Abstract
The WW-domain containing protein KIBRA has recently been identified as a new member of the Salvador/Warts/Hippo (SWH) pathway in Drosophila and is shown to act as a tumor suppressor gene in Drosophila. This pathway is conserved in humans and members of the pathway have been shown to act as tumor suppressor genes in mammalian systems. We determined the methylation status of the 5′ CpG island associated with the KIBRA gene in human cancers. In a large panel of cancer cell lines representing common epithelial cancers KIBRA was unmethylated. But in pediatric acute lymphocytic leukemia (ALL) cell lines KIBRA showed frequent hypermethylation and silencing of gene expression, which could be reversed by treatment with 5-aza-2′-deoxycytidine. In ALL patient samples KIBRA was methylated in 70% B-ALL but was methylated in <20% T-ALL leukemia (p = 0.0019). In B-ALL KIBRA methylation was associated with ETV6/RUNX1 [t(12;21) (p13;q22)] chromosomal translocation (p = 0.0082) phenotype, suggesting that KIBRA may play an important role in t(12;21) leukemogenesis. In ALL paired samples at diagnosis and remission KIBRA methylation was seen in diagnostic but not in any of the remission samples accompanied by loss of KIBRA expression in disease state compared to patients in remission. Hence KIBRA methylation occurs frequently in B-cell acute lymphocytic leukemia but not in epithelial cancers and is linked to specific genetic event in B-ALL.
Key words: KIBRA, methylation, ALL, SWH pathway, ETV6/RUNX1 translocation
Introduction
The Salvador/Warts/Hippo (SWH) network was originally identified in D. melanogastor as a pathway responsible for regulating cellular proliferation and apoptosis through upregulation of cyclin E, diap1 and bantam.1 The core components of the fly network, Hippo, Sav, Wts, Mats are conserved in mammals as MST1/2, WW45, LATS1/2 and MOB1. The core of the hippo pathway has been well studied.2 The sterile-20 kinase Hippo forms a complex with Salvador, a WW-repeat scaffolding protein. This in turn phosphorylates and activates the DBF family kinase Warts. Activated Warts in association with MATS phosphorylates and inhibits the transcriptional co-activator Yorkie. This in turn leads to a reduction in cell number owing to the downregulation of transcriptional targets of Yorkie, the anti-apoptotic molecules diap1 and the microRNA bantam and the cell cycle regulator cyclin E. Hence the SWH pathway activation leads to smaller organs with fewer cells, and the reverse happens, increase in cell number and tissue size when the pathway is downregulated. Upstream components of the pathway include Merlin and Expanded. Very recently three groups identified the WW-domain containing protein Kibra as an upstream component of the SWH pathway and demonstrated that it acts as a tumor suppressor gene in Drosophila.3–5
RASSF1A tumor suppressor gene belongs to a family of proteins containing a RAS-association domain in either their C (RASSF1-RASSF6) or N-terminals (RASSF7-RASSF10).6–8 It has been demonstrated that in mammalian cells RASSF1A forms a complex similar to the SWH network in Drosophila.9 RASSF1A interacts with MST1 and MST2 and this binding is important for some of its functional properties.10–12 We previously undertook a methylation screen of all RASSF members (RASSF1A-RASSF10) in acute lymphocytic leukemia. We demonstrated that RASSF6 and RASSF10 were the only members that were frequently and specifically methylated in ALL. RASSF6 was very frequently methylated in B-ALL (94%) and in 41% T-ALL, while RASSF10 was frequently methylated in 88% T-ALL but showed <20% methylation in B-ALL.13 In this report we demonstrate that the latest member of the SWH pathway, KIBRA is frequently methylated in B-ALL and this methylation is associated with the most common genetic event occurring in B-ALL.
Results
Methylation analysis in common epithelial cancers.
A large panel of tumor cell lines (n = 49) consisting of major epithelial cancers was analyzed for methylation in the 5′CpG island associated with the KIBRA gene. We designed primers encompassing part of the KIBRA 5′CpG island and used COBRA assay to detect methylation. The cell line panel consisted of breast, colorectal, glioma, kidney, lung and prostate cancer cell lines. One out of seven glioma cell lines showed partial methylation, rest of the cancer cell lines in the panel showed no methylation (Sup. Fig. S1). We went on to analyze 20 glioma tumors, KIBRA was unmethylated in all cases (Sup. Fig. S1). Hence KIBRA is unmethylated in these epithelial cancers.
