Abstract
De novo DNA methylation in Arabidopsis thaliana is catalyzed by the methyltransferase DRM2, a homolog of the mammalian de novo methyltransferase DNMT3. DRM2 is targeted to DNA by small interfering RNAs (siRNAs) in a process known as RNA-directed DNA Methylation (RdDM). While several components of the RdDM pathway are known, a functional understanding of the underlying mechanism is far from complete. We employed both forward and reverse genetic approaches to identify factors involved in de novo methylation. We utilized the FWA transgene, which is methylated and silenced when transformed into wild-type plants, but unmethylated and expressed when transformed into de novo methylation mutants. Expression of FWA is marked by a late-flowering phenotype, which is easily scored in mutant versus wild-type plants. By reverse genetics we discovered the requirement for known RdDM effectors AGO6 and NRPE5a for efficient de novo methylation. A forward genetic approach uncovered alleles of several components of the RdDM pathway, including alleles of clsy1, ktf1 and nrpd/e2, which have not been previously shown to be required for the initial establishment of DNA methylation. Mutations were mapped and genes cloned by both traditional and whole genome sequencing approaches. The methodologies and the mutant alleles discovered will be instrumental in further studies of de novo DNA methylation.
Key words: DNA methylation, Arabidopsis, de novo, genetic screen, whole-genome sequencing
Introduction
Cytosine DNA methylation is an ancient form of epigenetic information found in all kingdoms of eukaryotes. Functions for DNA methylation-mediated gene silencing include imprinting and repression of transposable elements. In mammals, methylation occurs almost exclusively in the CG context. Maintenance of the methyl-mark persists through function of the DNA methyltransferase, DNMT1, whereas the initial, or de novo, methylation of unmodified DNA occurs through DNMT3 activity. The importance of DNA methylation for proper development is underscored by embryonic lethality in knockout mice for either type of methyltransferase.1,2
The flowering plant Arabidopsis thaliana serves as a powerful model to analyze the function of DNA methylation since it contains orthologs to both DNMT1 and DNMT3, termed METHYLTRANSFERASE1 (MET1) and DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2), respectively.3,4 Moreover, unlike mammals, Arabidopsis plants that contain homozygous null mutations for either gene are viable. In addition, Arabidopsis contains a third methyltransferase gene: the plant-specific CHROMOMETHYLASE 3 (CMT3). Unlike mammals, plants are able to methylate cytosines in three contexts: CG, CHG (where H is A, C or T) and CHH. Although there is a degree of redundancy, MET1 largely carries out CG methylation, CMT3 performs CHG and DRM2 is the primary CHH methyltransferase.3–5
The CHH context is of particular note because it lacks inherent symmetry across the DNA strand. Whereas methylation in symmetric contexts can be recognized as hemimethylated DNA on both daughter strands of DNA after replication and therefore be maintained passively,6 maintenance of asymmetric methylation must operate via an active signal. In the last decade a flood of studies have implicated a 24-nucleotide species of siRNAs as guiding DRM2 in a process known as RNA-directed DNA Methylation (RdDM).7–10
In addition to maintaining pre-existing CHH methylation, RdDM via DRM2 is needed to establish DNA methylation on unmethylated sequences, such as an incoming (FLOWERING WAGENINGEN) FWA transgene.11,12 FWA is a maternally imprinted homeodomain transcription factor. In vegetative tissue FWA is silenced as a result of methylation in its promoter and 5′ UTR.13,14 Heritable unmethylated fwa-1 epialleles are dominant and ectopic expression causes a late-flowering phenotype. RdDM mutant plants are incapable of de novo methylating FWA upon transformation, thus flower late.
Several aspects of the RdDM process remain a mystery. For example it is unclear what downstream effectors contribute to the silent state following de novo methylation. Additionally, it is uncertain how a 24 nt siRNA-loaded ARGONAUTE4 physically targets DRM2 to specific loci. However, perhaps the most compelling question is how RdDM machinery is able to recognize particular unmethylated DNA sequences. Although repetitive elements such as transposons are associated with small RNAs and DNA methylation, there is no known sequence specificity or secondary structure that is a hallmark of recruitment, let alone an endogenous factor that serves to respond to invasive DNA.15
In order to further clarify the process of the initial establishment of methylation, we employed both forward and reverse genetic screens taking advantage of the FWA late-flowering phenotype. In the first forward mutagenesis screen specifically designed to discover mutations that block the initial establishment of methylation, a broad picture of the RdDM pathway emerged. We utilized both traditional and emerging whole-genome sequencing approaches to identify mutations from the screen. Of particular note, the recently described SUPPRESSOR OF TY INSERTION 5-LIKE (SPT5-LIKE)/KOW DOMAIN-CONTAINING TRANSCRIPTION FACTOR 1 (KTF1)16–18 and CLASSY1 (CLSY1)19 genes were discovered by our approaches to be required for initial establishment of methylation.
Results and Discussion
A forward screen to discover de novo methylation mutants. In an effort to increase our knowledge of de novo methylation in Arabidopsis, we took an unbiased approach to screen for mutations that block the establishment of DNA methylation (Fig. 1). This was accomplished by transforming plants with an FWA transgene and screening for mutants that block the establishment of silencing of FWA and therefore produce a late flowering phenotype. Columbia-0 (Col-0) seeds were treated with ethyl methanesulfonate (EMS) and roughly 900 mutagenized M1 lines were allowed to self-pollinate to produce individual M2 families. Due to concerns that unrelated flowering time mutations would mimic the FWA hypomethylation phenotype, approximately 110 M2 lines that exhibited late flowering were discarded. The remaining M2 lines were transformed with the FWA transgene and the first generation of Basta-resistant transformants (T1s) was screened for late flowering plants. Roughly 300 T1 lines displayed a late-flowering phenotype upon transformation. The large number of late-flowering plants is likely due to liberal thresholds used to score the phenotype, incomplete silencing of FWA that occurs even in wild-type plants, natural variation and the segregation of flowering time mutants in a host of genes. RT-PCR from leaf material confirmed that FWA was expressed above wild-type levels in 128 lines.
