical activity in non-Hispanics. Physical activity is one of the few modifiable behaviors and may affect the risk of cancer through changing DNA methylation. Future studies with a larger sample size and a longitudinal design are warranted to further investigate its impact on DNA methylation.
Patients and Methods
Study population.
The North Texas Healthy Heart Study was initiated in 2006 to assess racial/ethnic differences in cardiovascular serum markers and calcium scores. Briefly, study participants were recruited from the general population and primary care clinics through the NorTex network (www.hsc.unt.edu/Nortex), using either public advertisement or physician's referral.25–27 The inclusion criteria include (1) males and females over the age of 45, (2) being Caucasian, Hispanic or African American and (3) no current or previous self-reported history of stroke, peripheral arterial disease, renal failure, heart failure or coronary heart disease and cancer.
A baseline survey was conducted at enrollment to elicit information on demographics, smoking, alcohol drinking, medication use, use of oral contraceptives and hormone replacement therapy for women and life stress. For alcohol drinking, participants were asked the frequency of alcoholic drinks (never, monthly or less, 2–4 times a month, 2–3 times a week and 4 or more times a week). For cigarette smoking, participants were asked whether they smoke cigarettes every day, some days or not at all and whether they have stopped smoking for one day or longer during the past 12 months because they were trying to quit smoking. Body weight, height, waist and hip circumferences and percentage of body fat were measured at enrollment. A self-administrated Block food frequency questionnaire was distributed to the study participants asking their usual dietary intake during the last 12 months. Daily physical activity was measured using accelerometers. A 25 ml sample of whole blood was collected from study participants at the time of enrollment.
Informed consent was obtained from all study participants. The study protocol was approved by the Institutional Review Board of the University of North Texas Health Science Center. Study participants who completed the baseline survey were included in the present study.
Physical activity.
Study participants (N = 161) were asked to wear an Actigraph GT1M Monitor (Actigraph, LLC; Ft. Walton Beach, FL) accelerometer over the right hip on an elasticized belt for the three consecutive weekdays and one weekend. They were asked to wear the device while they were awake and to take it off for swimming or bathing. Accelerometers were returned to the study coordinator when study participants returned for their scheduled clinic visits at the University of North Texas Health Science Center Patient Care Center where the accelerometer data were downloaded. The accelerometers collect and record physical activity as “counts,” which measure vertical accelerations during the epoch period. Data were recorded in 1 min epochs for up to four days.
Accelerometer data were obtained from 158 participants. Participants whose accelerometers were not returned were excluded (N = 3). For the analysis of 158 participants, we adapted the criteria used in NHANES 2003–2004 to define a valid day.17 Specifically, a valid day was defined as having 10 or more hours of accelerometer wear. Wear time was determined by subtracting non-wear time from 24 h. Non-wear was defined by an interval of at least 60 consecutive minutes of zero activity intensity counts, with allowance for 1–2 min of counts between 0 and 100. This resulted in 131 participants who had two or more valid days of accelerometer data. Study findings presented in this paper were from the 131 participants with two or more valid days of accelerometer data.
The amount of physical activity as measured by accelerometers is estimated as the time (minutes/day) spent in moderate and vigorous physical activity according to count thresholds. The threshold for moderate physical activity was 2,020 mean counts per minute (equivalent to three METs).17 Mean counts per minute were calculated by dividing the sum of activity counts for a valid day by the number of minutes of wear time in that day across all valid days. The threshold for vigorous physical activity was 5,999 mean counts per minute (equivalent to six METs).17 A combined amount of physical activity was estimated as the sum of the amount of moderate and vigorous physical activities. The accelerometer data collected from four days were converted to the amount of physical activity per week by multiplying the amount of physical activity by a factor of 7/4.
Global DNA methylation.
Global DNA methylation was measured using bisulfite conversion of DNA and real-time PCR (MethyLight) for LINE-1 in the peripheral blood as described previously in reference 28. Briefly, DNA was bisulfite-treated using an EZ DNA Methylation Kit (Zymo Research) following the manufacturer's recommendations. Samples were amplified using the reported primers for LINE1 and AluC4 on an Applied Biosystems 7900. Controls included whole genome-amplified and fully methylated DNA. Quantitative data on % methylation was calculated by the method of Livak and Schmittgen using threshold cycles for both the gene of interest and the control gene for the sample and fully the methylated control.29
Statistical analysis.