Methylation and expression analysis in leukemia cell lines.
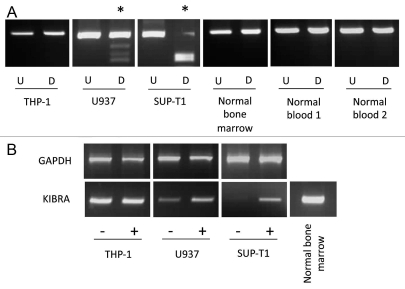
We next examined a panel of acute lymphocytic leukemia cell lines for KIBRA methylation. Among six cell lines, three were completely and one was partially methylated for KIBRA and two cell lines were unmethylated (Fig. 1A). In order to demonstrate biological significance of this methylation, the leukemia cell lines were treated with 5-aza-2′-deoxycytidine. KIBRA gene expression could be seen after 5-aza-2′-deoxycytidine treatment in the methylated cell lines, while no change was seen in unmethylated lines (Fig. 1B).
Figure 1.
Leukemia cell line methylation and expression status. KIBRA methylation and re-expression in leukemia cell lines. (A) Undigested PCR product (U) is shown next to digested PCR product (D) for three leukemia cell lines, one normal bone marrow sample and two normal blood samples. (B) GAPDH and KIBRA RT-PCR results are shown for unmethylated cell line THP-1, partially methylated cell line U937 and completely methylated cell line SUP-T1 pre (−) and post (+) treatment with 5-aza-2′-deoxycitidine. * indicates methylated samples.
Methylation analysis in primary ALL leukemia samples.
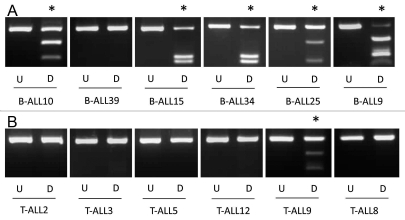
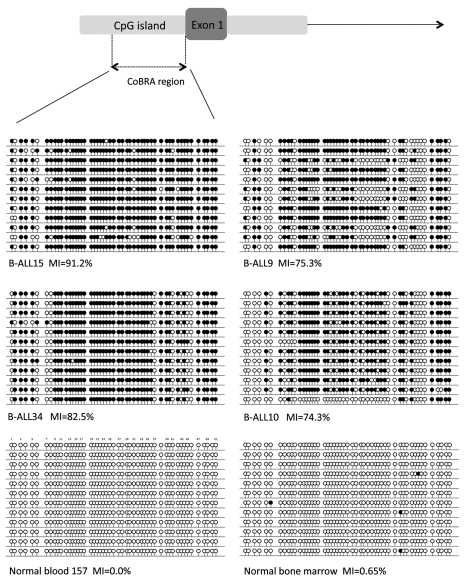
In acute lymphocytic leukemia KIBRA methylation was detected in 33/47 (70%) B-cell ALL and much less frequently in T-cell ALL, 2/12 (16.7%) (Fig. 2A and B) (p = 0.0019). In order to confirm the COBRA results and to determine the extent and pattern of KIBRA CpG island methylation, we sequenced the DNA from bisulfite-modified ALL samples. As seen in Figure 3 the CpG region analyzed was heavily methylated throughout giving high values for the methylation index (MI). No methylation was detected in DNA from age matched peripheral blood lymphocytes from healthy individuals or in healthy bone marrow (Fig. 3). In B-cell ALL patient samples KIBRA methylation was strongly associated with ETV6/RUNX1 translocation (p = 0.0082), the most common translocation occurring in this leukemia sub-type. Our data suggests that KIBRA may play an important role in t(12;21) leukemogenesis. Survival analysis was not possible in the ALL cohort since very few individuals underwent relapse. We also analyzed other members of the SWH pathway (FAT1, LATS1 and LATS2, MOBs, MST1 and MST2 and SAV1) for epigenetic inactivation in acute lymphocytic leukemia. Only FAT1 was found to be frequently methylated in B-ALL (65%) and less so in T-ALL (25%) (Sup. Figs. 2 and 3). FAT1 methylation in B-ALL associated with ETV6/RUNX1 translocation (p < 0.05).
