Abstract
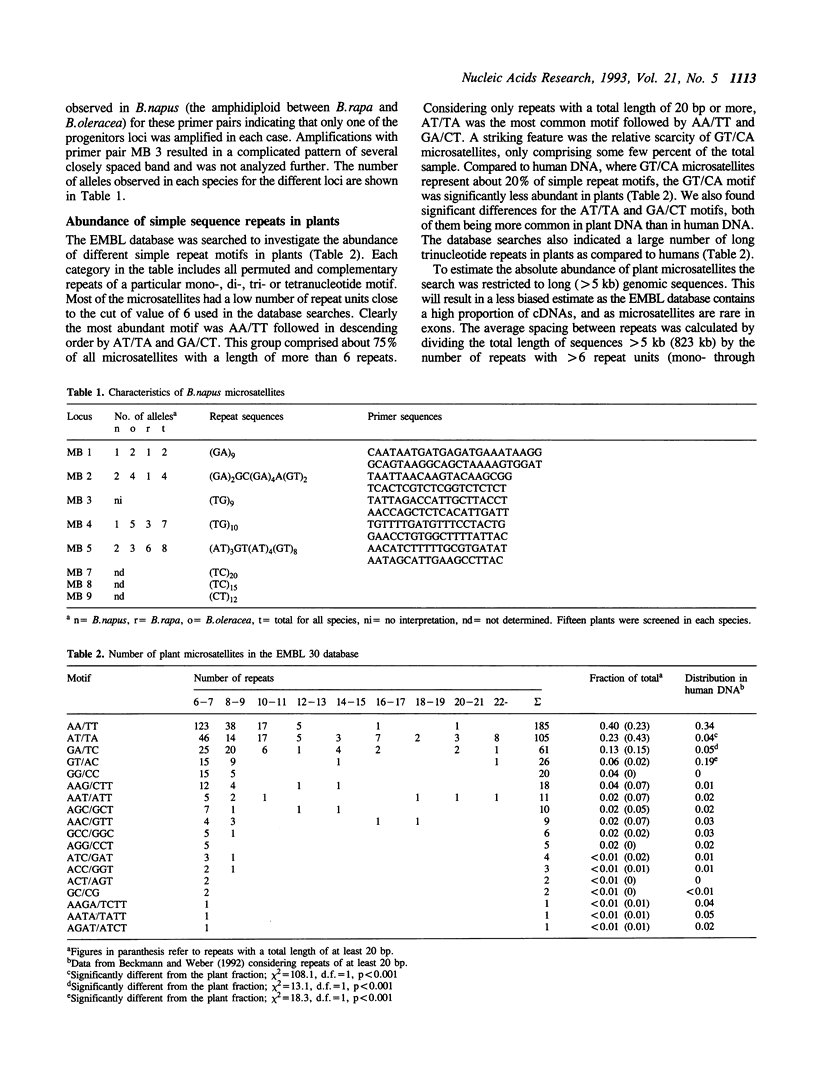
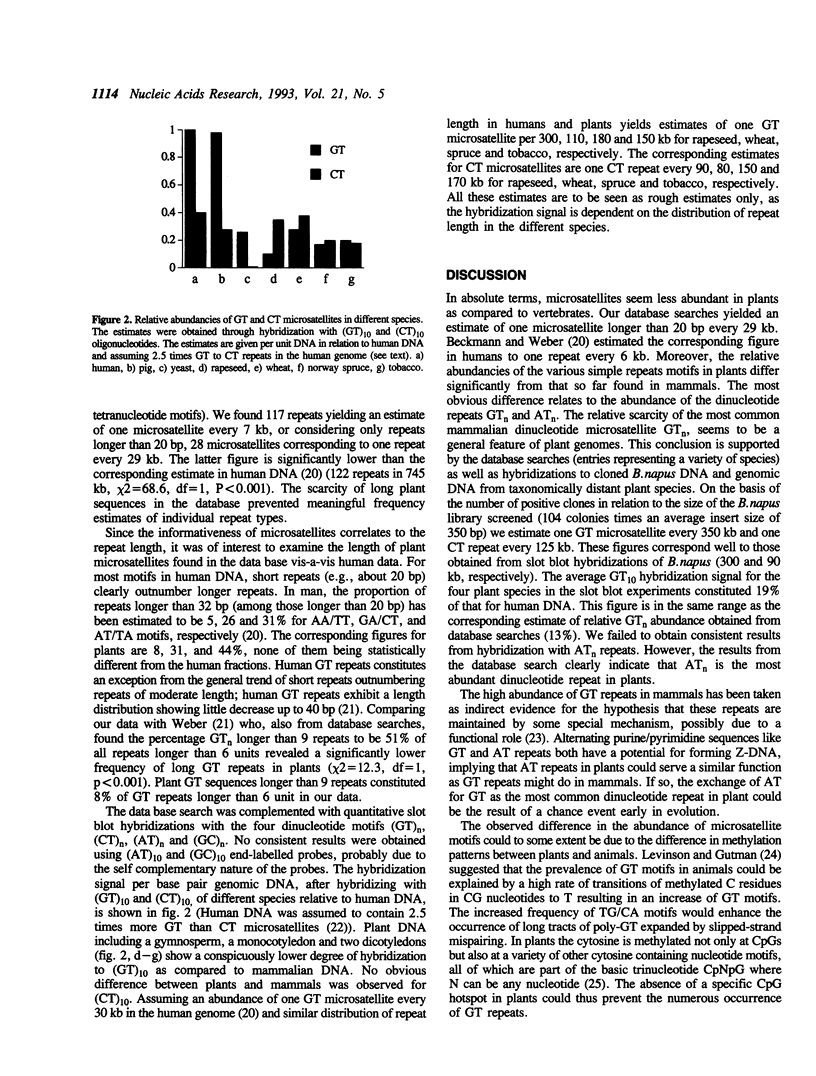
The abundance of different simple sequence motifs in plants was accessed through data base searches of DNA sequences and quantitative hybridization with synthetic dinucleotide repeats. Database searches indicated that microsatellites are five times less abundant in the genomes of plants than in mammals. The most common plant repeat motif was AA/TT followed by AT/TA and CT/GA. This group comprised about 75% of all microsatellites with a length of more than 6 repeats. The GT/CA motif being the most abundant dinucleotide repeat in mammals was found to be considerably less frequent in plants. To address the question if plant simple repeat sequences are variable as in mammals, (GT)n and (CT)n microsatellites were isolated from B.napus. Five loci were investigated by PCR-analysis and amplified products were obtained for all microsatellites from B. oleracea, B.napus and B.rapa DNA, but only for one primer pair from B.nigra. Polymorphism was detected for all microsatellites.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akkaya M. S., Bhagwat A. A., Cregan P. B. Length polymorphisms of simple sequence repeat DNA in soybean. Genetics. 1992 Dec;132(4):1131–1139. doi: 10.1093/genetics/132.4.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman J. S., Weber J. L. Survey of human and rat microsatellites. Genomics. 1992 Apr;12(4):627–631. doi: 10.1016/0888-7543(92)90285-z. [DOI] [PubMed] [Google Scholar]
- Condit R., Hubbell S. P. Abundance and DNA sequence of two-base repeat regions in tropical tree genomes. Genome. 1991 Feb;34(1):66–71. doi: 10.1139/g91-011. [DOI] [PubMed] [Google Scholar]
- Economou E. P., Bergen A. W., Warren A. C., Antonarakis S. E. The polydeoxyadenylate tract of Alu repetitive elements is polymorphic in the human genome. Proc Natl Acad Sci U S A. 1990 Apr;87(8):2951–2954. doi: 10.1073/pnas.87.8.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H., Johansson M., Sandberg K., Andersson L. Cloning of highly polymorphic microsatellites in the horse. Anim Genet. 1992;23(2):133–142. doi: 10.1111/j.1365-2052.1992.tb00032.x. [DOI] [PubMed] [Google Scholar]