First, we compared the characteristics between participants who had more than two valid days of accelerometer data (N = 131) and those who did not (N = 30) using Chi-square test and analysis of variance (ANOVA). We then compared the level of moderate, vigorous and combined physical activities according to various characteristics of study participants including age, race/ethnicity, education, body mass index, smoking, alcohol drinking and dietary folate intake. Because levels of physical activity were significantly different between men and women, these comparisons were performed in men and women separately. Second, we examined the association between physical activity and global genomic DNA methylation in linear regression models since the distribution of global methylation was approximately normal. Outliers of physical activity (i.e., 107.7 min/day and 120.7 min/day) were removed in all linear regression models. We first tested whether a linear trend exists for the association between levels of physical activity and global methylation. The results are compatible with a linear relationship between physical activity and global methylation but the estimates are too imprecise to be more assertive. We therefore chose to model physical activity as a categorical variable in future regression models. In order to control for the potential confounding effect of age, gender, race/ethnicity, education, body composition, cigarette smoking, alcohol drinking and dietary folate intake, we performed two multivariate linear regression models: age, gender, race/ethnicity were adjusted in multivariate linear regression model 1 (i.e., the partially adjusted model); three body composition measures (central obesity, ratio of fat mass to fat-free mass, ratio of subcutaneous fat to visceral fat), cigarette smoking, alcohol drinking, dietary folate intake and education level were additionally adjusted in multivariate linear regression model 2 (i.e., the fully adjusted model). In addition, we explored potential effect modification by gender and race/ethnicity in all three models. All analyses were performed using SAS (version 9.1; SAS Institute, Cary, NC).
Acknowledgements
This research received supported from University of North Texas School of Public Health and Institute for Cancer Research and Joe and Jessie Crump Fund for Medical Education. This research was also supported by NIH grant P20MD001633 to Roberto Cardarelli and Joan Carroll and NIH grant ES009089 to Regina M. Santella.
Abbreviations
- LINE-1
interspersed repeat sequences
- METs
metabolic equivalents
References
- 1.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jacob RA, Gretz DM, Taylor PC, James SJ, Pogribny IP, Miller BJ, et al. Moderate folate depletion increases plasma homocysteine and decreases lymphocyte DNA methylation in postmenopausal women. J Nutr. 1998;128:1204–1212. doi: 10.1093/jn/128.7.1204. [DOI] [PubMed] [Google Scholar]
- 3.Pufulete M, Al-Ghnaniem R, Khushal A, Appleby P, Harris N, Gout S, et al. Effect of folic acid supplementation on genomic DNA methylation in patients with colorectal adenoma. Gut. 2005;54:648–653. doi: 10.1136/gut.2004.054718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rampersaud GC, Kauwell GP, Hutson AD, Cerda JJ, Bailey LB. Genomic DNA methylation decreases in response to moderate folate depletion in elderly women. Am J Clin Nutr. 2000;72:998–1003. doi: 10.1093/ajcn/72.4.998. [DOI] [PubMed] [Google Scholar]
- 5.Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, Cavallo D, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67:876–880. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- 6.Pavanello S, Bollati V, Pesatori AC, Kapka L, Bolognesi C, Bertazzi PA, et al. Global and gene-specific promoter methylation changes are related to anti-B[a]PDE-DNA adduct levels and influence micronuclei levels in polycyclic aromatic hydrocarbon-exposed individuals. Int J Cancer. 2009;125:1692–1697. doi: 10.1002/ijc.24492. [DOI] [PubMed] [Google Scholar]
- 7.Pilsner JR, Liu X, Ahsan H, Ilievski V, Slavkovich V, Levy D, et al. Genomic methylation of peripheral blood leukocyte DNA: influences of arsenic and folate in Bangladeshi adults. Am J Clin Nutr. 2007;86:1179–1186. doi: 10.1093/ajcn/86.4.1179. [DOI] [PubMed] [Google Scholar]
- 8.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong CC, Caspi A, Williams B, Craig IW, Houts R, Ambler A, et al. A longitudinal study of epigenetic variation in twins. Epigenetics. 2010;5:516–526. doi: 10.4161/epi.5.6.12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann MJ, Schulz WA. Causes and consequences of DNA hypomethylation in human cancer. Biochem Cell Biol. 2005;83:296–321. doi: 10.1139/o05-036. [DOI] [PubMed] [Google Scholar]