Figure 2.
KIBRA methylation in ALL samples. CoBRA results are shown for six childhood B-ALL samples (A) and six childhood T-ALL samples (B). Undigested products (U) are shown next to BstUI digested products (D) for each sample. * indicates methylated samples.
Figure 3.
Sequencing of ALL samples and control blood and bone marrow. A schematic of the CpG island encompassing the first exon of KIBRA is shown with the CpG island in light grey, exon 1 in dark grey and the region analyzed for methylation by CoBRA and clone sequencing represented by dashed lines. Clone sequencing results are shown for four childhood B-ALL samples (B-ALL15, B-ALL9, B-ALL34 and B-ALL10), one normal blood sample and one normal bone marrow sample. Black circles represent methylated CpGs and white circles represent unmethylated CpGs. The methylation index (MI) is given as a percentage for each sample.
Methylation and expression analysis in paired ALL samples at diagnosis and remission.
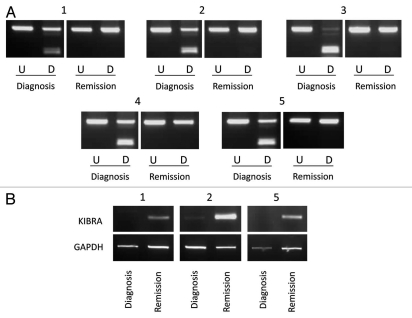
We analyzed five paired ALL samples with the ETV6/RUNX1 translocation for KIBRA methylation in matched DNA at diagnosis and in remission. KIBRA was methylated in all five diagnostic samples but was unmethylated in all five matched remission samples (Fig. 4A). This provides further evidence that KIBRA methylation is cancer specific and is associated with the malignant state. We also examined KIBRA expression in the above matched samples. KIBRA expression was reduced or absent in the disease state compared to patients in remission (Fig. 4B). Hence loss of KIBRA is highly associated with the disease state in these patients and is biologically relevant.
Figure 4.
Paired diagnosis and remission analysis. (A) CoBRA results are shown for five childhood ALL patients at diagnosis and remission stages of disease. All samples are shown with undigested product (U) next to digested product (D). (B) Expression results are shown for three of the five patients shown in (A) at diagnosis and remission stages of disease for KIBRA and GAPDH.
Discussion
Acute lymphocytic leukemia is the most common form of childhood leukemia in the western World. ALL is characterized by specific translocations, including the most common translocation ETV6/RUNX1 [t(12;21)(p13;q22)], occurring in approximately 50% of childhood cases. ETV6/RUNX1 translocation is associated with good prognosis in individuals with this disease.14 Aberrant DNA methylation has been well documented for epithelial and hematological cancers and shown to play a strong role in tumorigenesis. Hypermethylation of tumor suppressor gene promoters are a well characterized hallmark of many cancers.15,16 There have been several recent genome-wide studies to identify hypermethylated genes in acute lymphocytic leukemia.17–19 Our laboratory recently carried out a comprehensive analysis of all RASSF gene family members (RASSF1-RASSF10) with an associated 5′ CpG island in acute lymphocytic leukemia. We demonstrated that RASSF6 was the most frequently methylated family member in B-cell ALL (94%), while the newly characterized RASSF10 was most frequently methylated in T-cell ALL (88%).13 RASSF1A has recently been linked with a highly conserved pathway between Drosophila and mammals, Salvador-Warts-Hippo (SWH) tumor suppressor network. RASSF1A binds to a core component of the pathway, MST1 and MST2 (Hippo in Drosophila) and the subsequent biological role of this association has been determined in various cellular contexts.10,11,20,21 The SWH pathway was originally identified in the fly to control organ size. The core components of the pathway (Hippo, Salvador, Warts/Lats, Mats, Yorkie and very recently Kibra) are evolutionally conserved in mammals (MST1 and 2, SAV1, LATS1 and 2, MOB1, YAP and KIBRA). Recent studies have demonstrated a key role for the SWH pathway in regulating cell contact inhibition, organ size control and development of cancer in mammals.22–26 Kibra was recently identified as upstream member of this pathway and a potential tumor suppressor in Drosophila. Human KIBRA has been reported to play a role in cell migration and is known to be phosphorylated by protein kinase C (PKC) zeta.27–29
