Abstract
Glycolysis and glyconeogenesis play crucial roles in the ATP supply and synthesis of glycoconjugates, important for the viability and virulence, respectively, of the human-pathogenic stages of Trypanosoma brucei, Trypanosoma cruzi, and Leishmania spp. These pathways are, therefore, candidate targets for antiparasite drugs. The glycolytic/gluconeogenic enzyme enolase is generally highly conserved, with similar overall fold and identical catalytic residues in all organisms. Nonetheless, potentially important differences exist between the trypanosomatid and host enzymes, with three unique, reactive residues close to the active site of the former that might be exploited for the development of new drugs. In addition, enolase is found both in the secretome and in association with the surface of Leishmania spp. where it probably functions as plasminogen receptor, playing a role in the parasite's invasiveness and virulence, a function possibly also present in the other trypanosomatids. This location and possible function of enolase offer additional perspectives for both drug discovery and vaccination.
1. Introduction
All known organisms belonging to the Trypanosomatidae, a family of protists within the order Kinetoplastida, are parasites of vertebrates, invertebrates, or plants. Species of two trypanosomatid genera, Trypanosoma and Leishmania, are responsible for several serious diseases of humans and domestic animals in the tropical and subtropical regions of the world [1].
Trypanosoma brucei is the causative agent of human sleeping sickness, a disease that threatens millions of mainly impoverished people in 36 countries of the African continent [2]. This parasite is transmitted by several species of the genus Glossina, insects commonly known as tsetse flies, which are endemic in the affected sub-Saharan countries. The two human infective subspecies of T. brucei, T. b. rhodesiense, and T. b. gambiense, live extracellularly, multiplying as so-called long-slender trypanosomes in the bloodstream [3]. By a population-density-dependent mechanism, the slender trypanosomes may transform into nonproliferative stumpy bloodstream forms. Trypanosomes are passed on to the insect vector when a tsetse fly takes blood from an infected host. Once in the tsetse fly gut, the stumpy parasites differentiate into so-called procyclic-form trypanosomes, which subsequently undergo several successive differentiations while migrating from the gut via the proboscis to the salivary glands. From there, the parasites, then called metacyclic forms, are transmitted to a new host [1, 4]. Without treatment, human sleeping sickness is considered fatal. Vaccines or adequate drugs for effective treatment of this disease are currently not available [2].
Trypanosoma cruzi is responsible for Chagas' disease in South and Central America, a form of trypanosomiasis that threatens 15 million people on the American continent. This parasite is mainly transmitted to humans via the feces of infected triatomine bugs; however, transfusions with infected blood, organ transplantations, and use of contaminated food are also important ways of transmission [5]. T. cruzi is an intracellular parasite of humans and many other vertebrates; the parasite is dedicated to invade cardiac and digestive tissues, where infective trypomastigotes differentiate into replicative amastigote forms. The amastigotes differentiate again into trypomastigotes in the cytosol before they burst from the infected host cell, thus liberating parasites that perform a new round of cell infection. Blood trypomastigotes, taken up again by triatomine bugs when they feed on an infected host, differentiate then into replicative epimastigotes in the insect's intestine. A next differentiation occurs in the intestine, producing vertebrate infective, metacyclic trypomastigotes. The parasites are transmitted again to the vertebrate host when the infected bug takes a blood meal, depositing the contaminated feces on the skin of the new host [1]. People infected with T. cruzi can only be effectively treated when such treatment is applied shortly after the infection has occurred.
Several species of the genus Leishmania are the etiological agents of leishmaniasis, a disease characterized by clinical manifestations with a wide range of symptoms. The disease is distributed in 88 countries of tropical and subtropical areas around the world. It is estimated that currently 350 million people are at risk. The parasite is transmitted by the bite of a female phlebotomine sand fly, which feeds from vertebrate blood [1].
Leishmaniasis can occur as three different forms: cutaneous leishmaniasis is the most common and mild form of the disease characterized by the presence of ulcers that appear at the places of parasite inoculation; mucocutaneous leishmaniasis is manifested by destruction of the mucosa membranes of mouth, nose, and throat after parasite spreading; visceral leishmaniasis is the most dangerous form of the disease in which organs such as liver and spleen are compromised. Leishmania species multiply mainly inside host macrophages as amastigote forms; this form of the parasite is present in the phagolysosome where the parasite divides until they burst from the host cells. Subsequently, the released amastigotes invade new cells, in that way spreading the infection. The parasite is taken up by the sand fly when a female phlebotomine feeds on an infected host. The amastigotes then differentiate into proliferative metacyclic promastigote forms in the sand fly gut before migrating to the salivary glands in order to be transmitted to a new host [1]. Cutaneous and mucocutaneous leishmaniases are disabling diseases that affect the quality of life of infected people; however, visceral leishmaniasis is a fatal disease when no treatment is applied.
Although several drugs are available to be used for sleeping sickness, Chagas' disease and leishmaniasis, they do not offer adequate treatment. None has been rationally designed to specifically treat these diseases. As a consequence, toxic side effects, sometimes very serious, are associated with the use of such drugs, and increasing resistance has been reported against some of them. Additionally, the application of some of the treatments requires highly qualified staff and medical equipment, which are usually not available in the regions where most of the people affected by these diseases live [1]. Consequently, there exists an urgent need to develop new drugs that will inhibit the viability of the parasites while causing minimal side effects to the infected people and which can be easily administered and will be affordable for these people usually living in resource-poor areas of the world. In that context, efforts have been made in recent decades to identify, in the parasites, crucial metabolic pathways or cellular structures with features sufficiently different from those in the host in order to exploit them as chemotherapeutic targets.
Several features distinguish members of the Trypanosomatidae family from other eukaryotic cells. A unique feature of all kinetoplastids is that the majority of glycolytic and gluconeogenic enzymes are compartmentalized in unique peroxisome-like organelles called glycosomes [6, 7]. Several lines of evidence have shown that this compartmentalization is absolutely essential for survival of the organisms. Metabolic studies on T. brucei, T. cruzi, and Leishmania spp. have shown that in the human-pathogenic stages of each of these parasites glycolysis and/or gluconeogenesis are crucial—or at least major processes in the provision of free energy and carbon—and consequently essential for parasite viability. Moreover, the peculiar compartmentalization of these pathways in kinetoplastids has endowed the enzymes with specific structural and functional properties that offer potential for development of trypanocidal drugs [8].
Trypanosomatid parasites are also equipped with several potent mechanisms that allow them to evade the immune response by the host, as well as to migrate through several tissue barriers to reach their final destination. T. brucei is present in the bloodstream of the mammalian host during the initial phase of the infection. A sophisticated mechanism of antigenic variation at the level of their surface coat allows the parasites to escape continuously from the host's immune defense system [4]. At a second stage of the disease, the parasites reach the central nervous system by crossing the blood-brain barrier via a multistep mechanism of which the details remain to be established [9]. In contrast, T. cruzi lives only briefly in the bloodstream of the vertebrate host before invading the cardiac muscle and cells of the digestive system. It is known that metacyclic trypomastigotes initially adhere to the host cell through several proteins exposed on the parasites' cell surface that function as ligands and adhering molecules. Then, the parasites control their invasion by exploiting phagocytic and nonphagocytic mechanisms that lead to their internalization into the parasitophorous vacuole. Once there, the parasites differentiate into amastigotes, which excrete potent pore-forming peptides and transsialidases/neuraminidases that mediate the vacuolar membrane's disintegration to let the parasites free in the cytosol in order to multiply [10]. Metacyclic promastigotes of Leishmania spp. are exposed during a very short time to the lytic factors present in the bloodstream. Surviving parasites are able to adhere through different mechanisms to the host cell, which belongs to the monocyte/macrophage family. The presence on the parasites' surface of lipophosphoglycans, metalloproteases, and specific ligands for macrophage receptors allows the parasites to invade the host cells, as well as migrate through the extracellular matrix [11]. Once in the phagolysosome, promastigotes differentiate into amastigotes that are better equipped with molecules that provide resistance against the stress conditions encountered in the phagolysosome, such as the presence of hydrolases and the acidic pH. The surface molecules of the amastigotes play an important role in inhibiting the mechanism that triggers the macrophage's activation as well as the proper antigen presentation through the MHCI and II complexes, which eventually would activate a host protective TH1-cell response [12].
Studies of the metabolic pathways essential for parasite survival and the mechanisms by which the trypanosomatids establish an infection in the human host are important both for future chemotherapy and vaccination. Such studies may identify molecules—unique or at least highly different from host molecules—that are interesting as candidate drug targets and the subsequent discovery of selective inhibitors of these molecules to combat the parasitic diseases, as well as molecules that could be exploited as potential antigens to induce immunoprotection against the trypanosomiases and leishmaniases.
In this paper, we will argue that enolase, a protein involved both as enzyme in glycolysis and gluconeogenesis, metabolic processes essential for T. brucei, T. cruzi, and Leishmania spp., and probably also as a virulence factor in the pathogenesis caused by Leishmania spp., T. cruzi and possibly also T. brucei, represents both an interesting drug target and vaccine candidate.
2. General Aspects of Enolase
Enolase is the enzyme responsible for the reversible conversion of D-2-phosphoglycerate (2PGA) and phosphoenolpyruvate (PEP) in glycolysis and gluconeogenesis, two metabolic pathways that are often vital for cellular function [13]. Enolase is a highly conserved protein found in Archaea, Bacteria, and eukaryotes with catalytic properties that are similar among divergent organisms. The ubiquitous presence of the enzyme and the sequence homology between enolases from extant organisms belonging to different phyla indicate that an enolase gene has already been present in the common ancestor and diversified by speciation of organisms and gene duplication within organisms [14].