- Goff D. J., Galvin K., Katz H., Westerfield M., Lander E. S., Tabin C. J. Identification of polymorphic simple sequence repeats in the genome of the zebrafish. Genomics. 1992 Sep;14(1):200–202. doi: 10.1016/s0888-7543(05)80309-x. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y., Naveh-Many T., Cedar H., Razin A. Sequence specificity of methylation in higher plant DNA. Nature. 1981 Aug 27;292(5826):860–862. doi: 10.1038/292860a0. [DOI] [PubMed] [Google Scholar]
- Hamada H., Petrino M. G., Kakunaga T. A novel repeated element with Z-DNA-forming potential is widely found in evolutionarily diverse eukaryotic genomes. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6465–6469. doi: 10.1073/pnas.79.21.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H., Seidman M., Howard B. H., Gorman C. M. Enhanced gene expression by the poly(dT-dG).poly(dC-dA) sequence. Mol Cell Biol. 1984 Dec;4(12):2622–2630. doi: 10.1128/mcb.4.12.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson G., Gutman G. A. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987 May;4(3):203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- Litt M., Luty J. A. A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle actin gene. Am J Hum Genet. 1989 Mar;44(3):397–401. [PMC free article] [PubMed] [Google Scholar]
- Luty J. A., Guo Z., Willard H. F., Ledbetter D. H., Ledbetter S., Litt M. Five polymorphic microsatellite VNTRs on the human X chromosome. Am J Hum Genet. 1990 Apr;46(4):776–783. [PMC free article] [PubMed] [Google Scholar]
- Moore S. S., Sargeant L. L., King T. J., Mattick J. S., Georges M., Hetzel D. J. The conservation of dinucleotide microsatellites among mammalian genomes allows the use of heterologous PCR primer pairs in closely related species. Genomics. 1991 Jul;10(3):654–660. doi: 10.1016/0888-7543(91)90448-n. [DOI] [PubMed] [Google Scholar]