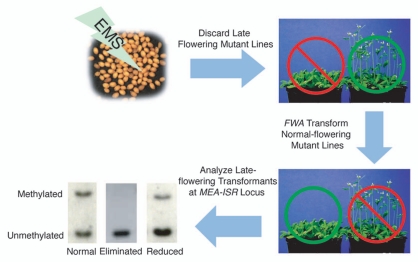
Figure 1.
Schematic representation of the FWA screen approach. EMS mutagenized Col-0 plants were transformed with the FWA transgene. Prior to transformation, any mutagenized line that exhibited a late-flowering phenotype was discarded. The first generation of FWA-transformed mutant plants were screened for flowering time. All late-flowering lines were deemed potential de novo methylation mutants. As a secondary screen, late-flowering lines were divided into three classes based on their phenotype at the MEA-ISR locus. Genomic DNA was digested with the methylation sensitive enzyme MspI and digestion was analyzed by Southern blot.
As mentioned previously, all known mutants that block de novo methylation are components of RdDM. Therefore, to identify an assay for confirming and classifying our mutants, we searched for an endogenous locus that has an siRNA-dependent methylation state that can be examined with relative facility. The MEDEA-INTERGENIC SUBTELOMERIC REPEATS (MEA-ISR) are a set of tandem repeats downstream of the MEDEA gene that are targeted by DRM2.4 Using the MspI restriction endonuclease, non-CG methylation at MEA-ISR was examined by Southern blot analysis. Wild-type plants display roughly a 1:1 ratio of methylated to unmethylated bands. In strong mutants, such as null drm2 and ago4 alleles, non-CG methylation is virtually eliminated (Fig. 2A). Weaker mutants, such as null mutations in dicer-like 3 (dcl3)—the dicer protein most strongly associated with RdDM—exhibit an intermediate banding pattern (Fig. 2A). Thus, the 128 de novo methylation mutants were sub-classified into three categories based on their MEA-ISR phenotype: MEA-ISR normal (wild type), MEA-ISR eliminated (phenocopy of drm2) and MEA-ISR reduced (phenocopy of dcl3) (Fig. 1).
Figure 2.
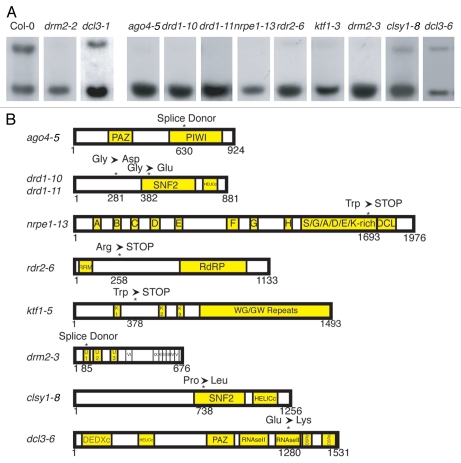
Characterization of mutant lines, and mutation identification. (A) MEA-ISR Southern blot phenotype for nine de novo mutant lines. Seven lines were sub-categorized MEA-ISR eliminated and two as MEA-ISR reduced. The allele resulting in each mutant phenotype is indicated. (B) Protein models for each mutation identified from the screen. The mutation is denoted above the model. drd1 is the only gene for which we obtained multiple alleles: drd1-10 is a Gly→Asp missense and drd1-11 is a Gly→Glu missense.
Based on the Southern blot phenotypes, seven lines fell into the MEA-ISR eliminated class, and ten lines fell into the MEA-ISR reduced class; the remaining 111 de novo mutant lines are termed MEA-ISR normal. Mutations mapped in the first two classes will be discussed.
Map based approach to discover mutations.
In order to positionally clone the mutations that block de novo methylation, we took advantage of the maintenance methylation defect in the MEA-ISR eliminated and MEA-ISR reduced classes. Mutant lines were outcrossed to Landsberg erecta (Ler) and individual plants from segregating F2 populations were assayed for a MEA-ISR Southern blot loss of methylation phenotype. We then utilized a series of PCR based molecular markers to map each of the mutations. We included markers that are tightly linked to the genes already known or suspected to affect de novo methylation in order to most efficiently identify the genes harboring mutations.
Linkage analysis followed by sequencing led to the discovery of seven mutations in six different genes that phenocopy drm2 (MEA-ISR eliminated class), and two mutations in two different genes that phenocopy dcl3 (MEA-ISR reduced class) (Fig. 2A and B, and described below). Allelism crosses with null T-DNA alleles were used to confirm gene identities. Included in the list are alleles of genes that heretofore have not been previously shown to be required for the initial establishment of methylation: KTF1 and CLSY1.
Analysis of de novo methylation components upstream of small RNA production.