MSTs and LATs members of the SWH pathway have previously been shown to be epigenetically inactivated in certain human cancers. Frequent methylation of MST2 (96%) was found in head and neck squamous cell carcinoma and less frequently for LATS1 (24%) and LATS2 (8%).30 MST1 and MST2 were also methylated in soft tissue sarcomas.31 LATS1 and LATS2 were frequently methylated in human astrocytomas and in breast cancer.32,33 LATS2 was also methylated in 24% of acute lymphocytic leukemia.34 A very recent study highlighted the importance of the hippo signaling in the pathogenesis of B-cell lymphomas and demonstrated for the first time the Hippo pathway as tumor suppressor genes contributing to lymphoma tumorigenesis.35 Decreased expression of Hippo members MOBKL2A, MOBKL2B and LATS2 was associated with inferior survival in mantle cell lymphoma (MCL); furthermore, loss of the genomic regions where the three genes reside was observed in 38% of MCLs. MOBKL2A and MOBKL2B are homologs of the MOB1 gene that interacts with LATS in inhibition of YAP, a growth promoter.
There has been no comprehensive analysis of the SWH pathway genes in both epithelial and hematological malignancies. Therefore, we have undertaken a detailed epigenetic analysis of KIBRA a newly identified member of the SWH pathway in both epithelial and hematological cancers. We describe for the first time frequent methylation of KIBRA in B-cell ALL, but not in major epithelial cancers. KIBRA gene methylation in childhood B-cell ALL is associated with the most common translocation (ETV6/RUNX1) found in this subtype of leukemia and leads to loss of KIBRA expression in the malignant state. Roman-Gomez et al. have previously shown that a set of four genes (DKK3, sFRP2, PTEN and P73) show methylation specificity for ETV6/RUNX1 positive B-ALL and suggest that these genes may play important roles in t(12;21) leukemogenesis. Furthermore they demonstrated that patients carrying the ETV6/RUNX1 translocation showed a higher degree of genes simultaneously methylated compared to ETV6/RUNX1 negative cases.36 This suggests that epigenetic inactivation plays a more important role in t(12;21) leukemogenesis compared to other forms of B-ALL. Morrow et al. demonstrated that ETV6/RUNX1 translocation is necessary but insufficient for leukemia development.37 The above data suggests that subsequent molecular events occurring in early childhood are required for leukemogenesis, one such mechanism could be epigenetic inactivation.
Our results further indicate that KIBRA is a potential tumor suppressor gene not only in Drosophila but also in mammalian cells and plays an important role in B-cell leukemogenesis and may provide a potential new therapeutic target for clinical applications especially due to the reversible nature of promoter hypermethylation. It will be interesting to see if KIBRA is methylated in other B-cell leukemias and lymphomas and to understand what role does KIBRA play in B-cell development/differentiation. Our results add further evidence of the Hippo tumor suppressor pathway dysregulation in hematological cancers.
Materials and Methods
Cell lines and primary patient samples.
A panel of 49 epithelial cancer cell lines (five breast, seven colorectal, seven glioma, ten kidney, 15 lung and five prostate) and seven leukemia cell lines were used for initial analysis [Jurkat (JKT), NALM6, U937, SUP-T1, CCRF-CEM (CEM), DND-41 and THP-1], in addition 20 glioma tumors were also analyzed. A total of 70 childhood ALL samples were used comprising 52 B-ALL, 1 pre-B-ALL, 12 T-ALL and five unclassified. Characteristics of the ALL samples have been described previously in reference 17. Five diagnosis and remission paired samples were also used for childhood ALL methylation and expression analyses. Ethical guidelines were followed for all collected samples.
Methylation analysis.
