Abstract
The mitochondrial tyrosyl-tRNA synthetases (mt TyrRSs) of Pezizomycotina fungi are bifunctional proteins that aminoacylate mitochondrial tRNATyr and are structure-stabilizing splicing cofactors for group I introns. Studies with the Neurospora crassa synthetase (CYT-18 protein) showed that splicing activity is dependent upon Pezizomycotina-specific structural adaptations that form a distinct group I intron-binding site in the N-terminal catalytic domain. Although CYT-18’s C-terminal domain also binds group I introns, it has been intractable to X-ray crystallography in the full-length protein. Here, we determined an NMR structure of the isolated C-terminal domain of the Aspergillus nidulans mt TyrRS, which is closely related to but smaller than CYT-18’s. The structure shows an S4 fold like that of bacterial TyrRSs, but with novel features, including three Pezizomycontia-specific insertions. 15N-1H two-dimensional NMR showed that C-terminal domains of the full-length A. nidulans and GeoBacillus stearothermophilus synthetases do not tumble independently in solution, suggesting restricted orientations. Modeling onto a CYT-18/group I intron co-crystal structure indicates that the C-terminal domains of both subunits of the homodimeric protein bind different ends of the intron RNA, with one C-terminal domain having to undergo a large shift on its flexible linker to bind tRNATyr or the intron RNA on either side of the catalytic domain. The modeling suggests that the C-terminal domain acts together with the N-terminal domain to clamp parts of the intron’s catalytic core, that at least one C-terminal domain insertion functions in group I intron binding, and that some C-terminal domain regions bind both tRNATyr and group I intron RNAs.
The Neurospora crassa mitochondrial tyrosyl-tRNA synthetase (mt TyrRS; CYT-18 protein) and those of other fungi of the subphylum Pezizomycotina are bifunctional proteins that both aminoacylate mt tRNATyr and promote the splicing of mt group I introns (1, 2). Previous studies showed that CYT-18 recognizes conserved structural features of the group I intron catalytic core and promotes splicing by stabilizing the catalytically active RNA structure (3–5). The group I intron catalytic core consists of two extended helical domains: P4–P6 consisting of stacked helices P5, P4, P6, and P6a/b, and P3–P9 consisting of helices P9, P7, P3, and P8 (6). The two domains interact via a series of tertiary contacts, with the P3–P9 domain wrapping around the P4–P6 domain forming a cleft that contains the intron RNA’s active site. This active site binds the splice sites and guanosine cofactor and uses specifically bound Mg2+ ions to catalyze splicing via guanosine-initiated transesterification reactions. Biochemical and genetic experiments suggested that CYT-18 binds first to the P4–P6 domain to promote its assembly and then makes additional contacts with the P3–P9 domain that stabilize the active RNA structure relative to alternative non-native structures (3–5, 7). The structural stabilization of the group I intron core afforded by CYT-18 compensates for RNA structural defects that impair self-splicing (3).
Bacterial TyrRSs are comprised of an N-terminal nucleotide-binding fold or catalytic domain, an intermediate α-helical domain, and a C-terminal tRNA-binding domain. The latter has a fold similar to that of ribosomal protein S4 and is attached to the remainder of the protein via a flexible linker (8, 9). The functional TyrRS is a homodimer, which binds tRNATyr asymmetrically across the two subunits (10, 11). The catalytic domain of one subunit (subunit A) binds the tRNA’s acceptor stem and catalyzes aminoacylation, while the intermediate α-helical and C-terminal domains of the other subunit (subunit B) bind the tRNA’s anticodon and variable arms. Although TyrRS homodimers contain two active sites, only a single tRNATyr is bound and charged, a phenomenon known as “half-sites reactivity” (12).