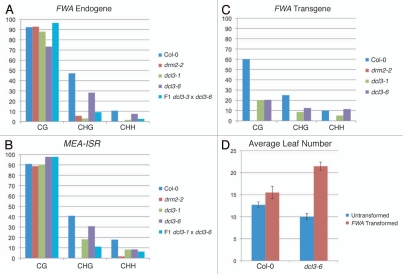
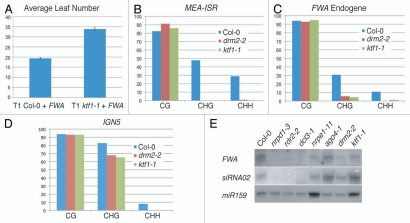
The species of small RNAs most frequently associated with RdDM are 24 nucleotides in length.7,20 Three components required to synthesize this class of siRNAs were isolated in the screen: rdr2-6, dcl3-6 and clsy1-8. RNA-DEPENDENT RNA Polymerase 2 (RDR2) likely produces double-stranded RNA from transcripts generated by the plant-specific RNA Polymerase IV (Pol IV).10 Here, we identified an allele of rdr2 that contains a premature stop codon well upstream of the RNA-dependent RNA Polymerase (RdRP) domain (Fig. 2B), which therefore is likely a null allele. DCL3 is proposed to process the RDR2 double stranded RNA product into 24-mer siRNAs.21,22 We identified a glutamate-to-lysine missense mutation in a highly conserved DCL3 residue in one of the Ribonuclease III (RNAseIII) domains (Fig. 2B). The allele recovered in the screen appears to have a weaker effect on RdDM than a dcl3 null allele (Fig. 3A and B). This perhaps is due to the fact that DCL3 contains two RNAseIII domains, so while one is impaired, the second retains its enzymatic activity (Fig. 2B). Despite this, the mutation still affects de novo methylation of the FWA transgene such that it causes significantly reduced transgene methylation causing retarded flowering time compared to the wild-type control (Fig. 3C and D). Genetic complementation with a null dcl3 allele confirms that the missense in dcl3-6 causes the impaired methylation phenotype (Fig. 3A and B). The values for FWA transgene methylation are much lower than the endogenous copies, even in wild-type plants, due to incomplete methylation that is observed in the first generation of transformants.
Figure 3.
Analysis of the dcl3 allele recovered from the screen. (A–C) Sodium bisulfite treatment and analysis of methylation state of (A) FWA endogene (B) MEA-ISR and (C) FWA transgene. Y axis denotes percent methylation. (D) FWA flowering-time assay. Average number of leaves upon flowering for both untransformed and FWA transformed lines.
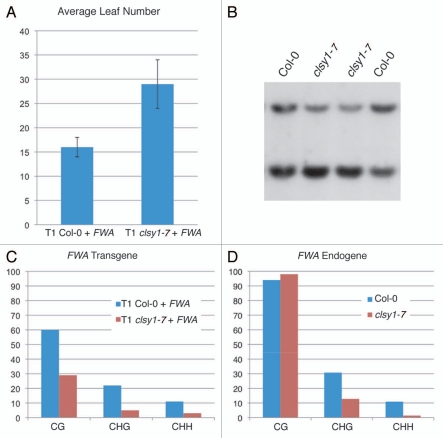
A third mutation in a gene required for siRNA synthesis is a proline-to-leucine missense in clsy1-8 (Fig. 2A and B). CLSY1 is a member of a SNF2-like domain-containing class of proteins that also contain a helicase domain near the carboxyl-terminus, and structural modeling predicts a putative chromatin remodeling function for the CLSY1 protein.19 Genetic data suggests that CLSY1 is required for proper localization of both NRPD1—the largest subunit of Pol IV—and RDR2, suggesting it acts upstream of both of these components.19 The mutation discovered here is adjacent to both the putative ATP binding site and DNA back-bone binding residues within the SNF2-like domain (Fig. 2B).19 The proline is a conserved residue among Arabidopsis chromatin remodeling proteins, and its mutation in clsy1-8 may result in an altered structural conformation that affects either or both of these functions. We found that a null clsy1 T-DNA insertion mutant, clsy1-7, did not completely block de novo methylation, as assayed by FWA transformation (Fig. 4A and C). Concordantly, the maintenance methylation phenotype at the MEA-ISR locus was also incomplete, with residual non-CG methylation present at the MspI restriction site (Fig. 4B). In order to confirm that CLSY1 is required for RdDM maintenance methylation, we examined the FWA endogene in the mutant (Fig. 4D). Indeed, in clsy1 the non-CG methylation is sharply reduced. A possible explanation for the weaker phenotype of clsy1 mutants compared to other components in the pathway may be partial redundancy with another member of the subfamily of SNF2-like proteins. A likely candidate is CLSY2, which contains 81% sequence homology with CLSY1.
Figure 4.
Analysis of clsy1 de novo methylation phenotype. (A) FWA flowering-time assay. (B) MEA-ISR Southern blot from two individual Col-0 plants and two clsy1-7 plants to show the reproducibility of the weak phenotype. (C and D) Sodium bisulfite analysis of the FWA transgene (C) and endogene (D).
Analysis of de novo methylation components downstream of siRNA production.
Subsequent to biogenesis, 24-nt siRNAs target RdDM machinery to homologous DNA sequences for DRM2-mediated methylation. Mutations in genes downstream of siRNA production do not exhibit severe defects in small RNA accumulation at many RdDM target loci; however, non-CG methylation is sharply reduced or eliminated.23–25 The FWA screening approach produced six alleles in five genes required for de novo DNA methylation but not strictly required for siRNA biogenesis: ago4-5, nrpe1-13, drd1-10, drd1-11, drm2-3 and ktf1-5 (Fig. 2B).