DNA was bisulfite-modified as described previously in reference 38. We analyzed a 368 bp region of the KIBRA CpG island just upstream of the transcription start site using the following semi-nested primers, forward: 5′-GTA TTT GGY GGA GGT AGA AGT TAT AAA TT-3′, reverse nested: 5′-CAT AAT CCR AAA AAT AAC RCC CRC AAA TAA-3′ and reverse: 5′-TAT CCC RCR AAT CRA CCA ACT AAT AAT A-3′. Primary and secondary PCR reactions were touchdown PCRs with a final annealing temperature of 55°C and total cycle number of 42 and 47 respectively with 5 µl primary PCR product transferred to the secondary reaction. 15–20 µl of PCR product was digested with BstUI (CGCG) at 37°C overnight prior to visualization on a 2% agarose gel.
Clone sequencing was used to quantify selected CoBRA results. Selected PCR products were cloned into the pGEM-T-Easy vector according to manufacturers' instructions. Single colony PCR was carried out on up to 12 colonies for each cloned sample using the following forward primer: 5′-TAA TAC GAC TCA CTA TAG GG-3′; and reverse primer: 5′-ACA CTA TAG AAT ACT CAA GC-3′. PCR products were then sequenced and methylation indexes (MIs) calculated for each sample as a percentage of the number of methylated CpGs out of the total number of CpGs analyzed per sample.
Expression analysis.
Leukemia cell lines were maintained in RPMI1640 (Sigma) with 10% FCS, 2 mM glutamine, 20 mM HEPES, 1 mM sodium pyruvate, 12.6 mM glucose monohydrate at 37°C, 5% CO2. Genome demethylation was achieved by treatment with 5 µM 5-aza-2′-deoxycitidine over five days with daily media changes. RNA-bee (AMS biotechnology) was used to extract RNA according to manufacturers' instructions. Control bone marrow RNA was purchased from AMS biotechnology. cDNA was synthesized from 1 µg total RNA using Superscript III (Invitrogen) and random hexamer primers (Fermentas). Expression was assessed using a touchdown PCR with a total cycle number of 40 and final annealing temperature of 56°C using the following primers: F: 5′-AAA CAG AGC AGG GAG CTC AA-3′ and R: 5′-CCC ATC CAT ATC AGG TGA GG-3′. GAPDH analysis was carried out concurrently with a lower cycle number (32 instead of 40) using the forward primer: 5′-TGA AGG TCG GAG TCA ACG-3′ and reverse primer: 5′-CAT GTG GGC CAT GAG GTC C-3′.
Statistical analysis.
Statistical analysis was performed as indicated. p < 0.05 was considered significant.
Acknowledgements
F. Latif laboratory funded in part by Cancer Research UK, Breast Cancer Campaign and Sport Aiding Medical Research for Kids (SPARKS).
Supplementary Material
References
- 1.Harvey K, Tapon N. The Salvador-Warts-Hippo pathway—an emerging tumour-suppressor network. Nat Rev Cancer. 2007;7:182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- 2.Zeng Q, Hong W. The emerging role of the hippo pathway in cell contact inhibition, organ size control and cancer development in mammals. Cancer Cell. 2008;13:188–192. doi: 10.1016/j.ccr.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 3.Yu J, Zheng Y, Dong J, Klusza S, Deng WM, Pan D. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev Cell. 2010;18:288–299. doi: 10.1016/j.devcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Genevet A, Wehr MC, Brain R, Thompson BJ, Tapon N. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev Cell. 2010;18:300–308. doi: 10.1016/j.devcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumgartner R, Poernbacher I, Buser N, Hafen E, Stocker H. The WW domain protein Kibra acts upstream of Hippo in Drosophila. Dev Cell. 2010;18:309–316. doi: 10.1016/j.devcel.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 6.Richter AM, Pfeifer GP, Dammann RH. The RASSF proteins in cancer; from epigenetic silencing to functional characterization. Biochim Biophys Acta. 2009;1796:114–128. doi: 10.1016/j.bbcan.