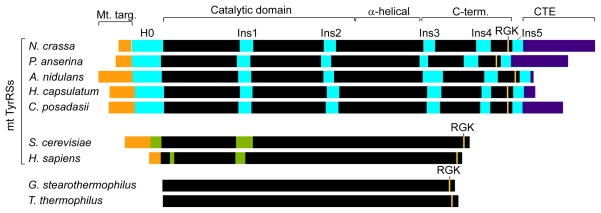
Mt TyrRSs are structurally homologous to the bacterial enzymes, and CYT-18 uses both its N-terminal catalytic and C-terminal tRNA-binding domains to bind group I intron RNAs (13–15). However, group I intron splicing activity has been found only for the mt TyrRSs of Pezizomycotina, filamentous fungi that includes the model organisms Neurospora crassa, Aspergillus nidulans, and Podospora anserina, as well as important human pathogens, such as Histoplasma capsulatum, Coccidioides posadasii, and Aspergillus fumigatus (2). The acquisition of group I intron splicing activity by the mt TyrRSs of these fungi can be traced to a series of structural adaptations in different regions of the protein, including a number of small “insertions”, which occurred during or after the divergence of Pezizomycotina from yeast (2, 16). As illustrated in Figure 1 for several representative examples, these Pezizomycotina-specific insertions include an α-helical N-terminal extension (H0), two small insertions in the catalytic domain (Ins1 and Ins2), three additional insertions in the C-terminal domain (Ins3, 4, and 5), and a variable length C-terminal extension (CTE). Studies with CYT-18 showed that the N-terminal domain insertions H0, Ins1 and Ins2 are required for group I intron splicing but not TyrRS activity and form part of a new group I intron-binding site distinct from that which binds tRNATyr (14, 16, 17). A co-crystal structure of a splicing active C-terminally truncated CYT-18 protein (CYT-18/Δ424–669) with a group I intron RNA (the bacteriophage Twort orf142-I2 ribozyme) revealed key features of the RNA-protein interface and showed that H0, Ins1, and Ins2 bind directly to the group I intron catalytic core in position to stabilize key tertiary interactions (18).
Figure 1.
Comparison of splicing-active Pezizomycotina mt TyrRSs with non-splicing bacterial and mt TyrRSs. The Pezizomycotina mt TyrRSs are distinguished by a series of insertions, including an α-helical N-terminal extension H0, Ins1 and Ins2 in the catalytic domain, and Ins3–5 in the C-terminal domain. Other fungal mt TyrRSs contain an Ins1 and sporadic insertions at the positions of Ins2 and 3, but with no significant sequence similarity to the Pezizomycotina mt TyrRS insertions. Black, regions of the canonical TyrRSs fold; orange, mt targeting sequences; cyan, Pezizomycotina-specific insertions; violet, C-terminal extensions (CTE); green, insertions relative to bacterial sequences in the yeast and human mt TyrRSs.
Thus far, there has been relatively little information about how CYT-18’s C-terminal domain contributes to group I intron splicing or about the structure and function of the C-terminal domain insertions. Although a C-terminally truncated CYT-18 protein consisting of the N-terminal catalytic and intermediate α-helical domains can splice many group I introns, the C-terminal domain contributes to group I intron binding and is essential for splicing some introns, e.g., the N. crassa mt large ribosomal subunit (Nc mt LSU) intron (14). Genetic studies showed that CYT-18’s C-terminal domain is needed to compensate for certain structural mutations that impair self-splicing of group I intron RNAs, including mutations that weaken two key long-range tertiary interactions: L9-P5, which helps establish the correct relative orientation of the two catalytic core domains, and L2-P8, which helps position the P1 helix containing the 5’-splice site at the RNA’s active site (15). Additionally, small deletions within the catalytic domain’s Ins2, which binds near the L9-P5 tetraloop-tetraloop receptor interaction, have more severe effects in the C-terminally truncated CYT-18/Δ424–669 protein than in the full-length protein, suggesting that C-terminal domain binding can compensate for loss of some N-terminal domain interactions (17). Together, these findings suggest that CYT-18’s N- and C-terminal domains both contribute to group I intron binding, with greater or lesser dependence on the C-terminal domain reflecting different structural defects that must be compensated for in each intron. Site-directed hydroxyl-radical cleavage experiments showed that CYT-18’s C-terminal domain binds near P6–P6a, P3–P8 and P5 in the Nc ND1 intron and near P6–P6a, P2, P4 and P5 in the Nc mt LSU intron (19). While CYT-18’s C-terminal domain contributes to the splicing of a number of group I introns, it inhibits the second step of splicing in some heterologous group I introns, suggesting that it co-evolved to function optimally with N. crassa mt group I introns (20).