- Naylor L. H., Clark E. M. d(TG)n.d(CA)n sequences upstream of the rat prolactin gene form Z-DNA and inhibit gene transcription. Nucleic Acids Res. 1990 Mar 25;18(6):1595–1601. doi: 10.1093/nar/18.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordheim A., Rich A. The sequence (dC-dA)n X (dG-dT)n forms left-handed Z-DNA in negatively supercoiled plasmids. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1821–1825. doi: 10.1073/pnas.80.7.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardue M. L., Lowenhaupt K., Rich A., Nordheim A. (dC-dA)n.(dG-dT)n sequences have evolutionarily conserved chromosomal locations in Drosophila with implications for roles in chromosome structure and function. EMBO J. 1987 Jun;6(6):1781–1789. doi: 10.1002/j.1460-2075.1987.tb02431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlötterer C., Amos B., Tautz D. Conservation of polymorphic simple sequence loci in cetacean species. Nature. 1991 Nov 7;354(6348):63–65. doi: 10.1038/354063a0. [DOI] [PubMed] [Google Scholar]
- Stallings R. L., Ford A. F., Nelson D., Torney D. C., Hildebrand C. E., Moyzis R. K. Evolution and distribution of (GT)n repetitive sequences in mammalian genomes. Genomics. 1991 Jul;10(3):807–815. doi: 10.1016/0888-7543(91)90467-s. [DOI] [PubMed] [Google Scholar]
- Tautz D., Renz M. Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Res. 1984 May 25;12(10):4127–4138. doi: 10.1093/nar/12.10.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahls W. P., Wallace L. J., Moore P. D. The Z-DNA motif d(TG)30 promotes reception of information during gene conversion events while stimulating homologous recombination in human cells in culture. Mol Cell Biol. 1990 Feb;10(2):785–793. doi: 10.1128/mcb.10.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J. L. Informativeness of human (dC-dA)n.(dG-dT)n polymorphisms. Genomics. 1990 Aug;7(4):524–530. doi: 10.1016/0888-7543(90)90195-z. [DOI] [PubMed] [Google Scholar]
- Weber J. L., May P. E. Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction. Am J Hum Genet. 1989 Mar;44(3):388–396. [PMC free article] [PubMed] [Google Scholar]
- Williams J. G., Kubelik A. R., Livak K. J., Rafalski J. A., Tingey S. V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990 Nov 25;18(22):6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]