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Sherwood V, Manbodh R, Sheppard C, Chalmers AD. RASSF7 is a member of a new family of RAS association domain-containing proteins and is required for completing mitosis. Mol Biol Cell. 2008;19:1772–1782. doi: 10.1091/mbc.E07-07-0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Underhill-Day N, Hill V, Latif F. N-terminal RASSF family (RASSF7-RASSF10) Epigenetics. 2011;6:284–292. doi: 10.4161/epi.6.3.14108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo C, Tommasi S, Liu L, Yee JK, Dammann R, Pfeifer GP. RASSF1A is part of a complex similar to the Drosophila Hippo/Salvador/Lats tumor-suppressor network. Curr Biol. 2007;17:700–705. doi: 10.1016/j.cub.2007.02.055. [DOI] [PubMed] [Google Scholar]
- 10.Matallanas D, Romano D, Yee K, Meissl K, Kucerova L, Piazzolla D, et al. RASSF1A elicits apoptosis through an MST2 pathway directing proapoptotic transcription by the p73 tumor suppressor protein. Mol Cell. 2007;27:962–975. doi: 10.1016/j.molcel.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dallol A, Hesson LB, Matallanas D, Cooper WN, O'Neill E, Maher ER, et al. RAN GTPase is a RASSF1A effector involved in controlling microtubule organization. Curr Biol. 2009;19:1227–1232. doi: 10.1016/j.cub.2009.05.064. [DOI] [PubMed] [Google Scholar]
- 12.Dallol A, Kolch W, Latif F. When RASSF1A RAN into tumor suppression: Ran GTPase is a RASSF1A effector involved in controlling microtubule organization. Cell Cycle. 2009;8:3796–3797. doi: 10.4161/cc.8.23.10024. [DOI] [PubMed] [Google Scholar]
- 13.Hesson LB, Dunwell TL, Cooper WN, Catchpoole D, Brini AT, Chiaramonte R, et al. The novel RASSF6 and RASSF10 candidate tumour suppressor genes are frequently epigenetically inactivated in childhood leukaemias. Mol Cancer. 2009;8:42. doi: 10.1186/1476-4598-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pui CH, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006;354:166–178. doi: 10.1056/NEJMra052603. [DOI] [PubMed] [Google Scholar]
- 15.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 16.Galm O, Herman JG, Baylin SB. The fundamental role of epigenetics in hematopoietic malignancies. Blood Rev. 2006;20:1–13. doi: 10.1016/j.blre.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 17.Dunwell T, Hesson L, Rauch TA, Wang L, Clark RE, Dallol A, et al. A genome-wide screen identifies frequently methylated genes in haematological and epithelial cancers. Mol Cancer. 2010;9:44. doi: 10.1186/1476-4598-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin-Subero JI, Ammerpohl O, Bibikova M, Wickham-Garcia E, Agirre X, Alvarez S, et al. A comprehensive microarray-based DNA methylation study of 367 hematological neoplasms. PLoS One. 2009;4:6986. doi: 10.1371/journal.pone.0006986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuang SQ, Tong WG, Yang H, Lin W, Lee MK, Fang ZH, et al. Genome-wide identification of aberrantly methylated promoter associated CpG islands in acute lymphocytic leukemia. Leukemia. 2008;22:1529–1538. doi: 10.1038/leu.2008.130. [DOI] [PubMed] [Google Scholar]
- 20.Avruch J, Xavier R, Bardeesy N, Zhang XF, Praskova M, Zhou D, et al. Rassf family of tumor suppressor polypeptides. J Biol Chem. 2009;284:11001–11005. doi: 10.1074/jbc.R800073200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vichalkovski A, Gresko E, Cornils H, Hergovich A, Schmitz D, Hemmings BA. NDR kinase is activated by RASSF1A/MST1 in response to Fas receptor stimulation and promotes apoptosis. Curr Biol. 2008;18:1889–1895. doi: 10.1016/j.cub.2008.10.060. [DOI] [PubMed] [Google Scholar]