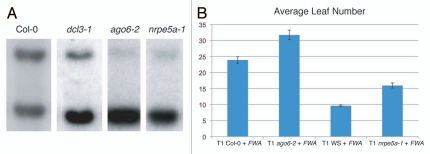
Small RNAs associated with RdDM are primarily loaded into AGO4.9,26 AGO4 contains two conserved domains termed PAZ and PIWI. The PIWI domain contains a triad of catalytic residues (D660, D742 and H874) that are conserved in argonaute proteins in plants, humans and S. pombe, and have been previously shown to be required for AGO4 slicer activity and RdDM.26 An allele of ago4 identified from this screen contains a splice donor mutation that causes a frame shift and eventual premature stop downstream of amino acid 630 (Fig. 2B). Given that the three catalytic triad residues all lie downstream of the mutation, this allele likely renders the protein catalytically inactive, explaining the strong de novo methylation phenotype and the strong loss of methylation MEA-ISR phenotype (Fig. 2A). AGO4 acts partially redundantly with its close family member AGO6 in maintenance DNA methylation, and the ago6-2 mutant shows a reduced MEA-ISR methylation phenotype (Fig. 5A).27 Therefore, we wanted to confirm that ago6 similarly affects de novo DNA methylation. Analysis of FWA-transformed ago6-2 plants confirmed that indeed there is a late-flowering phenotype (Fig. 5C) (Note: the variation in flowering time for Col-0 + FWA from different experiments is likely due to differences in growth conditions).
Figure 5.
Reverse genetics showing that nrpe5a and ago6 are required for de novo DNA methylation. (A) MEA-ISR Southern blot. (B) Flowering-time assay.
Previous studies have shown that AGO4 colocalizes with NRPE1, the largest subunit of the plant-specific RNA Polymerase V (Pol V) in the nucleus.28 Pol V produces a transcript that is necessary to recruit AGO4 to chromatin of methylated loci.29 Additionally, NRPE1 contains a hydrophilic (S/G/A/D/E/K-rich) domain in its C-terminal that interacts with AGO4.28,30 The interaction is dependent on conserved tryptophan-glycine/glycine-tryptophan (WG/GW) repeats within NRPE1.30 An allele of nrpe1 arose from the FWA screen containing a premature stop codon at amino acid position 1693 (Fig. 2B). RT-PCR analysis indicated that the nrpe1-13 mutant transcript is also significantly reduced, possibly by nonsense-mediated mRNA decay (data not shown). This is consistent with the strong loss-of-function mutant phenotype of this allele.
Because NRPE1 is required for de novo methylation, we also performed FWA transformation on an allele of the recently described Pol V-specific subunit NRPE5a.31,32 The nrpe5a-1 mutant indeed showed a partial late flowering phenotype after transformation, showing that NRPE5a is also required for establishment of methylation (Fig. 5B). Consistent with this effect, the nrpe5a-1 mutant also showed a partial maintenance methylation phenotype at MEA-ISR (Fig. 5A). A possible explanation for why this mutant does not have as severe a methylation defect as nrpe1 may be because of partial redundancy with its paralogs, NRPE5b and NRPE5c.32
Pol V transcriptional activity is dependent on the SNF2-like putative chromatin remodeler DEFECTIVE IN RNA-DIRECTED DNA METHYLATION 1 (DRD1).33 Null alleles of drd1 display a severe decrease in non-CG methylation at RdDM target loci.34,35 Two alleles of drd1 emerged from the FWA screen, both of which exhibited a strong loss of methylation at MEA-ISR (Fig. 2A). drd1-11 contains a missense mutation in a highly conserved glycine within the SNF2-like domain (Fig. 2B). The second allele, drd1-10, is also a glycine missense; however, it occurs in a non-conserved region of DRD1 (Fig. 2B). This glycine is therefore either critical for the function of DRD1, or it is possible that the mutation causes structural changes rendering the protein non-functional.
WG/GW repeats, such as those contained in NRPE1, have been shown to interact with argonaute proteins, forming a motif termed the “AGO-hook.”36 Bioinformatic, biochemical and forward genetic approaches all have converged on an Arabidopsis AGO-hook-containing protein that is required for RdDM.16–18 The protein, named KTF1, also contains homology with the yeast transcription elongation factor SPT5.16 The FWA screen approach produced an allele of ktf1 that contains a premature stop in amino acid 378 demonstrating its requirement for de novo methylation, observed by late-flowering FWA transformants (Fig. 6A). Additional analysis at endogenous loci indicates a substantial loss in non-CG methylation at DRM2-dependent sites in the ktf1-5 mutant (Fig. 6B–D).
Figure 6.
Molecular characterization of ktf1. (A) FWA flowering-time assay. (B–D) Sodium bisulfite analysis of MEA-ISR (B), FWA (C) and IGN5 (D). IGN5 is a third locus targeted by RdDM machinery. (E) Small RNA blots showing the abundance of various small RNAs in different mutant backgrounds. miRNA159 serves as a loading control. ktf1 mutant does not abolish 24 nt species of siRNAs, suggesting it is acting downstream of siRNA biosynthesis.
KTF1 does not appear to be required for the production of 24 nucleotide siRNAs at all RdDM-targeted loci (Fig. 6E).16,17 This characteristic places it in the RdDM pathway downstream of Pol IV-RDR2 activity. Although KTF1 exhibits homology to RNA Polymerase II (Pol II) elongation factors in other eukaryotes, it does not appear to affect the accumulation of Pol V transcripts showing that it is not required for Pol V transcription.17 However, KTF1 does interact with both AGO4 and Pol V, and has RNA-binding capability.16–18 A potential function could be recruiting AGO4 to chromatin at sites of Pol V transcription.