Since enolase catalyzes the reversible conversion of 2PGA into PEP, the forward reaction being the ninth step of glycolysis and the reverse reaction being the first one of gluconeogenesis, an organism, tissue, or cell may need to optimize the properties of this enzyme in such a way that the flux in either one or the other direction is facilitated, dependent on whether it should adapt its metabolism to performing the catabolic or anabolic process. Indeed, in many organisms (vertebrates, Saccharomyces cerevisiae, Toxoplasma gondii) the presence of different enolase isoforms has been reported, often with kinetic properties favouring a flux in the glycolytic or gluconeogenic direction. In vertebrates, it has been shown that expression of the different enolase isoforms, formed by homodimers and heterodimers of α, β, and γ subunits, is regulated in a developmentally and/or tissue-specific manner [13, 15]. In yeast, the expression of the two enolase isoforms is dependent on the carbon source used to propagate the cells, glucose, or a nonglycolytic substrate; however, the kinetic properties of homodimeric enzymes of both isoforms are very similar [16]. In the case of T. gondii, the specific expression of enolase genes is linked to life-stage conversion. While enolase 1 is expressed during encystations where anaerobic glycolysis is promoted, enolase 2 is expressed in the actively dividing and invasive tachyzoite [17].
Some highly conserved proteins perform multiple functions that are different from their “classical”, well-known activities, and, therefore, are called “moonlighting” functions. These latter functions are often revealed when these proteins are found at cellular localizations different from where the primary function is exerted. A considerable number of glycolytic enzymes, including enolase, exhibit nonglycolytic functions in several organisms, both bacteria and eukaryotes. Enolase is located mainly in the cytosol of all eukaryotic cells where, besides its function in glycolysis and gluconeogenesis, it is also involved in regulation of cell morphology and material trafficking by interacting with the cytoskeleton system [18]. Enolase has also been detected in the nucleus of mammalian cells where it participates in transcriptional regulation of genes involved in morphological transformation and cell proliferation [19]. Bacterial enolase has been reported as a main component of the degradosome where it functions, in a still poorly understood manner, in the regulation of mRNA stability [20]. The mammalian α-enolase is also found as a main structural component of the eye lens, where it is found as an inactive monomer [13].
Additionally, a considerable number of reports described that enolase can be expressed at the cell surface of several nonpathogenic and pathogenic organisms. The enzyme has been found at the cell wall of S. cerevisiae and also at the surface of the pathogenic yeast Candida albicans where it constitutes an immunodominant antigen during invasive candidiasis [21]. In some bacterial pathogens, such as Bacillus anthracis, Streptococcus pneumoniae, and Streptococcus mutans [22–24], surface enolase has been highlighted as an important virulence factor. In Apicomplexan parasites like Plasmodium spp. [25], cell surface enolase has been suggested to participate in the tissue invasion process. Additionally, in Entamoeba invadens, enolase expression is induced by environmental signals, and it shows association with cytoplasmic vesicle-like structures that transport the protein to the cyst wall where it plays an essential but so far unknown function [26]. Human and animal parasites of the genus Schistosoma have been reported to express enolase also on the surface of the adult worms [27, 28], where it could exert an important role in the inhibition of clot formation during host infection [28].
Enolase may also be excreted to the extracellular environment where it mediates degradation of host tissues and immune evasion, such as has been observed for the human pathogenic Streptococcus pyogenes and the insect parasite Aphidius ervi [29, 30].
3. Enolase of Trypanosomatids
African trypanosomes living in their mammalian host are entirely dependent on glucose, abundantly available in the blood. Metabolic studies performed on bloodstream-form T. brucei have shown that glycolysis represents the only process through which ATP is synthesized by the parasite. Inhibition of glycolysis, therefore, leads to rapid death of these parasites. The same is probably true for the human pathogenic stages of T. cruzi. Axenically cultured amastigotes have an essentially glycolytic metabolism, fermenting glucose to succinate and acetate [32], strongly suggesting that also in these cells enolase might be a candidate drug target. Glycolysis is probably also essential in the trypomastigote form of these parasites, as it lives in a glucose-rich bloodstream where it shows a highly upregulated glucose uptake activity [33]. Interestingly, the pathogenic stage of Leishmania, the amastigotes living intracellularly in phagolysosomes of macrophages, utilize amino sugars as a source of carbon and energy, indicating that glycolysis is important [34], while even during hexose uptake gluconeogenesis has been shown to be an essential pathway for the synthesis of glycoconjugates and β-mannan, both required for the virulence of the parasite [35, 36]; therefore, enolase could also be considered as vital, and thus a drug target, in this organism. The fact that even during uptake of (amino) sugars gluconeogenesis is required has been interpreted as an indication that the sugar levels in the phagolysosome are generally too low to sustain fluxes through essential pathways such as N-glycosylation, the pentose-phosphate pathway, inositol synthesis, and the catabolism of major carbohydrate reserve material [34].
In these three parasitic trypanosomatid species, as in all kinetoplastids, the majority of the glycolytic enzymes—dependent on the life-cycle stage, the 6 or 7 enzymes from hexokinase to glyceraldehyde-3-phosphate dehydrogenase or phosphoglycerate kinase, converting glucose into 1,3-bisphosphoglycerate or 3-phosphoglycerate, respectively—are compartmentalized in peroxisome-like organelles called glycosomes [6, 37, 38]. Only the last enzymes of the pathway—phosphoglycerate mutase, enolase, and pyruvate kinase—were not found to be associated with these organelles. More detailed studies specifically devoted to enolase have confirmed that its activity is exclusively present in the cytosol of T. brucei bloodstream forms [39], T. cruzi epimastigotes (W. Quiñones, and J. L. Concepción, unpublished results), and L. mexicana promastigotes [40]. In T. cruzi, the enzyme was detected by peptide mass fingerprinting in the different developmental stages with a higher expression in trypomastigotes and amastigotes compared to epimastigotes [41]. In contrast, mass-spectrometric analysis of differentiating L. mexicana showed that the glycosomal glycolytic enzymes were 1.4-fold upregulated during axenic differentiation of promastigotes to amastigotes whereas the cytosolic glycolytic enzymes including enolase were 2-fold downregulated [42].
In bloodstream-form T. brucei, enolase has been genetically validated as a drug target by RNA interference (RNAi) [43]. Upon induction of RNAi, enolase activity gradually decreased during the first 24 h to 16%, leading to growth arrest followed by trypanosome death after two days.
The genes coding for enolases of T. brucei, T. cruzi, L. mexicana, and L. major have been identified in the genome sequence database (TriTrypDB: http://tritrypdb.org/tritrypdb/) for a variety of African trypanosomes (T. brucei, T. gambiense, T. congolense, T. vivax), the American trypanosome T. cruzi, and several Leishmania species (L. major, L. mexicana, L. infantum, L. braziliensis). In each case, a single gene copy per haploid genome has been found, except in T. gambiense where two tandemly linked genes coding for identical proteins were found, clearly the result of a recent gene duplication in this species. They all encode polypeptides of 429 amino acids (including the initiator methionine) with relative molecular masses of 46 kDa. Only the predicted T. congolense enolase polypeptide has 430 residues due to the presence of an additional Lys residue at the C-terminus. The Trypanosoma amino-acid sequences show approximately 80% identity with the Leishmania sequences. Comparison of the three isoenzymes of the parasite's human host and trypanosomatid enolases reveals between 60 and 63% identity. Residues essential for substrate binding, catalytic activity, as well as those constituting the binding sites of substrates, and two Mg2+ ions are invariably present in all sequences (see the multiple sequence alignment in Figure 1).
Figure 1.
Characteristics of the enolase sequences of trypanosomatids. Alignment of the amino-acid sequences of enolase from L. mexicana (accession number ABA64522), T. brucei (accession number XP_822542), T. cruzi (accession number XP_819700), human α-enolase (accession number NP_001419), and S. pneumoniae (accession number Q97QS2). Open circles indicate residues involved in ligand binding (PEP, metals), closed circles unique reactive active-site residues found in enolases of trypanosomatids. The bar indicates the plasminogen-binding motif in S. pneumoniae. This plasminogen-binding peptide contains both positively and negatively charged residues flanked by hydrophobic amino acids, important for plasminogen binding [31].
The protein has been more extensively studied in T. brucei and L. mexicana [39, 40, 44]. Both enzymes have been purified from the parasites and expressed as N-terminally His-tagged proteins in Escherichia coli. The kinetic properties of both the natural and bacterially expressed enzymes were determined for the forward (glycolytic) and reverse (gluconeogenic) reactions. In each case, the enzyme displayed standard Michaelis-Menten kinetics for both substrates with very similar Km and Vmax values (Table 1). Mg2+ is essential for the activity of all enolases (with Ka values varying between 0.26 and 0.45 mM; optimal concentration for activity between 1 and 2 mM), from trypanosomatids and other organisms, but inhibits the enzymes at high concentrations. The Kiapp values of the trypanosomatid and yeast enzymes for Mg2+ are similar (43–50 mM), but the mammalian enzyme is more susceptible to inhibition; the inhibition of this latter enzyme, but not that of trypanosomatids and yeast, is pH dependent and seems to involve two processes with Kiapp values of 7.5 (40% inhibition) and >100 mM. Furthermore, other divalent metals such as Co2+, Mn2+, and Cu2+ inhibit all enolases tested. Enolases are also generally inhibited by the monovalent ions Li+ and Na+; rabbit muscle enolase is activated by K+, but the trypanosomatid and yeast enzymes are not. Furthermore, the L. mexicana enzyme is inhibited by fluoride with Ki values of 1.9 mM (versus 2PGA) and 6.3 mM (versus PEP) for the natural enzyme and 13.0 mM (versus 2PGA) and 10.0 mM (versus PEP) for the bacterially expressed enzyme, in agreement with reports about other enolases. Similarly, enolase purified from T. cruzi epimastigotes is inhibited by fluoride with a Ki value of 2.6 mM (versus 2PGA; L. González-González, W. Quiñones, and J. L. Concepción, unpublished results). The L. mexicana enolase is also inhibited by inorganic pyrophosphate (PPi): Ki = 0.232 mM (versus 2PGA) and 0.238 mM (versus PEP) for the natural enzyme and Ki = 0.127 mM (versus 2PGA) and 0.260 mM (versus PEP) for the recombinant enzyme. For the natural T. cruzi enzyme, inhibition by PPi with a Ki = 0.21 mM (versus 2PGA) was found (L. González-González, W. Quiñones, and J. L. Concepción, unpublished results). The susceptibility of T. brucei enolase to PPi has not been tested. It should be noted that, previously, several other Leishmania and T. cruzi glycolytic enzymes have been shown to be inhibited by PPi [45, 46] whereas no such inhibition was found for T. brucei enzymes.