The C-terminal domain of CYT-18, like those of most bacterial TyrRSs, has been intractable to X-ray crystallography in the full-length protein, presumably due to its attachment via a flexible linker that impedes crystallization in a specific orientation (8). Here, we used heteronuclear multidimensional NMR to determine the solution structure of the closely related but smaller C-terminal domain of the splicing-active Aspergillus nidulans (An) mt TyrRS. The structure confirmed the S4-like fold, but with Pezizomycotina-specific features, including the C-terminal domain insertions, which potentially contribute to novel functions. Modeling indicated that flexible attachment of the C-terminal domain is critical for its ability to interact with mt tRNATyr and group I intron RNAs on opposite sides of the catalytic domain. Surprisingly, however, NMR experiments showed that the C-terminal domains of the full-length An mt TyrRS and GeoBacillus stearothermophilus (Gs) TyrRSs do not tumble independently, implying that their attachment to the remainder of the protein is less flexible than was believed previously.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The DNA coding sequence of the A. nidulans mt TyrRS C-terminal domain (residues 455–617 with an additional initiating methionine) was PCR amplified from the previously described expression construct for the full-length An mt TyrRS (2) and cloned between the NcoI and BamHI sites of pET11d (Novagen). For protein expression, the resulting plasmid was transformed into E. coli strain BL21(DE3), grown to O.D.550 = ~0.5, and induced with 0.3 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). The polypeptide was purified using cation exchange and size-exclusion chromatography, as described for the full-length An mt TyrRS and CYT-18 proteins (2). The full-length An mt TyrRS used for NMR was expressed and purified as described (2). The Gs TyrRS ORF (a gift of Dr. Eric First, Louisiana State University Health Sciences Center, Shreveport, LA) was cloned as an N-terminal maltose-binding protein (MBP) fusion in the vector pMAL-C2T, expressed in E. coli BL21(DE3), and purified by amylose-affinity chromatography. The fusion protein was cleaved overnight at 4 °C with tobacco etch virus (TEV) protease, and the Gs TyrRS was separated from MBP by size-exclusion chromatography.
To prepare 15N and 13C/15N isotope-enriched proteins, Cells were grown as described above, but with M9 minimal media containing 0.5 g/L 15N ammonium chloride and/or 2.0 g/L 13C glucose (Cambridge Isotope Laboratories) as the source of nitrogen and carbon, respectively. The Cells grew more slowly in M9 minimal medium than in LB broth and were typically induced for 24–30 h.
NMR Spectroscopy
NMR spectra were recorded at 25 °C using a 500 MHz Varian Inova spectrometer equipped with a triple-resonance probe and a z-axis pulsed field gradient. NMR samples typically contained 0.5–1.0 mM of protein in 10 mM potassium phosphate buffer (pH 7.0), 100 mM KCl and 90% H2O/10% D2O. Backbone resonance assignments were obtained using standard triple-resonance NMR methods. Specifically, three-dimensional HNCA, HNCACB and HN(CO)CACB, HNCO and HACACBCO spectra were used to correlate the backbone amide protons with the N, Cα, C, and Cβ signals of the same and adjacent amino acids. Side-chain resonance assignments were obtained using three-dimensional 15N-1H-1H HSQC-TOCSY and 13C-1H-1H HCCH-TOCSY spectra, and two-dimensional homonuclear 1H-1H 2QF-COSY and TOCSY spectra. NOE cross peaks were detected using two-dimensional 1H-1H NOESY spectra acquired with mixing times of 80 ms and 150 ms, and three-dimensional 1H-1H-15N NOESY-HSQC and 1H-1H-13C NOESY-HSQC spectra with mixing times of 100 ms. Although acquired at only 500 MHz proton frequency, the NOE spectra critical to the structure analysis were acquired with high digital resolution; the three-dimensional 1H-1H-15N NOESY-HSQC and 1H-1H-13C NOESY-HSQC spectra contained 320 and 112 real points in the second and third dimensions, respectively, with total acquisition times of 120 h each. NMR data were processed using NMR-Pipe (24). 15N-1H heteronuclear NOE and 15N T1 and T2 relaxation times for the C-terminal domain of the A. nidulans mt TyrRS were measured using pulse sequences that included gradient selection, sensitivity enhancement, and pulses for minimizing saturation of solvent water (21). The 15N-1H heteronuclear NOE was measured by a comparison of spectra acquired with either a 5-sec delay between each free induction decay or a 2-sec delay followed by a 3-sec series of 120° nonselective 1H pulses. For T1 relaxation measurements, two-dimensional spectra with relaxation delays of 10, 260, 510, 760, and 1010 msec were obtained; for T2 relaxation measurements, two-dimensional spectra with relaxation delays of 29, 58, 87, 116, and 145 msec were acquired. For both T1 and T2 measurements, the relaxation delay between each acquisition of the free induction decay was 3 sec. 1H, 15N and 13C chemical shifts were referenced as recommended (22), with 1H chemical shifts measured relative to internal 2,2-dimethyl-2-silapentane-5-sulfonate (DSS) at 0 ppm. The 0 ppm frequencies for 13C and 15N were determined by multiplying the 0 ppm 1H reference frequency by 0.251449530 and 0.101329118 respectively.