Finally, we isolated a strong allele of the de novo methyltransferase DRM2; a splice donor mutation at amino acid 85 predicted to cause a frameshift and premature stop well upstream of the DNA methyltransferase catalytic domain (Fig. 2B). The allele caused FWA late flowering and a strong loss of methylation at MEA-ISR. Although DRM2 is the methyltransferase targeted by RdDM machinery, no previous RdDM mutagenesis screen has isolated a mutant allele of the gene.
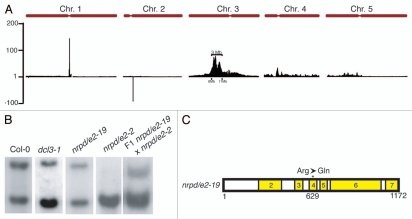
Whole-genome sequencing can efficiently identify mutants with weak phenotypes. Although MEA-ISR is a useful locus for examining defects in RdDM, we found that some late-flowering mutants from our forward screen had only a very subtle phenotype at MEA-ISR as assayed by Southern blot analysis. Therefore, scoring mutants in F2 mapping populations proved extremely difficult. For example, in a population of 56 Col/Ler F2 plants scored as being homozygous for the recessive m48 mutant, we found that the area showing linkage on chromosome 3 never fell below 25 cM, suggesting that some mutants were mis-scored and that the population was contaminated with wild-type plants. In order to map and clone m48 we therefore employed a whole-genome sequencing approach.
First, we reduced our initial mapping population of 56 recombinants to 38 plants that exhibited homozygosity for Col-0 specific PCR based molecular markers in a wide region of chromosome three, to reduce the frequency of mis-scored plants. The DNA from the set of 38 recombinants was then pooled, and we performed paired-end shotgun sequencing using Illumina GA II technology, giving 142 million high quality reads and 21.4 times coverage of the Arabidopsis genome.
In order to locate regions that are enriched for Col-0 sequences, we utilized the Mapping and Assembly with Short Sequences (MASS) approach described by Cuperus et al.37 The region exhibiting the highest enrichment for Col-0 SNPs fell in a 3 MB window on chromosome three (Fig. 7A), with the highest peaks between positions 8.25 and 8.8 MB. Within the 3 MB interval, 11 G-A or C-T mutations were identified, which are consistent with EMS mutagenesis (Sup. Table 1). From these 11, 2 fell into intergenic regions, one occurred in a miRNA, and the remaining 8 were in protein-coding genes.
Figure 7.
Identifcation of an nrpd/e2 mutant allele by a whole-genome sequencing approach. (A) Enrichment of Col-0 specific single nucleotide polymorphisms in pooled DNA from population of F2 Ler × m48 recombinant plants. Ratios were calculated in sliding 250 KB windows at 50 KB intervals. The greatest peak of Col-0 specific polymorphisms is observed between 8 and 11 MB on chromosome 3. (B) MEA-ISR Southern blot. m48 has a weaker phenotype than the nrpd/e2-2 null allele. However, a genetic complementation test confirms the mutation causes the observed phenotype. (C) Protein model for NRPD/E2. Numbered domains in yellow refer to conserved Rbp2 regions across the protein. The asterisk denotes a missense mutation recovered from screen.
One mutation occurred in NRPD/E2, a gene previously reported to be involved in RdDM.10,34 The mutation discovered here is an Arg-to-Gln missense mutation in the conserved Rbp2_4 domain (Fig. 7C). NRPD/E2 is a paralog of NRPB2, the second largest subunit of Pol II. It is shared by both Pol IV and Pol V, and null mutations cause substantial loss in 24nt siRNA accumulation and non-CG methylation.20,38 The allele identified from the screen does not have as severe a phenotype as a null allele at the MEA-ISR locus (Fig. 7B). In all likelihood, this is due to the missense mutation impairing the protein function without rendering it completely inactive. A genetic complementation test confirmed that the mutation in the nrpd/e2 gene does cause the MEA-ISR methylation phenotype in the m48 line (Fig. 7B). Its FWA flowering-time phenotype provides the first evidence that NRPD/E2 is required for the initial establishment of DNA methylation.
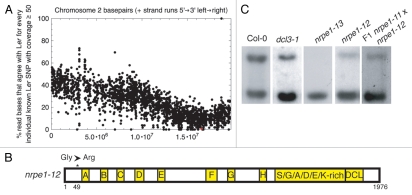
The mutant m61 also exhibited a reduced phenotype at the MEA-ISR locus (Fig. 8C), and we utilized whole-genome sequencing to both map and identify the gene affected in this mutant, this time with no pre-selection by traditional mapping. From a mapping population of 200 plants, we identified 42 that displayed an apparent weak MEA-ISR methylation phenotype by Southern blot. Pooled DNA from the 42 apparent recombinants was sequenced in the same manner as described above, giving 33.5 times coverage of the genome. Depletion of Ler SNPs was observed only on chromosome two (Fig. 8A), identifying an 8 MB region as the likely location of this mutation. Mutations were analyzed from this 8 MB region (Sup. Table 2), and the most likely mutation to cause the phenotype was a Glycine-to-Arginine missense occurring at amino acid position 49 in NRPE1 (nrpe1-12) (Fig. 8B). Genetic complementation confirmed that indeed this mutation causes the observed phenotype (Fig. 8C). The mutation was caused by a G-to-A base change consistent with EMS mutagenesis. Interestingly, out of 42 reads for this nucleotide, 38 were mutant and four were wild type, showing that roughly 10% of the pooled DNA was contaminated by wild-type sequences due to mis-scoring of the weak mutant phenotype. Importantly, the whole genome sequencing approach was able to compensate for the mis-scoring of this mutant phenotype and allow us to efficiently identify the affected gene. Identification of this weak mutation would have been nearly impossible by a traditional mapping strategy. Such results indicate the promise of using a whole-genome approach to identify even weak mutants with recombinant DNA from a relatively limited number of individual samples.