Table 1.
Kinetic characteristics of the natural and recombinant enolases of T. brucei, T. cruzi, and L. mexicana.
| Source of enzyme | Km | Vmax | ||
|---|---|---|---|---|
| (μM) | (μmol·min−1·mg−1) | |||
| 2PGA | PEP | 2PGA | PEP | |
| T. brucei (natural) | 54 | 244 | ND | ND |
| T. brucei (recombinant) | 49 | 289 | 63 | 6.3 |
| T. cruzi (natural) | 55 | 147 | 97 | 42 |
| T. cruzi (recombinant) | 50 | 410 | 0.87 | 12.35 |
| L. mexicana (natural) | 80 | 216 | 16.0 | 3.38 |
| L. mexicana (recombinant) | 51 | 200 | 80.0 | 3.64 |
| Rabbit muscle | 16 | 238 | 31 | 6.4 |
| Yeast | 57 | 264 | 65 | 7.8 |
Data of T. brucei, rabbit muscle, and yeast enolases were taken from Hannaert et al. [39], those of L. mexicana enolases from Quiñones et al. [40], and the T. cruzi data have not yet been reported before (L. González-González, W. Quiñones, and J. L. Concepción, unpublished results). Abbreviations: 2PGA: D-2-phosphoglycerate; PEP: phosphoenolpyruvate; ND: not determined.
4. Enolase Three-Dimensional Structure: Unique Features of the Trypanosomatid Enzyme
Crystal structures of enolase—the enzyme in its apo-form and with bound ligands—are available for a variety of organisms: S. cerevisiae, lobster, and E. coli. The structural analysis of these different enolases has shown that the enzyme usually folds as a homodimer, although, for enolase of some bacteria, an octameric quaternary structure has been reported. The overall structure and catalytic site are highly conserved, explaining the very similar kinetic properties between the enzymes from highly different organisms, as described in the previous section. Each subunit of the dimer is composed of two domains, a small N-terminal domain containing three α helices and four β sheets and a C-terminal domain which folds as an atypical eight-fold α/β barrel at the end of which the active site is found. Although the catalytic site lies at the dimer interface, each site involves residues of only a single subunit, explaining that the monomeric form is catalytically active, despite earlier claims of the contrary [47].
The early studies, particularly with the yeast enzyme, had shown that the enzyme is naturally activated by two divalent metal ions that bind to each monomer in two distinct metal-binding sites (I and II) near the active site. These studies also revealed the importance of sequential binding of a first metal ion, the substrate and a second metal ion, and associated local conformational changes, notably movements of three loops near the active site, for catalysis to occur. The loops define “open,” “closed,” and “incompletely closed” active-site structures [48]. Structures of the apo-enzyme and the enzyme in complex with its substrate or product indicate that, in the “open” conformation, the active site is totally exposed to solvent. When the substrate or product and the divalent ions are bound, an important conformational change occurs, bringing the loops one (comprising residues 37–44), two (150–166), and three (248–272) to the catalytic active site producing the “closed” enzyme conformation [48]. The catalytic mechanism of the reaction implies an acid-base chemistry involving a Lys-Glu dyad.
It had also already initially been found that enolase activity is inhibited by high metal concentrations, which suggested the presence of a third inhibitory metal binding site. Notwithstanding the agreement about the kinetic data, no structural data were available to support this hypothesis.
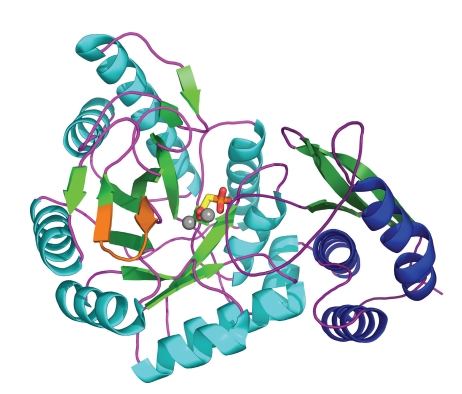
More recently, seven structural determinations have been performed for the T. brucei enzyme in ligation states with substrates, inhibitors, sulphate, phosphate, and metal ions– and in various conformations [49, 50]. A subunit structure of T. brucei enolase is shown in Figure 2. All these structures together have provided a profound insight into the structure-function relationship and the catalytic mechanism of the enzyme, and they solved some questions raised by the earlier structure determinations.
Figure 2.
Cartoon view of T. brucei enolase in complex with the enolase inhibitor phosphonoacetohydroxamate (PAH; PDB code 2ptz [50]). Secondary structure is coloured blue (α-helices), green (β-strands) or purple (loops) with the N-terminal domain to the right of the figure drawn in darker hues. Ligand is shown as sticks and bound metals as isolated spheres. The putative plasminogen-binding site is shown in orange.
First, the different structures of the T. brucei enzyme and complementary molecular dynamics simulations revealed a strikingly larger conformational variability for the catalytic loops than initially thought. Secondly, in the studies of the parasite's enzyme, also an unexpected diversity of metal binding was found, and two additional metal-binding sites were detected near the active site. The occupancy of one of these sites, site no. III, explained the inhibitory effect of Mg2+ at high concentrations; the contribution of site IV to this inhibition remains to be determined.
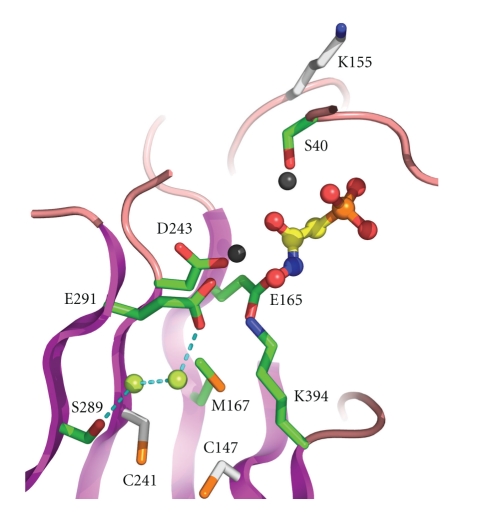
Importantly, all available structures together, from trypanosomes and other organisms, and a sequence comparison of trypanosomatid and human enolases have revealed that, despite the high similarity (approximately 60% positional amino-acid identity), similar overall fold and active-site architecture and identical catalytic residues, potentially important differences exist between parasite and host enzymes that might be exploited for drug discovery. First, each of the trypanosomatid enolases contains three unique residues, absent from the enolases of mammals and almost all other organisms studied (Figure 1), which might be targets for irreversible reaction with active-site inhibitors [39]. A pair of cysteine residues, numbered 147 and 241 (see Figure 3), is present in a water-filled cavity near the active site. Despite their proximity, no disulphide bond between these residues was observed in the structure, nor is expected to be formed in vivo because of the low redox potential in the cytosol. One of these residues, and probably both, could be chemically modified by iodoacetamide with concomitant loss of enzyme activity indicating their accessibility to potentially reactive ligands [49]. Substrate analogues extended with reactive groups reaching either one of these Cys residues underneath the catalytic site may thus result in trypanosomatid-enzyme specific, irreversible inhibitors. In addition, a unique Lys, at position 155, was located on one of the active-site loops (Figure 3) and could also potentially interact with ligands in the active site. Although the side chain of this Lys residue points away from the catalytic site in original T. brucei enolase crystal structures, later structures, backed up by molecular dynamics simulations, show the Lys to approach the catalytic site more closely; it may, therefore, be available for irreversible binding of active-site inhibitors. Despite the closed catalytic site being small, the studies showed that occupation of the site by ligands does not need to lead to full closure, leaving open a tunnel near which Lys 155 is closely located. This offers thus a possibility for the design of enlarged ligands that interact with this potentially modifiable residue and inhibit the enzyme irreversibly.
Figure 3.
Close-up view of the catalytic site and neighbourhood of T. brucei enolase in complex with the inhibitor phosphonoacetohydroxomate (PAH) (PDB code 2ptz) [50]. Ligand is shown in ball and stick representation and bound metal as grey spheres. Bound water molecules near the Cys residues are shown as lime green spheres. Selected secondary structure is shown as magenta (β-strands) or pink (loops). Selected residues are shown as sticks and labeled with those discussed in the text as potentially relevant to trypanosomatid-specific drug design coloured white.
5. Plasma-Membrane Bound and Secreted Enolase in Leishmania Parasites
A study of enolase localization in L. mexicana by differential centrifugation detected this protein, as expected from its function in glycolysis/gluconeogenesis, mainly in the cytosol. However, some enolase was also present in the microsomal fraction that contains predominantly the plasma membrane [40]. Similar results were obtained upon selective permeabilization of cells with digitonin; Western blot analysis showed that enolase was present in both the supernatant and pellet of each cell suspension, independent of the digitonin concentration added, indicating that the detergent was unable to fully release the enzyme from the permeabilized cells. These results suggested that enolase might be associated with membrane constituents of the cell. The presence of enolase at the plasma membrane was further assessed by immunofluorescence experiments using nonpermeabilized parasites [40]. These experiments not only confirmed the plasma-membrane localization of enolase but also showed that it is associated to the external surface of the parasite. In agreement with these results is the recent visualization of enolase in the proteome of the plasma membrane from Leishmania chagasi, prepared by two different approaches: live cell biotinylation followed by streptavidin affinity purification and octyl glucoside extraction [51]. Prior to these studies, subcellular fractionation of L. infantum with digitonin followed by proteomic analysis of the different fractions had predicted that enolase could have an additional localization, apart from the cytosol, also in this species [52].