- 22.Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y, et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009;16:425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu Q, et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci USA. 2010;107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song H, Mak KK, Topol L, Yun K, Hu J, Garrett L, et al. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc Natl Acad Sci USA. 2010;107:1431–1436. doi: 10.1073/pnas.0911409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee KP, Lee JH, Kim TS, Kim TH, Park HD, Byun JS, et al. The Hippo-Salvador pathway restrains hepatic oval cell proliferation, liver size and liver tumorigenesis. Proc Natl Acad Sci USA. 2010;107:8248–8253. doi: 10.1073/pnas.0912203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kremerskothen J, Plaas C, Büther K, Finger I, Veltel S, Matanis T, et al. Characterization of KIBRA, a novel WW domain-containing protein. Biochem Biophys Res Commun. 2003;300:862–867. doi: 10.1016/s0006-291x(02)02945-5. [DOI] [PubMed] [Google Scholar]
- 28.Rosse C, Formstecher E, Boeckeler K, Zhao Y, Kremerskothen J, White MD, et al. An aPKC-exocyst complex controls paxillin phosphorylation and migration through localised JNK1 activation. PLoS Biol. 2009;7:1000235. doi: 10.1371/journal.pbio.1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Büther K, Plaas C, Barnekow A, Kremerskothen J. KIBRA is a novel substrate for protein kinase Czeta. Biochem Biophys Res Commun. 2004;317:703–707. doi: 10.1016/j.bbrc.2004.03.107. [DOI] [PubMed] [Google Scholar]
- 30.Steinmann K, Sandner A, Schagdarsurengin U, Dammann RH. Frequent promoter hypermethylation of tumor-related genes in head and neck squamous cell carcinoma. Oncol Rep. 2009;22:1519–1526. doi: 10.3892/or_00000596. [DOI] [PubMed] [Google Scholar]
- 31.Seidel C, Schagdarsurengin U, Blümke K, Würl P, Pfeifer GP, Hauptmann S, et al. Frequent hypermethylation of MST1 and MST2 in soft tissue sarcoma. Mol Carcinog. 2007;46:865–871. doi: 10.1002/mc.20317. [DOI] [PubMed] [Google Scholar]
- 32.Jiang Z, Li X, Hu J, Zhou W, Jiang Y, Li G, et al. Promoter hypermethylation-mediated downregulation of LATS1 and LATS2 in human astrocytoma. Neurosci Res. 2006;56:450–458. doi: 10.1016/j.neures.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 33.Takahashi Y, Miyoshi Y, Takahata C, Irahara N, Taguchi T, Tamaki Y, et al. Downregulation of LATS1 and LATS2 mRNA expression by promoter hypermethylation and its association with biologically aggressive phenotype in human breast cancers. Clin Cancer Res. 2005;11:1380–1385. doi: 10.1158/1078-0432.CCR-04-1773. [DOI] [PubMed] [Google Scholar]
- 34.Jiménez-Velasco A, Román-Gómez J, Agirre X, Barrios M, Navarro G, Vázquez I, et al. Downregulation of the large tumor suppressor 2 (LATS2/KPM) gene is associated with poor prognosis in acute lymphoblastic leukemia. Leukemia. 2005;19:2347–2350. doi: 10.1038/sj.leu.2403974. [DOI] [PubMed] [Google Scholar]
- 35.Hartmann EM, Campo E, Wright G, Lenz G, Salaverria I, Jares P, et al. Pathway discovery in mantle cell lymphoma by integrated analysis of high-resolution gene expression and copy number profiling. Blood. 2010;116:953–961. doi: 10.1182/blood-2010-01-263806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roman-Gomez J, Jimenez-Velasco A, Agirre X, Castillejo JA, Navarro G, Calasanz MJ, et al. CpG island methylator phenotype redefines the prognostic effect of t(12;21) in childhood acute lymphoblastic leukemia. Clin Cancer Res. 2006;12:4845–4850. doi: 10.1158/1078-0432.CCR-05-2592. [DOI] [PubMed] [Google Scholar]
- 37.Morrow M, Horton S, Kioussis D, Brady HJ, Williams O. TEL-AML1 promotes development of specific hematopoietic lineages consistent with preleukemic activity. Blood. 2004;103:3890–3896. doi: 10.1182/blood-2003-10-3695. [DOI] [PubMed] [Google Scholar]
- 38.Hesson L, Biíche I, Krex D, Criniere E, Hoang-Xuan K, Maher ER, et al. Frequent epigenetic inactivation of RASSF1A and BLU genes located within the critical 3p21.3 region in gliomas. Oncogene. 2004;23:2408–2419. doi: 10.1038/sj.onc.1207407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.