Figure 8.
Identification of nrpe1 mutant allele by a whole-genome sequencing approach. (A) Depletion in percentage of Ler specific single nucleotide polymorphisms across chromosome 2. Note that percentage of Ler SNPs never reaches zero, suggesting that pooled DNA contains wild-type (non-mutant) contaminants. The red dot indicates the physical location of NRPE1. (B) Protein model of NRPE1. Asterisk denotes missense mutation recovered from screen. (C) MEA-ISR Southern blot. The mutation discovered by the whole-genome approach has a weaker phenotype than the null allele of nrpe1 recovered from the screen.
Materials and Methods
Plant materials.
We used the following Arabidopsis strains: the wild-type WS, Ler and Col-0; the recessive nrpe5a-1 (FLAG_607D12) allele in the WS background; the recessive ago4-1 allele in Ler background (See Zilberman et al.46); recessive alleles clsy1-7 (SALK_018319), dcl3-1 (SALK_005512), drd1-6 (See Kanno et al.35), drm2-2 (SALK_150863), ktf1-1 (SALK_001294), nrpd/e2-2 (SALK_046208), nrpe1-11 (SALK_029919) and rdr2-2 (SALK_059661) in the Col-0 background.
EMS mutagenesis, FWA transformation and flowering time analysis.
10,000 Col-0 seeds were incubated in 0.3% EMS in a volume of 15 mL for 12 hours. Roughly 900 M2 families were then screened for flowering-time abnormalities. Any family containing late flowering mutants were discarded. The remaining lines were transformed with the FWA transgene. We performed FWA transformation using an AGL0 Agrobacterium tumefaciens strain carrying a pCAMBIA3300 vector with an engineered version of FWA in which an EcoRI site was converted into a BglII site. For selection, we sprayed the resultant T1 population with a 1:1,000 dilution of Finale™. We measured flowering time of resistant plants as the total number of leaves (rosette and cauline leaves) developed by a plant.
Southern blotting.
We performed Southern blotting at the MEA-ISR and Ta3 loci as described in Johnson et al.41 and Cao et al.4 respectively.
Bisulfite analysis.
We performed sodium bisulfite sequencing using EZ DNA Methylation Gold (Zymo Research) by following the manufacturer's instructions. Following amplication of bisulfite treated DNA, we cloned the resulting PCR fragments into pCR2.1-TOPO (Invitrogen) and analyzed 15 to 22 clones per sample. All primers are listed in Supplemental Table 3.
Whole genome mutation identification.
Recombinant mutants were pooled as described in the text. nrpd/e2-19 was identified by methods described by Cuperus et al.37 nrpe1-12 was identified as follows:
A list of 258,838 high-quality known homozygous Ler SNPs relative to Col-0 was composed by filtering:
ftp://ftp.arabidopsis.org/Polymorphisms/Ecker_ler.homozygous_snp.txt (downloaded 2009-06-19).
All 163,907,331 raw reads from 7 lanes from one GA II RTA SCS 2.4 55-cycle single end flow cell run were aligned with bowtie 0.10.1 (using qualities, allowing ≤2 mismatches in seed of first 28 cycles, and reporting all alignments of best stratum; additional parameters: maqerr = 60 without rounding, maxbts = 10,00,000, tryhard) to the TIGR5 Col-0 reference genome with mitochondrion and chloroplast; the 88,181,158 reads with exactly one alignment were retained.
Only the 4,842,337,521 read bases discretebasecalled A/C/G/T with quality score ≥20 aligning to a genomic A/C/G/T were retained, giving ∼42.1-fold average pooled-strand coverage of those genomic bases to which filtered alignments are possible.
All but 32 of the 258,838 known filtered Ler SNPs were aligned to at least one read base. In the 8,705,359 base-pair linkage region of interest (all chr2 bases from 11,000,001 onward), ∼99.2% of genomic base-pairs were aligned to at least one read base. Candidate mutant SNPs were taken to be genomic base-pairs with at least one and ≥ 49% non-Col-0 observations, ordered for priority by descending number of non-Col-0 observations. Of the top 40, one in NRPE1 (#13 on the list) was the most likely candidate, and was validated experimentally.
Conclusion
We present the first forward genetic screen that specifically searches for mutations that block the establishment of methylation of an incoming transgene. The mutations that have been characterized to date help to depict the components required for both de novo DNA methylation and RdDM maintenance at endogenous loci (Fig. 9). In addition to this work, other recent studies have reported genes that are required for RdDM, including INVOLVED IN DE NOVO 1/DEFECTIVE IN MERISTEM SILENCING 3 (IDN1/DMS3),39,40 INVOLVED IN DE NOVO 2 (IDN2),39 SU(VAR)3–9 HOMOLOG 2 (SUVH2), SU(VAR)3–9 HOMOLOG 9 (SUVH9),41 HISTONE DEACETYLACE 6 (HDA6),42,43 and DEFECTIVE IN MERISTEM SILENCING 4/RNA DIRECTED DNA METHYLATION 4 (DMS4/RDM4) (Fig. 9).44,45 Thus far, all RdDM mutations that have been tested, including all of those in the current study as well as mutations in IDN2, DMS3, SUVH2 and SUVH9 affect FWA de novo DNA methylation. These results strongly support the view that components of the RdDM DNA methylation maintenance pathway are also components of the pathway that establishes de novo DNA methylation.
Figure 9.