Measurement of the enolase activity in the fractions obtained by differential centrifugation of an L. mexicana lysate showed that it was all present in the cytosolic fraction; no activity whatsoever was found in the microsomal fraction [40], indicating that the enolase on the parasite surface is inactive. This was corroborated by the digitonin solubilization experiment: all enolase activity was released with the cytosolic marker and no activity was detected in the pellets [40]. Another indication for the lack of activity of this surface enolase was recently provided by the report that its localization in L. donovani and L. major is regulated by oligopeptidase B (OPB), a serine-protease that is highly expressed in the amastigote form [53]. Although loss of this peptidase in OPB−/− parasites resulted in the accumulation of enolase at the cell surface of the parasites, no increased enolase activity was detected in total cell lysates. The authors hypothesized that oligopeptidase B causes clearance of surface enolase, once in the macrophage, to avoid macrophage activation [53]. This hypothesis is consistent with enolase having multiple functions.
Enolase is also present at the cell surface of several mammalian cells [54–56]. In addition, enolase can be found at the cell surface of numerous prokaryotic and eukaryotic pathogenic organisms including, among others, group A streptococci [57], S. pneumoniae [58], L. mexicana [40, 53, 59], C. albicans [60], Schistosoma bovis [61], and Plasmodium falciparum [25]. In the majority of these cases, enolase could act as a plasminogen-binding protein. Plasminogen is the zymogenic form of the serine-protease plasmin that is part of the fibrinolytic system. This molecule possesses five kringle domains that provide lysine-binding sites responsible for its binding to protein substrates and cell surfaces [62]. Surface-located enolases of mammalian cells and some pathogens bind plasminogen through a C-terminal lysine that is recognized by the lysine-binding sites of the plasminogen molecule [63]. However, in the amino-acid sequence of enolase from S. pneumoniae, an internal motif (FYDKERKVYD) was identified as being responsible for the plasminogen recognition [23]. Similar internal motifs were identified in the enolases of L. mexicana (AYDAERKMY) and Aeromonas hydrophila (FYDAEKKEY) [59, 64]. None of these two latter enolases possesses a C-terminal lysine residue. Enolases from other trypanosomatids (Figures 1 and 2) as well as from other pathogenic organisms have similar internal motifs at the same position that could contribute to plasminogen binding [65]. A similar motif has been shown to be responsible for plasminogen binding by another L. protein, the parasite's homologue of receptors for activated C-kinase [66].
In addition to its localization on the membrane surface, enolase is secreted by several pathogenic microorganisms, such as the bacteria S. pyogenes [30], S. mutans [24], and Lactobacillus crispatus [67]. In Leishmania parasites, enolase has been found as a part of the secretome/exosome of L. donovani [68, 69], L. major [69], and L. braziliensis [70]. Enolase has also been found in the secretome of T. brucei [71, 72] and as a protein secreted by other parasites: Eimeria tenella [73], Fasciola hepatica [74], Echinostoma caproni [75], Giardia lamblia [76], and Schistosoma japonicum [77]. Different exosome proteomes from mammalian cell types indicate that enolase is commonly present in this structure [78].
Enolase does not contain a detectable secretion signal or membrane anchor region that can explain its membrane localization. Studies with S. pneumonia showed that soluble enolase can bind to the pneumococcal surface, suggesting that one possibility for the surface localization of enolase in this bacterium is the reassociation of secreted enolase with the plasma membrane [58]. One may thus speculate that, in the case of Leishmania parasites, enolase, after its secretion via exosomes, might bind to the cell surface. Further studies are necessary to elucidate the mechanism of translocation of enolase to the cell surface in these parasites.
6. The Role of Plasminogen/Plasmin and Enolase in the Virulence of Leishmania Parasites
The binding of plasminogen to the cell surface has been demonstrated for several mammalian cells [54, 63] including metastatic tumor cells. Equally, over the last 15 years, a steadily increasing number of reports have been published about plasminogen binding by both prokaryotic and eukaryotic pathogens, and even viruses [79]. This interaction has been implicated as part of their mechanisms of invasion and establishment in the host. However, in contrast to the large number of reports about plasminogen binding by mammalian cells and unicellular organisms, only few analyses have been reported thus far that provide evidence for the importance and physiological role of this interaction. Evidence for the physiological role and importance in infection and pathogenesis of the plasminogen-pathogen interaction has been provided by studies of animal models infected with group A streptococci [80], Yersinia pestis [81], Borrelia spp. [82–84], and S. pneumoniae [23]. The binding of plasminogen through their plasminogen-binding proteins and its activation made the pathogens acquire a potent protease for their own purpose by kidnapping the fibrinolytic system of the host. Plasmin is involved in several physiological processes, such as the degradation of fibrin and other extracellular matrix proteins. Acquisition of this host protease allows the pathogens to invade and disseminate in the host. In the case of the streptococci, Staphylococcus aureus, and Y. pestis, the conversion of plasminogen to plasmin is achieved by their own plasminogen activators [85]. The bound plasminogen can also be activated by the host plasminogen activators, the tissue plasminogen activator (t-PA), and urokinase (u-PA). Once the plasminogen is activated, the surface-bound plasmin cannot be inhibited by the host plasmin inhibitors (for reviews on bacteria-plasminogen interaction see [30, 85–89]).
Besides L. mexicana, surface plasminogen binding has also been demonstrated in several other parasites: T. cruzi [90], Trichomonas vaginalis [91], S. bovis [28, 61], and S. japonicum [27]. In most of them, enolase has been proposed as the plasminogen receptor. For L. mexicana, it was demonstrated that plasminogen and plasmin bind to both the promastigote and amastigote forms of this parasite [92, 93]. The Kd for this interaction in promastigotes is 2.4 μM for plasminogen and 1.2 μM for plasmin. These values are consistent with an in vivo interaction since the concentration of plasminogen in plasma and the extracellular milieu is around 2 μM [94]. Moreover, enolase seems to contribute for 60% to this binding, suggesting that it is the principal plasminogen receptor on the cell surface [59]. This parasite does not appear to have its own plasminogen activator, thus requiring an exogenous source of activators similar to several other plasminogen-binding pathogens such as Borrelia burgdorferi [95]. t-PA, urokinase, and streptokinase are able to activate the bound plasminogen of L. mexicana. Indeed, the activation of plasminogen by t-PA is facilitated in the presence of the parasite (up to nine-fold enhancement of this activation was observed) [92]. Interestingly, there are different plasminogen-binding capacities among the morphotypes present in axenic cultures of L. mexicana promastigotes. Round forms bind more plasminogen than the slender forms. The proportion of the former is higher in heat-shocked and aged cultures [93].
Although the role of the interaction with plasminogen is considered to be important in bacterial pathogenesis, it is not clear what the function of this interaction is in the case of Leishmania parasites, particularly because this organism thrives within macrophages. However, this parasite can find plasminogen, as promastigotes, at the moment of host inoculation or as amastigotes when it is liberated from a macrophage to infect other cells. A clue about a possible function of the interaction of Leishmania with plasminogen was provided by studies of cutaneous lesions caused by L. mexicana in plasminogen-deficient mice [96]. In the lesion of plg−/−, mice, the parasites were limited to isolated foci in contrast to the plg+/+ mice lesion where a scattered pattern was observed. The parasites were less infectious in the plasminogen-deficient mice. In the lesion of plg−/−, increased deposits of fibrin could be observed. These deposits are present in some organs in plasminogen-deficient mice [97, 98]. Thus, in natural conditions, fibrin could be limiting the encounter between Leishmania and macrophages. Fibrin is part of the host defense mechanism providing a barrier limiting invasion and dissemination and is also linked to inflammatory response [81, 99]. In addition, fibrin is one of the most important targets for plasmin in vivo [100]. The inflammatory response initiated by Leishmania parasites may increase capillary permeability, a process associated with inflammation [101], resulting in the deposition of fibrin matrix within the lesion. This fibrin matrix would provide, together with other extracellular matrix proteins, the medium for parasite-macrophage interaction. Activated plasminogen on the parasite surface, or on secreted plasminogen-binding proteins, might thus help to degrade this fibrin matrix favoring the encounter with macrophages. In the latter case, secreted plasminogen-binding proteins would act as “torpedo mines” mediating clearance of fibrin. Indeed, secreted proteins of L. mexicana are able to enhance the plasminogen activation by t-PA [66]. Macrophages have also plasminogen receptors [102] and produce plasminogen activator [103]. The plasmin formed on these cells, together with metalloproteases, mediate macrophage migration during the inflammatory process [104, 105]. This relation plasminogen-Leishmania-macrophage might even be more complex. It is known that plasmin can have a cytoprotective effect on macrophages by inhibiting apoptosis in these cells [106] and could so support more infection by the parasite. It is clear, however, that more information is needed to assess the function of the interaction of Leishmania parasites with plasminogen. Equally, more studies of the role and dynamics of fibrin in the leishmaniasis lesion are needed as well as of the effect of this molecule and plasminogen on the inflammation process within the lesion.
Although fibrinolysis seems to be the most important function of plasmin [100], one may not discard the possibility that other known functions of this enzyme may play a role in the case of Leishmania infection and pathogenesis. Plasmin can function in procollagenase activation and release of peptides for nutrition [85]. Since both Leishmania parasites and macrophages have plasminogen receptors, it is tempting to speculate that plasminogen might act as a bridge between these two cells. Moreover, it has been demonstrated previously that the pathogen Mycoplasma fermentans is able to adhere and invade HeLa cells in a plasminogen-dependent manner [107]. Although the real function of the interaction plasminogen-Leishmania is not known, this interaction suggests that plasminogen-binding proteins, such as enolase, could be virulence factors.