- 11.Hsiung DT, Marsit CJ, Houseman EA, Eddy K, Furniss CS, McClean MD, et al. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2007;16:108–114. doi: 10.1158/1055-9965.EPI-06-0636. [DOI] [PubMed] [Google Scholar]
- 12.Moore LE, Pfeiffer RM, Poscablo C, Real FX, Kogevinas M, Silverman D, et al. Genomic DNA hypomethylation as a biomarker for bladder cancer susceptibility in the Spanish Bladder Cancer Study: a case-control study. Lancet Oncol. 2008;9:359–366. doi: 10.1016/S1470-2045(08)70038-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi JY, James SR, Link PA, McCann SE, Hong CC, Davis W, et al. Association between global DNA hypomethylation in leukocytes and risk of breast cancer. Carcinogenesis. 2009;30:1889–1897. doi: 10.1093/carcin/bgp143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou L, Wang H, Sartori S, Gawron A, Lissowska J, Bollati V, et al. Blood leukocyte DNA hypomethylation and gastric cancer risk in a high-risk Polish population. Int J Cancer. 2010;127:1866–1874. doi: 10.1002/ijc.25190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim U, Flood A, Choi SW, Albanes D, Cross AJ, Schatzkin A, et al. Genomic methylation of leukocyte DNA in relation to colorectal adenoma among asymptomatic women. Gastroenterology. 2008;134:47–55. doi: 10.1053/j.gastro.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pufulete M, Al-Ghnaniem R, Leather AJ, Appleby P, Gout S, Terry C, et al. Folate status, genomic DNA hypomethylation and risk of colorectal adenoma and cancer: a case control study. Gastroenterology. 2003;124:1240–1248. doi: 10.1016/s0016-5085(03)00279-8. [DOI] [PubMed] [Google Scholar]
- 17.Troiano RP, Berrigan D, Dodd KW, Masse LC, Tilert T, McDowell M. Physical activity in the United States measured by accelerometer. Med Sci Sports Exerc. 2008;40:181–188. doi: 10.1249/mss.0b013e31815a51b3. [DOI] [PubMed] [Google Scholar]
- 18.Kampert JB, Blair SN, Barlow CE, Kohl HW., 3rd Physical activity, physical fitness and all-cause and cancer mortality: a prospective study of men and women. Ann Epidemiol. 1996;6:452–457. doi: 10.1016/s1047-2797(96)00059-2. [DOI] [PubMed] [Google Scholar]
- 19.US Department of Health and Human Services, author; USDoHaH, editor. Services. Atlanta, GA: US Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Chronic Disease Prevention and Health Promotion; 1996. Physical Activity and Health: A Report of the Surgeon General. [Google Scholar]
- 20.WCRF/AICR, author. Food, nutrition, physical activity and the prevention of cancer: a global perspective. Washington, DC: World Cancer Research Fund; American Institute for Cancer Research; 2007. [Google Scholar]
- 21.Hursting SD, Lashinger LM, Colbert LH, Rogers CJ, Wheatley KW, Nunez NP, et al. Energy balance and carcinogenesis: underlying pathways and targets for intervention. Curr Cancer Drug Targets. 2007;7:484–491. doi: 10.2174/156800907781386623. [DOI] [PubMed] [Google Scholar]
- 22.Flynn MG, McFarlin BK, Phillips MD, Stewart LK, Timmerman KL. Toll-like receptor 4 and CD14 mRNA expression are lower in resistive exercise-trained elderly women. J Appl Physiol. 2003;95:1833–1842. doi: 10.1152/japplphysiol.00359.2003. [DOI] [PubMed] [Google Scholar]
- 23.Nakajima K, Takeoka M, Mori M, Hashimoto S, Sakurai A, Nose H, et al. Exercise effects on methylation of ASC gene. Int J Sports Med. 2010;31:671–675. doi: 10.1055/s-0029-1246140. [DOI] [PubMed] [Google Scholar]
- 24.Waterland RA, Michels KB. Epigenetic epidemiology of the developmental origins hypothesis. Annu Rev Nutr. 2007;27:363–388. doi: 10.1146/annurev.nutr.27.061406.093705. [DOI] [PubMed] [Google Scholar]
- 25.Carroll JF, Chiapa AL, Rodriquez M, Phelps DR, Cardarelli KM, Vishwanatha JK, et al. Visceral fat, waist circumference and BMI: impact of race/ethnicity. Obesity (Silver Spring) 2008;16:600–607. doi: 10.1038/oby.2007.92. [DOI] [PubMed] [Google Scholar]
- 26.Carroll JF, Fulda KG, Chiapa AL, Rodriquez M, Phelps DR, Cardarelli KM, et al. Impact of race/ethnicity on the relationship between visceral fat and inflammatory biomarkers. Obesity (Silver Spring) 2009;17:1420–1427. doi: 10.1038/oby.2008.657. [DOI] [PubMed] [Google Scholar]
- 27.Kotsopoulos J, Sohn KJ, Kim YI. Postweaning dietary folate deficiency provided through childhood to puberty permanently increases genomic DNA methylation in adult rat liver. J Nutr. 2008;138:703–709. doi: 10.1093/jn/138.4.703. [DOI] [PubMed] [Google Scholar]
- 28.Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823–6836. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]