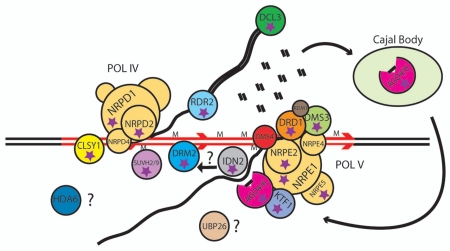
Model for de novo methylation. A repetitive invasive genetic element is marked in red. Pol IV together with CLSY1 generates a single stranded RNA, which is made double stranded by RDR2, and diced into 24 nt siRNAs which are primarily loaded into AGO4. DMS3 and DRD1 act upstream of Pol V, which creates a “scaffold” transcript that recruits AGO4 to chromatin. IDN2 binds to double stranded RNA, and may stabilize the siRNA-Pol V RNA hybrid. Possibly through an intermediate protein or chromatin modification, DRM2 is recruited to de novo methylate the invasive DNA. Proteins with stars have been shown to be required for FWA de novo DNA methylation.
Acknowledgements
We thank T. Lagrange for kindly sending homozygous nrpe5a-1 mutant seeds. We thank G. Shah, J. Chien and L. Evans for assistance with the forward screen. We thank members of the Jacobsen laboratory for supportive discussions. Jacobsen lab research was supported by US National Institutes of Health grant GM60398. M.V.C.G. was supported by USPHS National Research Service Award GM07104. I.A. was supported by a postdoctoral fellowship from the Ministerio de Educacion y Ciencia. S.F. is a Howard Hughes Medical Institute Fellow of the Life Sciences Research Foundation. J.A.L. was supported the National Institutes of Health National Research Service Award 5F32GM820453. M.P. was supported by NSF Plant Genome Research Program Grant 0701745. J.C.C. was supported by grants from the National Science Foundation (MCB-0618433 and MCB-0956526). S.E.J. is an investigator of the Howard Hughes Medical Institute.
Abbreviations
- CLSY1
CLASSY 1
- CMT3
CHROMOMETHYLASE 3
- Col-0
Columbia-0
- DCL3
DICER-LIKE 3
- DMS3
DEFECTIVE IN MERISTEM SILENCING 3
- DMS4
DEFECTIVE IN MERISTEM SILENCING 4
- DRD1
DEFECTIVE IN RNA-DIRECTED DNA METHYLATION 1
- DRM2
DOMAINS-REARRANGED METHYLATRANSFERASE 2
- EMS
ethyl methanesulfonate
- FWA
FLOWERING WAGENINGEN
- IDN1
INVOLVED IN DE NOVO 1
- IDN2
INVOLVED IN DE NOVO 2
- KTF1
KOW DOMAIN CONTAINING TRANSCRIPTION FACTOR 1
- Ler
Landsberg erecta
- MEA-ISR
MEDEA-INTERGENIC SUBTELOMERIC REPEATS
- MET1
METHYLTRANSFERASE 1
- Pol IV
RNA polymerase IV
- Pol V
RNA polymerase V
- RdDM
RNA-directed DNA methylation
- RDM4
RNA DIRECTED DNA METHYLATION 4
- RDR2
RNA-DEPENDENT RNA POLYMERASE 2
- SPT5L
suppressor of Ty insertion 5-LIKE
- SUVH2
SU(VAR)3-9 HOMOLOG 2
- SUVH9
SU(VAR)3-9 HOMOLOG 9
- WS
Wassilewskija
Supplementary Material
References
- 1.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 2.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 3.Saze H, Mittelsten Scheid O, Paszkowski J. Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat Genet. 2003;34:65–69. doi: 10.1038/ng1138. [DOI] [PubMed] [Google Scholar]
- 4.Cao X, Jacobsen SE. Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc Natl Acad Sci USA. 2002;99:16491–16498. doi: 10.1073/pnas.162371599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, et al. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science. 2001;292:2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- 6.Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 7.Aufsatz W, Mette MF, van der Winden J, Matzke AJ, Matzke M. RNA-directed DNA methylation in Arabidopsis. Proc Natl Acad Sci USA. 2002;99:16499–16506. doi: 10.1073/pnas.162371499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao X, Aufsatz W, Zilberman D, Mette MF, Huang MS, Matzke M, et al. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol. 2003;13:2212–2217. doi: 10.1016/j.cub.2003.11.052. [DOI] [PubMed] [Google Scholar]
- 9.Zilberman D, Cao X, Johansen LK, Xie Z, Carrington JC, Jacobsen SE. Role of Arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr Biol. 2004;14:1214–1220. doi: 10.1016/j.cub.2004.06.055. [DOI] [PubMed] [Google Scholar]
- 10.Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- 11.Chan SW, Zilberman D, Xie Z, Johansen LK, Carrington JC, Jacobsen SE. RNA silencing genes control de novo DNA methylation. Science. 2004;303:1336. doi: 10.1126/science.1095989. [DOI] [PubMed] [Google Scholar]
- 12.Cao X, Jacobsen SE. Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol. 2002;12:1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- 13.Soppe WJ, Jacobsen SE, Alonso-Blanco C, Jackson JP, Kakutani T, Koornneef M, Peeters AJ. The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol Cell. 2000;6:791–802. doi: 10.1016/s1097-2765(05)00090-0. [DOI] [PubMed] [Google Scholar]
- 14.Kinoshita T, Miura A, Choi Y, Kinoshita Y, Cao X, Jacobsen SE, et al. One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science. 2004;303:521–523. doi: 10.1126/science.1089835. [DOI] [PubMed] [Google Scholar]
- 15.Chan SW, Zhang X, Bernatavichute YV, Jacobsen SE. Two-step recruitment of RNA-directed DNA methylation to tandem repeats. PLoS Biol. 2006;4:363. doi: 10.1371/journal.pbio.0040363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bies-Etheve N, Pontier D, Lahmy S, Picart C, Vega D, Cooke R, et al. RNA-directed DNA methylation requires an AGO4-interacting member of the SPT5 elongation factor family. EMBO Rep. 2009;10:649–654. doi: 10.1038/embor.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He XJ, Hsu YF, Zhu S, Wierzbicki AT, Pontes O, Pikaard CS, et al. An effector of RNA-directed DNA methylation in arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell. 2009;137:498–508. doi: 10.1016/j.cell.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang L, Jones AM, Searle I, Patel K, Vogler H, Hubner NC, et al. An atypical RNA polymerase involved in RNA silencing shares small subunits with RNA polymerase II. Nat Struct Mol Biol. 2009;16:91–93. doi: 10.1038/nsmb.1539. [DOI] [PubMed] [Google Scholar]