7. Discussion and Conclusions
As described above, enolase may have one or more of three important functions in the different trypanosomatid parasites, in glycolysis, in gluconeogenesis, and/or as plasminogen receptor. In bloodstream-form T. brucei, enolase is involved in glycolysis, a process crucial for the ATP supply of the parasites. In intracellular Leishmania amastigotes, amino sugars are utilized as carbon and energy source, while also gluconeogenesis is an essential pathway for the synthesis of glycoconjugates and β-mannan, both required for virulence of the parasite; therefore, enolase should also be important for the parasitic life of this organism. Less information exists about the nature of the carbohydrate and energy metabolism of the human-pathogenic stages of T. cruzi, but the available data indicate that axenically cultured trypomastigotes and amastigotes are highly dependent on glucose catabolism, strongly suggesting that, also in these cells, enolase fulfills an essential function in metabolism. Therefore, in each of these trypanosomatid species, enolase may be considered as a validated or at least a highly likely candidate drug target.
In addition to this essential metabolic role played by enolase in the cytosol of the parasites, the enzyme was also shown to be present, in a catalytically inactive form, at the outer surface of L. mexicana and T. cruzi (W. Quiñones, and J. L. Concepción, and L. Avilan, unpublished results), where it probably acts as a plasminogen receptor. Although pathogens may have also other means to bind plasminogen, in L. mexicana, enolase was proposed to be the predominant receptor for this zymogen, responsible for 60% of its total binding. This plasminogen binding, and the subsequent activation of the plasminogen into the protease plasmin, could be important for the parasite's virulence.
Several moonlighting functions are known for the glycolytic/gluconeogenic enzyme enolase (discussed in Section 1). In some cases, organisms have multiple enolase isoforms, and some moonlighting functions have been attributed to specific isoforms. In each of the trypanosomatids, except in T. gambiense, only a single enolase gene copy is present, thus the encoded protein must be responsible for both the glycolytic/gluconeogenic activity and the plasminogen-binding function. The absence of activity of the surface-associated plasminogen-binding form may be due to processing of the enzyme during or after its translocation across the plasma membrane. This remains to be determined. How the enzyme arrives at the surface, and is bound to the membrane, is not clear yet. The protein does not possess a predicted transmembrane region or glycosylphosphatidylinositol (GPI) anchor site, nor a detectable N-terminal transit peptide, suggesting that it is not transported via the classical secretion pathway. Moreover, enolase was detected in the secretome of L. donovani [68], and also in that of T. brucei, both bloodstream-form and procyclic cells [71, 72]. Strong experimental evidence has been provided for both Leishmania and Trypanosoma that these parasites use multiple nonclassical secretion pathways, including active exocytosis via microvesicles or exosomes. This suggests that the enolase is first secreted and subsequently retrieved by Leishmania to associate in a still unknown manner with its outer surface.
In addition to a gene coding for a typical enolase, some trypanosomatids possess genes coding for hypothetical proteins with regions homologous to those of authentic enolases. It is unlikely that these proteins, which are usually considerably longer than typical enolases, possess enolase activity, because some catalytically important residues seem to be missing. Their location in the cell and their function—a receptor function or something different—remain to be determined.
Enolases from a variety of pathogenic organisms have the ability to bind plasminogen via a C-terminal Lys residue. However, in S. pneumoniae enolase an internal peptide located in a loop at the surface of the protein was identified as the primary site for interaction with plasminogen [31]. For L. mexicana that, like almost all other trypanosomatid enolases (except T. congolense), lacks a C-terminal Lys, a peptide was found with high sequence similarity to, and at the corresponding position as a plasminogen-binding peptide of, S. pneumoniae enolase. This sequence is highly conserved in the enolase of all other Leishmania species. The plasminogen binding of Leishmania enolase and the involvement of this peptide in this interaction, remain to be proved by biochemical and/or structural studies.
The peptide is also well conserved in the enolase of T. cruzi as well as, intriguingly, in that of all species of African trypanosomes. The necessity of T. cruzi to recruit host plasminogen can be understood in view of the fact that this parasite, like Leishmania spp., invades tissues and enters cells and may require extracellular proteases for these activities. However, the conservation of the candidate zymogen-binding peptide and the presence of enolase in the secretome of the extracellularly living T. brucei raises the question as to whether it plays also a role as plasminogen-receptor in African trypanosomes, and if so, what could be the function of recruiting a host protease. It should be noted that, whereas in L. mexicana the enolase is bound to the cell surface, it has not been detected in the proteome of the T. brucei plasma membrane [108]. The ability of African trypanosomes to degrade fibrinogen and fibrin has already been reported several decades ago [109]. It may be that the infection is accompanied with an increased synthesis of fibrinogen by the host and formation of microthrombi that have to be dealt with by the parasites. Alternatively, the parasites may recruit plasminogen from the host to enable them to cross the blood-brain barrier. This crossing is a still poorly understood process, but may occur in a paracellular way. In this case, plasmin, as a host-derived protease, can help to degrade adherens junctions and tight junctions in the trypanosome-induced blood-brain barrier dysfunction model as proposed by Grab et al. [110].
The facts that enolase plays an essential function in the metabolism of probably all trypanosomatids and is probably an important virulence factor for, at least, the intracellular parasites offer two possibilities for using this protein to combat or prevent an infection. First, the presence of the plasminogen-receptor form of enolase could be used in two different ways to interfere with the virulence of the parasites. Compounds may be developed as drugs that prevent or disrupt the receptor-plasminogen interaction. Alternatively, the trypanosomatid enolase, or unique parts of its sequence, may be used either as an antigen for an infected person to boost his immune system in combating the parasites, or it may be administered as a vaccine to protect people at risk of being infected. Secondly, as discussed above, the availability of the crystal structure of T. brucei enolase has opened the possibility to design parasite-enzyme selective inhibitors. Substrate analogues may be synthesized that exploit unique physicochemical and/or steric features of the catalytic site of the parasite's enzyme and, therefore, provide high affinity and selectivity. Moreover, incorporating substitutions that interact with the reactive Lys and Cys residues (Figure 3) specific of the trypanosomatid enolases will not only increase the selectivity but also render the inhibition irreversible. Irreversible and uncompetitive inhibitors are particularly attractive because, contrary to the much more often observed competitive inhibition, the increase of substrate concentration, resulting from this process, will not overcome the inhibition but may rather lead to an increase of metabolic intermediates to toxic levels [111]. Because of the high similarity between the enolases of the different trypanosomatids, it is likely that inhibitors developed in this way for the T. brucei enzyme may also affect the activity of the enzymes of T. cruzi and Leishmania spp. Alternatively, the availability of systems for the easy production of purified bacterially expressed enolases of all three trypanosomatids allows high-throughput screening of large libraries of drug-like compounds. Any hits obtained by such screenings will subsequently be optimized through successive cycles of structure-activity relationship (SAR) analysis. The next steps in the development of drug candidates against these diseases will involve the selection of compounds with potent and highly selective inhibitory activity on cultured parasites versus human cells, and the evaluation of their efficacy, bioavailability, and toxicity in infected animal models. Finally, lead compounds thus obtained may serve for their further development to trypanocidal or leishmanicidal drugs.
Acknowledgments
The authors would like to thank Dr. Daniel J. Rigden (University of Liverpool, UK) for critically reading the paper and preparing Figures 2 and 3. L. Avilan, W. Quiñones, and J. L. Concepción thank the Fondo Nacional de Ciencia, Tecnología e Investigación (FONACIT) for financial support, for projects MC-2007000960 (L. Avilan) and MC-2007001425, J. L. Concepción and W. Quiñones, respectively. M. Gualdrón-López, V. Hannaert, and P. A. M. Michels gratefully acknowledge the Fonds de la Recherche Scientifique (F.R.S-FNRS) and associated foundations (FRSM and FRIA) and the Belgian Interuniversity Attraction Poles-Federal Office for Scientific, Technical, and Cultural Affairs for financial support. L. Avilan and M. Gualdrón-López contributed equally to this paper.
Abbreviations
- GPI:
glycosylphosphatidylinositol
- OPB:
oligopeptidase B
- PAH:
phosphonoacetohydroxamate
- PEP:
phosphoenolpyruvate
- PGA:
phosphoglycerate
- PLG:
plasminogen
- PPi:
inorganic pyrophosphate
- RNAi:
RNA interference.
References
- 1.Stuart K, Brun R, Croft S, et al. Kinetoplastids: related protozoan pathogens, different diseases. Journal of Clinical Investigation. 2008;118(4):1301–1310. doi: 10.1172/JCI33945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simarro PP, Jannin J, Cattand P. Eliminating human African trypanosomiasis: where do we stand and what comes next? PLoS Medicine. 2008;5(2, article e55) doi: 10.1371/journal.pmed.0050055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steverding D. The history of African trypanosomiasis. Parasites and Vectors. 2008;1(1, article 3) doi: 10.1186/1756-3305-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sternberg JM. Human African trypanosomiasis: clinical presentation and immune response. Parasite Immunology. 2004;26(11-12):469–476. doi: 10.1111/j.0141-9838.2004.00731.x. [DOI] [PubMed] [Google Scholar]
- 5.Coura JR, Dias JCP. Epidemiology, control and surveillance of Chagas disease—100 years after its discovery. Memorias do Instituto Oswaldo Cruz. 2009;104(1):31–40. doi: 10.1590/s0074-02762009000900006. [DOI] [PubMed] [Google Scholar]
- 6.Opperdoes FR, Borst P. Localization of non glycolytic enzymes in a microbody like organelle in Trypanosoma brucei: the glycosome. FEBS Letters. 1977;80(2):360–364. doi: 10.1016/0014-5793(77)80476-6. [DOI] [PubMed] [Google Scholar]
- 7.Michels PAM, Bringaud F, Herman M, Hannaert V. Metabolic functions of glycosomes in trypanosomatids. Biochimica et Biophysica Acta. 2006;1763(12):1463–1477. doi: 10.1016/j.bbamcr.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 8.Verlinde CLMJ, Hannaert V, Blonski C, et al. Glycolysis as a target for the design of new anti-trypanosome drugs. Drug Resistance Updates. 2001;4(1):50–65. doi: 10.1054/drup.2000.0177. [DOI] [PubMed] [Google Scholar]
- 9.Kristensson K, Nygård M, Bertini G, Bentivoglio M. African trypanosome infections of the nervous system: parasite entry and effects on sleep and synaptic functions. Progress in Neurobiology. 2010;91(2):152–171. doi: 10.1016/j.pneurobio.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 10.de Souza W, de Carvalho TMU, Barrias ES. Review on Trypanosoma cruzi: host cell interaction. International Journal of Cell Biology. 2010;2010:18 pages. doi: 10.1155/2010/295394. Article ID 295394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yao C. Major surface protease of trypanosomatids: one size fits all? Infection and Immunity. 2010;78(1):22–31. doi: 10.1128/IAI.00776-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naderer T, McConville MJ. The Leishmania-macrophage interaction: a metabolic perspective. Cellular Microbiology. 2008;10(2):301–308. doi: 10.1111/j.1462-5822.2007.01096.x. [DOI] [PubMed] [Google Scholar]
- 13.Pancholi V. Multifunctional α-enolase: its role in diseases. Cellular and Molecular Life Sciences. 2001;58(7):902–920. doi: 10.1007/PL00000910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Day INM, Peshavaria M, Quinn GB. A differential molecular clock in enolase isoprotein evolution. Journal of Molecular Evolution. 1993;36(6):599–601. doi: 10.1007/BF00556365. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka M, Sugisaki K, Nakashima K. Switching in levels of translatable mRNAs for enolase isozymes during development of chicken skeletal muscle. Biochemical and Biophysical Research Communications. 1985;133(3):868–872. doi: 10.1016/0006-291x(85)91215-x. [DOI] [PubMed] [Google Scholar]
- 16.McAlister L, Holland MJ. Targeted deletion of a yeast enolase structural gene. Identification and isolation of yeast enolase isozymes. Journal of Biological Chemistry. 1982;257(12):7181–7188. [PubMed] [Google Scholar]
- 17.Bolten KE, Marsh AE, Reed SM, Dubey JP, Toribio RE, Saville WJA. Sarcocystis neurona: molecular characterization of enolase domain I region and a comparison to other protozoa. Experimental Parasitology. 2008;120(1):108–112. doi: 10.1016/j.exppara.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Keller A, Peltzer J, Carpentier G, et al. Interactions of enolase isoforms with tubulin and microtubules during myogenesis. Biochimica et Biophysica Acta. 2007;1770(6):919–926. doi: 10.1016/j.bbagen.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 19.Lung J, Liu K-J, Chang J-Y, Leu S-J, Shih N-Y. MBP-1 is efficiently encoded by an alternative transcript of the ENO1 gene but post-translationally regulated by proteasome-dependent protein turnover. FEBS Journal. 2010;277(20):4308–4321. doi: 10.1111/j.1742-4658.2010.07819.x. [DOI] [PubMed] [Google Scholar]
- 20.Kaberdin VR, Lin-Chao S. Unraveling new roles for minor components of the E. coli RNA degradosome. RNA Biology. 2009;6(4):402–405. doi: 10.4161/rna.6.4.9320. [DOI] [PubMed] [Google Scholar]
- 21.López-Villar E, Monteoliva L, Larsen MR, et al. Genetic and proteomic evidences support the localization of yeast enolase in the cell surface. Proteomics. 2006;6:S107–118. doi: 10.1002/pmic.200500479. [DOI] [PubMed] [Google Scholar]
- 22.Agarwal S, Kulshreshtha P, Bambah Mukku D, Bhatnagar R. α-enolase binds to human plasminogen on the surface of Bacillus anthracis. Biochimica et Biophysica Acta. 2008;1784(7-8):986–994. doi: 10.1016/j.bbapap.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 23.Bergmann S, Wild D, Diekmann O, et al. Identification of a novel plasmin(ogen)-binding motif in surface displayed α-enolase of Streptococcus pneumoniae. Molecular Microbiology. 2003;49(2):411–423. doi: 10.1046/j.1365-2958.2003.03557.x. [DOI] [PubMed] [Google Scholar]
- 24.Jones MN, Holt RG. Cloning and characterization of an α-enolase of the oral pathogen Streptococcus mutans that binds human plasminogen. Biochemical and Biophysical Research Communications. 2007;364(4):924–929. doi: 10.1016/j.bbrc.2007.10.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pal Bhowmick I, Kumar N, Sharma S, Coppens I, Jarori GK. Plasmodium falciparum enolase: stage-specific expression and sub-cellular localization. Malaria Journal. 2009;8(1, article 179) doi: 10.1186/1475-2875-8-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Segovia-Gamboa NC, Chávez-Munguía B, Medina-Flores Y, et al. Entamoeba invadens, encystation process and enolase. Experimental Parasitology. 2010;125(2):63–69. doi: 10.1016/j.exppara.2009.12.019. [DOI] [PubMed] [Google Scholar]
- 27.Yang J, Qiu C, Xia Y, et al. Molecular cloning and functional characterization of Schistosoma japonicum enolase which is highly expressed at the schistosomulum stage. Parasitology Research. 2010;107:667–677. doi: 10.1007/s00436-010-1913-z. [DOI] [PubMed] [Google Scholar]
- 28.de la Torre-Escudero E, Manzano-Román R, Pérez-Sánchez R, Siles-Lucas M, Oleaga A. Cloning and characterization of a plasminogen-binding surface-associated enolase from Schistosoma bovis. Veterinary Parasitology. 2010;173:73–84. doi: 10.1016/j.vetpar.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 29.Falabella P, Riviello L, De Stradis ML, et al. Aphidius ervi teratocytes release an extracellular enolase. Insect Biochemistry and Molecular Biology. 2009;39(11):801–813. doi: 10.1016/j.ibmb.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 30.Walker MJ, McArthur JD, McKay F, Ranson M. Is plasminogen deployed as a Streptococcus pyogenes virulence factor? Trends in Microbiology. 2005;13(7):308–313. doi: 10.1016/j.tim.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 31.Ehinger S, Schubert WD, Bergmann S, Hammerschmidt S, Heinz DW. Plasmin(ogen)-binding α-enolase from Streptococcus pneumoniae: crystal structure and evaluation of plasmin(ogen)-binding sites. Journal of Molecular Biology. 2004;343(4):997–1005. doi: 10.1016/j.jmb.2004.08.088. [DOI] [PubMed] [Google Scholar]
- 32.Engel JC, Franke de Cazzulo BM, Stoppani AOM, Cannata JJB, Cazzulo JJ. Aerobic glucose fermentation by Trypanosoma cruzi axenic culture amastigote-like forms during growth and differentiation to epimastigotes. Molecular and Biochemical Parasitology. 1987;26(1-2):1–10. doi: 10.1016/0166-6851(87)90123-x. [DOI] [PubMed] [Google Scholar]
- 33.Silber AM, Tonelli RR, Lopes CG, et al. Glucose uptake in the mammalian stages of Trypanosoma cruzi. Molecular and Biochemical Parasitology. 2009;168(1):102–108. doi: 10.1016/j.molbiopara.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 34.Naderer T, Heng J, McConville MJ. Evidence that intracellular stages of Leishmania major utilize amino sugars as a major carbon source. PLoS Pathogens. 2010;6(12) doi: 10.1371/journal.ppat.1001245. Article ID e1001245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naderer T, Ellis MA, Sernee MF, et al. Virulence of Leishmania major in macrophages and mice requires the gluconeogenic enzyme fructose-1,6-bisphosphatase. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(14):5502–5507. doi: 10.1073/pnas.0509196103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodríguez-Contreras D, Landfear SM. Metabolic changes in glucose transporter-deficient Leishmania mexicana and parasite virulence. Journal of Biological Chemistry. 2006;281(29):20068–20076. doi: 10.1074/jbc.M603265200. [DOI] [PubMed] [Google Scholar]
- 37.Taylor MB, Berghausen H, Heyworth P, Messenger N, Rees LJ, Gutteridge WE. Subcellular localization of some glycolytic enzymes in parasitic flagellated protozoa. International Journal of Biochemistry. 1980;11(2):117–120. doi: 10.1016/0020-711x(80)90243-8. [DOI] [PubMed] [Google Scholar]