- 19.Smith LM, Pontes O, Searle I, Yelina N, Yousafzai FK, Herr AJ, et al. An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell. 2007;19:1507–1521. doi: 10.1105/tpc.107.051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, Henderson IR, Lu C, Green PJ, Jacobsen SE. Role of RNA polymerase IV in plant small RNA metabolism. Proc Natl Acad Sci USA. 2007;104:4536–4541. doi: 10.1073/pnas.0611456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henderson IR, Zhang X, Lu C, Johnson L, Meyers BC, Green PJ, et al. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet. 2006;38:721–725. doi: 10.1038/ng1804. [DOI] [PubMed] [Google Scholar]
- 23.Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- 24.Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol. 2009;21:367–376. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 25.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Gen. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qi Y, He X, Wang XJ, Kohany O, Jurka J, Hannon GJ. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature. 2006;443:1008–1012. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
- 27.Zheng X, Zhu J, Kapoor A, Zhu JK. Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J. 2007;26:1691–1701. doi: 10.1038/sj.emboj.7601603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SW, et al. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell. 2006;126:93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 29.Wierzbicki AT, Ream TS, Haag JR, Pikaard CS. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet. 2009;41:630–634. doi: 10.1038/ng.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.El-Shami M, Pontier D, Lahmy S, Braun L, Picart C, Vega D, et al. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 2007;21:2539–2544. doi: 10.1101/gad.451207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lahmy S, Pontier D, Cavel E, Vega D, El-Shami M, Kanno T, et al. PolV(PolIVb) function in RNA-directed DNA methylation requires the conserved active site and an additional plant-specific subunit. Proc Natl Acad Sci USA. 2009;106:941–946. doi: 10.1073/pnas.0810310106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ream TS, Haag JR, Wierzbicki AT, Nicora CD, Norbeck AD, Zhu JK, et al. Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol Cell. 2009;33:192–203. doi: 10.1016/j.molcel.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, et al. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet. 2005;37:761–765. doi: 10.1038/ng1580. [DOI] [PubMed] [Google Scholar]
- 35.Kanno T, Mette MF, Kreil DP, Aufsatz W, Matzke M, Matzke AJ. Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr Biol. 2004;14:801–805. doi: 10.1016/j.cub.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 36.Till S, Lejeune E, Thermann R, Bortfeld M, Hothorn M, Enderle D, et al. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat Struct Mol Biol. 2007;14:897–903. doi: 10.1038/nsmb1302. [DOI] [PubMed] [Google Scholar]
- 37.Cuperus JT, Montgomery TA, Fahlgren N, Burke RT, Townsend T, Sullivan CM, et al. Identification of MIR390a precursor processing-defective mutants in Arabidopsis by direct genome sequencing. Proc Natl Acad Sci USA. 2010;107:466–471. doi: 10.1073/pnas.0913203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Onodera Y, Haag JR, Ream T, Nunes PC, Pontes O, Pikaard CS. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 39.Ausin I, Mockler TC, Chory J, Jacobsen SE. IDN1 and IDN2 are required for de novo DNA methylation in Arabidopsis thaliana. Nat Struct Mol Biol. 2009;16:1325–1327. doi: 10.1038/nsmb.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanno T, Bucher E, Daxinger L, Huettel B, Bohmdorfer G, Gregor W, et al. A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nat Genet. 2008;40:670–675. doi: 10.1038/ng.119. [DOI] [PubMed] [Google Scholar]
- 41.Johnson LM, Law JA, Khattar A, Henderson IR, Jacobsen SE. SRA-domain proteins required for DRM2-mediated de novo DNA methylation. PLoS Genet. 2008;4:1000280. doi: 10.1371/journal.pgen.1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He XJ, Hsu YF, Pontes O, Zhu J, Lu J, Bressan RA, et al. NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation. Genes Dev. 2009;23:318–330. doi: 10.1101/gad.1765209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aufsatz W, Mette MF, van der Winden J, Matzke M, Matzke AJ. HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J. 2002;21:6832–6841. doi: 10.1093/emboj/cdf663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He XJ, Hsu YF, Zhu S, Liu HL, Pontes O, Zhu J, et al. A conserved transcriptional regulator is required for RNA-directed DNA methylation and plant development. Genes Dev. 2009;23:2717–2722. doi: 10.1101/gad.1851809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kanno T, Bucher E, Daxinger L, Huettel B, Kreil DP, Breinig F, et al. RNA-directed DNA methylation and plant development require an IWR1-type transcription factor. EMBO Rep. 11:65–71. doi: 10.1038/embor.2009.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.