- 38.Hart DT, Opperdoes FR. The occurrence of glycosomes (microbodies) in the promastigote stage of four major Leishmania species. Molecular and Biochemical Parasitology. 1984;13(2):159–172. doi: 10.1016/0166-6851(84)90110-5. [DOI] [PubMed] [Google Scholar]
- 39.Hannaert V, Albert MA, Rigden DJ, et al. Kinetic characterization, structure modelling studies and crystallization of Trypanosoma brucei enolase. European Journal of Biochemistry. 2003;270(15):3205–3213. doi: 10.1046/j.1432-1033.2003.03692.x. [DOI] [PubMed] [Google Scholar]
- 40.Quiñones W, Peña P, Domingo-Sananes M, et al. Leishmania mexicana: molecular cloning and characterization of enolase. Experimental Parasitology. 2007;116(3):241–251. doi: 10.1016/j.exppara.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 41.Paba J, Santana JM, Teixeira ARL, Fontes W, Sousa MV, Ricart CAO. Proteomic analysis of the human pathogen Trypanosoma cruzi. Proteomics. 2004;4(4):1052–1059. doi: 10.1002/pmic.200300637. [DOI] [PubMed] [Google Scholar]
- 42.Rosenzweig D, Smith D, Opperdoes F, Stern S, Olafson RW, Zilberstein D. Retooling Leishmania metabolism: from sand fly gut to human macrophage. FASEB Journal. 2008;22(2):590–602. doi: 10.1096/fj.07-9254com. [DOI] [PubMed] [Google Scholar]
- 43.Albert MA, Haanstra JR, Hannaert V, et al. Experimental and in silico analyses of glycolytic flux control in bloodstream form Trypanosoma brucei. Journal of Biological Chemistry. 2005;280(31):28306–28315. doi: 10.1074/jbc.M502403200. [DOI] [PubMed] [Google Scholar]
- 44.Hannaert V, Brinkmann H, Nowitzki U, et al. Enolase from Trypanosoma brucei, from the amitochondriate protist Mastigamoeba balamuthi, and from the chloroplast and cytosol of Euglena gracilis: pieces in the evolutionary puzzle of the eukaryotic glycolytic pathway. Molecular Biology and Evolution. 2000;17(7):989–1000. doi: 10.1093/oxfordjournals.molbev.a026395. [DOI] [PubMed] [Google Scholar]
- 45.Pabón MA, Cáceres AJ, Gualdrón M, Quiñones W, Avilán L, Concepción JL. Purification and characterization of hexokinase from Leishmania mexicana. Parasitology Research. 2007;100(4):803–810. doi: 10.1007/s00436-006-0351-4. [DOI] [PubMed] [Google Scholar]
- 46.Cáceres AJ, Portillo R, Acosta H, et al. Molecular and biochemical characterization of hexokinase from Trypanosoma cruzi. Molecular and Biochemical Parasitology. 2003;126(2):251–262. doi: 10.1016/s0166-6851(02)00294-3. [DOI] [PubMed] [Google Scholar]
- 47.Kornblatt MJ, Lange R, Balny C. Can monomers of yeast enolase have enzymatic activity? European Journal of Biochemistry. 1998;251(3):775–780. doi: 10.1046/j.1432-1327.1998.2510775.x. [DOI] [PubMed] [Google Scholar]
- 48.Zhang E, Brewer JM, Minor W, Carreira LA, Lebioda L. Mechanism of enolase: the crystal structure of asymmetric dimer enolase- 2-phospho-D-glycerate/enolase-phosphoenolpyruvate at 2.0 Å resolution. Biochemistry. 1997;36(41):12526–12534. doi: 10.1021/bi9712450. [DOI] [PubMed] [Google Scholar]
- 49.da Silva Giotto MT, Hannaert V, Vertommen D, et al. The crystal structure of Trypanosoma brucei enolase: visualisation of the inhibitory metal binding site III and potential as target for selective, irreversible inhibition. Journal of Molecular Biology. 2003;331(3):653–665. doi: 10.1016/s0022-2836(03)00752-6. [DOI] [PubMed] [Google Scholar]
- 50.Navarro MVDAS, Gomes Dias SM, Mello LV, et al. Structural flexibility in Trypanosoma brucei enolase revealed by X-ray crystallography and molecular dynamics. FEBS Journal. 2007;274(19):5077–5089. doi: 10.1111/j.1742-4658.2007.06027.x. [DOI] [PubMed] [Google Scholar]
- 51.Yao C, Li Y, Donelson JE, Wilson ME. Proteomic examination of Leishmania chagasi plasma membrane proteins: contrast between avirulent and virulent (metacyclic) parasite forms. Proteomics—Clinical Applications. 2010;4(1):4–16. doi: 10.1002/prca.200900050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Foucher AL, Papadopoulou B, Ouellette M. Prefractionation by digitonin extraction increases representation of the cytosolic and intracellular proteome of Leishmania infantum. Journal of Proteome Research. 2006;5(7):1741–1750. doi: 10.1021/pr060081j. [DOI] [PubMed] [Google Scholar]
- 53.Swenerton RK, Zhang S, Sajid M, et al. The oligopeptidase B of Leishmania regulates parasite enolase and immune evasion. Journal of Biological Chemistry. 2011;286(1):429–440. doi: 10.1074/jbc.M110.138313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Plow EF, Felez J, Miles LA. Cellular regulation of fibrinolysis. Thrombosis and Haemostasis. 1991;66(1):32–36. [PubMed] [Google Scholar]
- 55.Redlitz A, Fowler BJ, Plow EF, Miles LA. The role of an enolase-related molecule in plasminogen binding to cells. European Journal of Biochemistry. 1995;227(1-2):407–415. doi: 10.1111/j.1432-1033.1995.tb20403.x. [DOI] [PubMed] [Google Scholar]
- 56.López-Alemany R, Suelves M, Muñoz-Cánoves P. Plasmin generation dependent on α-enolase-type plasminogen receptor is required for myogenesis. Thrombosis and Haemostasis. 2003;90(4):724–733. doi: 10.1160/TH03-04-0291. [DOI] [PubMed] [Google Scholar]
- 57.Pancholi V, Fischetti VA. α-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. Journal of Biological Chemistry. 1998;273(23):14503–14515. doi: 10.1074/jbc.273.23.14503. [DOI] [PubMed] [Google Scholar]
- 58.Bergmann S, Rohde M, Chhatwal GS, Hammerschmidt S. α-enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Molecular Microbiology. 2001;40(6):1273–1287. doi: 10.1046/j.1365-2958.2001.02448.x. [DOI] [PubMed] [Google Scholar]
- 59.Vanegas G, Quiñones W, Carrasco-López C, Concepción JL, Albericio F, Avilán L. Enolase as a plasminogen binding protein in Leishmania mexicana. Parasitology Research. 2007;101(6):1511–1516. doi: 10.1007/s00436-007-0668-7. [DOI] [PubMed] [Google Scholar]
- 60.Jong AY, Chen SHM, Stins MF, Kim KS, Tuan TL, Huang SH. Binding of Candida albicans enolase to plasmin(ogen) results in enhanced invasion of human brain microvascular endothelial cells. Journal of Medical Microbiology. 2003;52(8):615–622. doi: 10.1099/jmm.0.05060-0. [DOI] [PubMed] [Google Scholar]
- 61.Ramajo-Hernández A, Pérez-Sánchez R, Ramajo-Martín V, Oleaga A. Schistosoma bovis: plasminogen binding in adults and the identification of plasminogen-binding proteins from the worm tegument. Experimental Parasitology. 2007;115(1):83–91. doi: 10.1016/j.exppara.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 62.Castellino FJ, Ploplis VA. Structure and function of the plasminogen/plasmin system. Thrombosis and Haemostasis. 2005;93(4):647–654. doi: 10.1160/TH04-12-0842. [DOI] [PubMed] [Google Scholar]
- 63.Miles LA, Hawley SB, Baik N, Andronicos NM, Castellino FJ, Parmer RJ. Plasminogen receptors: the sine qua non of cell surface plasminogen activation. Frontiers in Bioscience. 2005;10(2):1754–1762. doi: 10.2741/1658. [DOI] [PubMed] [Google Scholar]
- 64.Sha J, Erova TE, Alyea RA, et al. Surface-expressed enolase contributes to the pathogenesis of clinical isolate ssu of Aeromonas hydrophilaa. Journal of Bacteriology. 2009;191(9):3095–3107. doi: 10.1128/JB.00005-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gandhi NS, Young K, Warmington JR, Mancera RL. Characterization of sequence and structural features of the Candida krusei enolase. In Silico Biology. 2008;8(5-6):449–460. [PubMed] [Google Scholar]
- 66.Gómez-Arreaza A, Acosta H, Barros-Álvarez X, Concepción JL, Albericio F, Avilan L. Leishmania mexicana: LACK (Leishmania homolog of receptors for activated C-kinase) is a plasminogen binding protein. Experimental Parasitology. 2011;124(4):752–761. doi: 10.1016/j.exppara.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 67.Hurmalainen V, Edelman S, Antikainen J, Baumann M, Lähteenmäki K, Korhonen TK. Extracellular proteins of Lactobacillus crispatus enhance activation of human plasminogen. Microbiology. 2007;153(4):1112–1122. doi: 10.1099/mic.0.2006/000901-0. [DOI] [PubMed] [Google Scholar]
- 68.Silverman JM, Chan SK, Robinson DP, et al. Proteomic analysis of the secretome of Leishmania donovani. Genome Biology. 2008;9(2):p. R35. doi: 10.1186/gb-2008-9-2-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Silverman JM, Clos J, de’Oliveira CC, et al. An exosome-based secretion pathway is responsible for protein export from Leishmania and communication with macrophages. Journal of Cell Science. 2010;123(6):842–852. doi: 10.1242/jcs.056465. [DOI] [PubMed] [Google Scholar]
- 70.Cuervo P, De Jesus JB, Saboia-Vahia L, Mendonça-Lima L, Domont GB, Cupolillo E. Proteomic characterization of the released/secreted proteins of Leishmania (Viannia) braziliensis promastigotes. Journal of Proteomics. 2009;73(1):79–92. doi: 10.1016/j.jprot.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 71.Atyame Nten CM, Sommerer N, Rofidal V, et al. Excreted/secreted proteins from trypanosome procyclic strains. Journal of Biomedicine and Biotechnology. 2010;2010:8 pages. doi: 10.1155/2010/212817. Article ID 212817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Geiger A, Hirtz C, Bécue T, et al. Exocytosis and protein secretion in Trypanosoma. BMC Microbiology. 2010;10, article 20 doi: 10.1186/1471-2180-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Labbé M, Péroval M, Bourdieu C, Girard-Misguich F, Péry P. Eimeria tenella enolase and pyruvate kinase: a likely role in glycolysis and in others functions. International Journal for Parasitology. 2006;36(14):1443–1452. doi: 10.1016/j.ijpara.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 74.Bernal D, de La Rubia JE, Carrasco-Abad AM, Toledo R, Mas-Coma S, Marcilla A. Identification of enolase as a plasminogen-binding protein in excretory-secretory products of Fasciola hepatica. FEBS Letters. 2004;563(1–3):203–206. doi: 10.1016/S0014-5793(04)00306-0. [DOI] [PubMed] [Google Scholar]
- 75.Marcilla A, Pérez-García A, Espert A, et al. Echinostoma caproni: identification of enolase in excretory/secretory products, molecular cloning, and functional expression. Experimental Parasitology. 2007;117(1):57–64. doi: 10.1016/j.exppara.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 76.Ringqvist E, Palm JED, Skarin H, et al. Release of metabolic enzymes by Giardia in response to interaction with intestinal epithelial cells. Molecular and Biochemical Parasitology. 2008;159(2):85–91. doi: 10.1016/j.molbiopara.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu F, Cui SJ, Hu W, Feng Z, Wang ZQ, Han ZG. Excretory/secretory proteome of the adult developmental stage of human blood fluke, Schistosoma japonicum. Molecular and Cellular Proteomics. 2009;8(6):1236–1251. doi: 10.1074/mcp.M800538-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Olver C, Vidal M. Proteomic analysis of secreted exosomes. Sub-cellular biochemistry. 2007;43:99–131. doi: 10.1007/978-1-4020-5943-8_7. [DOI] [PubMed] [Google Scholar]
- 79.Goto H, Wells K, Takada A, Kawaoka Y. Plasminogen-binding activity of neuraminidase determines the pathogenicity of influenza a virus. Journal of Virology. 2001;75(19):9297–9301. doi: 10.1128/JVI.75.19.9297-9301.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sun H, Ringdahl U, Momeister JW, et al. Plasminogen is a critical host pathogenicity factor for group A streptococcal infection. Science. 2004;305(5688):1283–1286. doi: 10.1126/science.1101245. [DOI] [PubMed] [Google Scholar]
- 81.Degen JL, Bugge TH, Goguen JD. Fibrin and fibrinolysis in infection and host defense. Journal of Thrombosis and Haemostasis. 2007;5(1):24–31. doi: 10.1111/j.1538-7836.2007.02519.x. [DOI] [PubMed] [Google Scholar]
- 82.Coleman JL, Gebbia JA, Piesman J, Degen JL, Bugge TH, Benach JL. Plasminogen is required for efficient dissemination of B. burgdorferi in ticks and for enhancement of spirochetemia in mice. Cell. 1997;89(7):1111–1119. doi: 10.1016/s0092-8674(00)80298-6. [DOI] [PubMed] [Google Scholar]
- 83.Nordstrand A, Shamaei-Tousi A, Ny A, Bergström S. Delayed invasion of the kidney and brain by Borrelia crocidurae in plasminogen-deficient mice. Infection and Immunity. 2001;69(9):5832–5839. doi: 10.1128/IAI.69.9.5832-5839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gebbia JA, Garcia Monco JC, Degen JL, Bugge TH, Benach JL. The plasminogen activation system enhances brain and heart invasion in murine relapsing fever borreliosis. Journal of Clinical Investigation. 1999;103(1):81–87. doi: 10.1172/JCI5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lähteenmäki K, Kuusela P, Korhonen TK. Bacterial plasminogen activators and receptors. FEMS Microbiology Reviews. 2001;25(5):531–552. doi: 10.1111/j.1574-6976.2001.tb00590.x. [DOI] [PubMed] [Google Scholar]
- 86.Lähteenmäki K, Edelman S, Korhonen TK. Bacterial metastasis: the host plasminogen system in bacterial invasion. Trends in Microbiology. 2005;13(2):79–85. doi: 10.1016/j.tim.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 87.Boyle MDP, Lottenberg R. Plasminogen activation by invasive human pathogens. Thrombosis and Haemostasis. 1997;77(1):1–10. [PubMed] [Google Scholar]
- 88.Coleman JL, Benach JL. Use of the plasminogen activation system by microorganisms. Journal of Laboratory and Clinical Medicine. 1999;134(6):567–576. doi: 10.1016/s0022-2143(99)90095-1. [DOI] [PubMed] [Google Scholar]
- 89.Bergmann S, Hammerschmidt S. Fibrinolysis and host response in bacterial infections. Thrombosis and Haemostasis. 2007;98(3):512–520. [PubMed] [Google Scholar]
- 90.Almeida L, Vanegas G, Calcagno M, Concepción JL, Avilan L. Plasminogen interaction with Trypanosoma cruzi. Memorias do Instituto Oswaldo Cruz. 2004;99(1):63–67. doi: 10.1590/s0074-02762004000100011. [DOI] [PubMed] [Google Scholar]
- 91.Mundodi V, Kucknoor AS, Alderete JF. Immunogenic and plasminogen-binding surface-associated α-enolase of Trichomonas vaginalis. Infection and Immunity. 2008;76(2):523–531. doi: 10.1128/IAI.01352-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Avilan L, Calcagno M, Figuera M, Lemus L, Puig J, Rodriguez AM. Interaction of Leishmania mexicana promastigotes with the plasminogen-plasmin system. Molecular and Biochemical Parasitology. 2000;110(2):183–193. doi: 10.1016/s0166-6851(00)00269-3. [DOI] [PubMed] [Google Scholar]
- 93.Calcagno M, Avilan L, Colasante C, Berrueta L, Salmen S. Interaction of different Leishmania mexicana morphotypes with plasminogen. Parasitology Research. 2002;88(11):972–978. doi: 10.1007/s00436-002-0688-2. [DOI] [PubMed] [Google Scholar]
- 94.Collen D, Verstraete M. Molecular biology of human plasminogen. II. Metabolism in physiological and some pathological conditions in man. Thromb Diath Haemorrh. 1975;34(2):403–408. [PubMed] [Google Scholar]
- 95.Klempner MS, Noring R, Epstein MP, et al. Binding of human plasminogen and urokinase-type plasminogen activator to the Lyme disease spirochete, Borrelia burgdorferi. Journal of Infectious Diseases. 1995;171(5):1258–1265. doi: 10.1093/infdis/171.5.1258. [DOI] [PubMed] [Google Scholar]
- 96.Maldonado J, Marina C, Puig J, Maizo Z, Avilan L. A study of cutaneous lesions caused by Leishmania mexicana in plasminogen-deficient mice. Experimental and Molecular Pathology. 2006;80(3):289–294. doi: 10.1016/j.yexmp.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 97.Ploplis VA, Carmeliet P, Vazirzadeh S, et al. Effects of disruption of the plasminogen gene on thrombosis, growth, and health in mice. Circulation. 1995;92(9):2585–2593. doi: 10.1161/01.cir.92.9.2585. [DOI] [PubMed] [Google Scholar]
- 98.Bugge TH, Flick MJ, Daugherty CC, Degen JL. Plasminogen deficiency causes severe thrombosis but is compatible with development and reproduction. Genes and Development. 1995;9(7):794–807. doi: 10.1101/gad.9.7.794. [DOI] [PubMed] [Google Scholar]
- 99.Sun H. The interaction between pathogens and the host coagulation system. Physiology. 2006;21(4):281–288. doi: 10.1152/physiol.00059.2005. [DOI] [PubMed] [Google Scholar]
- 100.Bugge TH, Kombrinck KW, Flick MJ, Daugherty CC, Danton MJS, Degen JL. Loss of fibrinogen rescues mice from the pleiotropic effects of plasminogen deficiency. Cell. 1996;87(4):709–719. doi: 10.1016/s0092-8674(00)81390-2. [DOI] [PubMed] [Google Scholar]
- 101.Arroyo AG, Iruela-Arispe ML. Extracellular matrix, inflammation, and the angiogenic response. Cardiovascular Research. 2010;86(2):226–235. doi: 10.1093/cvr/cvq049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Das R, Burke T, Plow EF. Histone H2B as a functionally important plasminogen receptor on macrophages. Blood. 2007;110(10):3763–3772. doi: 10.1182/blood-2007-03-079392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hart PH, Vitti GF, Burgess DR, Singleton DK, Hamilton JA. Human monocytes can produce tissue-type plasminogen activator. Journal of Experimental Medicine. 1989;169(4):1509–1514. doi: 10.1084/jem.169.4.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ploplis VA, French EL, Carmeliet P, Collen D, Plow EF. Plasminogen deficiency differentially affects recruitment of inflammatory cell populations in mice. Blood. 1998;91(6):2005–2009. [PubMed] [Google Scholar]
- 105.Wygrecka M, Marsh LM, Morty RE, et al. Enolase-1 promotes plasminogen-mediated recruitment of monocytes to the acutely inflamed lung. Blood. 2009;113(22):5588–5598. doi: 10.1182/blood-2008-08-170837. [DOI] [PubMed] [Google Scholar]
- 106.Mitchell JW, Baik N, Castellino FJ, Miles LA. Plasminogen inhibits TNFα-induced apoptosis in monocytes. Blood. 2006;107(11):4383–4390. doi: 10.1182/blood-2005-07-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yavlovich A, Rottem S. Binding of host extracellular matrix proteins to Mycoplasma fermentans and its effect on adherence to, and invasion of HeLa cells. FEMS Microbiology Letters. 2007;266(2):158–162. doi: 10.1111/j.1574-6968.2006.00534.x. [DOI] [PubMed] [Google Scholar]
- 108.Bridges DJ, Pitt AR, Hanrahan O, et al. Characterisation of the plasma membrane subproteome of bloodstream form Trypanosoma brucei. Proteomics. 2008;8(1):83–99. doi: 10.1002/pmic.200700607. [DOI] [PubMed] [Google Scholar]
- 109.Boreham PFL, Facer CA. Fibrinogen and fibrinogen/fibrin degradation products in experimental African trypanosomiasis. International Journal for Parasitology. 1974;4(2):143–151. doi: 10.1016/0020-7519(74)90097-6. [DOI] [PubMed] [Google Scholar]
- 110.Grab DJ, Garcia-Garcia JC, Nikolskaia OV, et al. Protease activated receptor signaling is required for African trypanosome traversal of human brain microvascular endothelial cells. PLoS Neglected Tropical Diseases. 2009;3(7, article e479) doi: 10.1371/journal.pntd.0000479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cornish-Bowden A. Why is uncompetitive inhibition so rare? A possible explanation, with implications for the design of drugs and pesticides. FEBS Letters. 1986;203(1):3–6. doi: 10.1016/0014-5793(86)81424-7. [DOI] [PubMed] [Google Scholar]