Structure Calculations
The structure of the C-terminal domain the A. nidulans mt TyrRS was determined using the restrained simulated annealing protocol within CNS version 1.2 (23), with structural restraints provided by the NMR data. Cross-peaks observed in the NOE spectra were used to assign inter-proton distance restraints of < 3.6 Å, < 4 Å, < 5.0 Å and < 6.0 Å, depending on the cross peak intensity. Pseudoatom corrections were applied to the distance restraints as follows: NOEs from valine or leucine methyl groups that were not stereospecifically assigned were measured from the center of the two methyl groups, and 2.5 Å was added to the interproton distance. For NOEs involving other methyl protons, distances were measured from the center of the methyl group and an additional 1.0 Å was added to the interproton distance. For NOEs involving methylene protons with no stereospecific assignment, distances were measured from the center of the methylene group and 0.7 Å was added to the interproton distance. For NOEs involving δ and ε protons of tyrosine rings, distances were measured from the center of the two δ protons (or ε protons) and 2.4 Å was added to the interproton distance. Backbone dihedral ϕ and ψ angle restraints (to a 60° range) were included for residues that are clearly within regions of regular α-helices and β-strands. The α-helices and β-strands were identified by their characteristic NOE cross-peak patterns, chemical-shift values of the Cα, Cβ, C, and Hα nuclei, and patterns of protection of the amide protons from exchange with the solvent upon transfer of the sample to D2O. The PREDITOR torsion angle analysis program (24) was used to assign ϕ and ψ angle restraints for backbone atoms outside of the regions of regular α/β structure, based on the chemical shifts of Cα, N, HN, Hα, Cβ and C. Hydrogen bonds were defined using distance bounds for amide protons that were clearly located within the regions of regular β-sheet and α-helical structure. To insure that the full range of structures consistent with the NMR restraints was sampled, an initial set of 24 different structures was generated, starting from an extended peptide conformation and using a simulated annealing protocol with dihedral angle and secondary structure restraints only. These diverse structures were then used as starting points for refinement via restrained simulated annealing with different initial trajectories, subject to the full set of NMR-derived restraints. A set of refined conformers having the lowest energy was retained for final analysis and evaluation using Procheck-NMR (25). These final structures are a fair representation of the full range of structures that are consistent with the experimental data. These structures also maintain reasonable molecular geometry and have no NOE-derived distance constraint violations greater than 0.5 Å. Structural statistics are summarized in Table S1. Order parameters were determined from the 15N-1H NOE, and 15N T1 and T2 relaxation data using the model free method (25, 26) implemented using the Modelfree 4.2 program (27). All structural visualizations and manipulations were done in PyMol (28).
RESULTS
Structure of the A. nidulans mt TyrRS C-terminal Domain
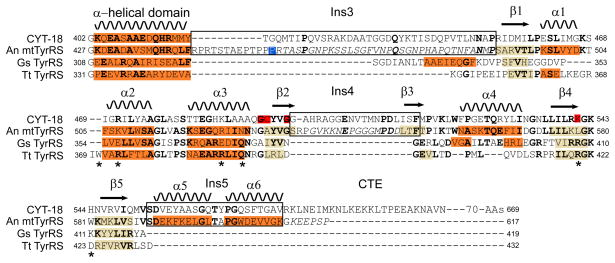
We chose the An mt TyrRS for NMR analysis because it has the smallest C-terminal domain of the Pezizomycotina mt TyrRSs that we have examined, reflecting that the CTE following Ins5 is much shorter in the An mt TyrRS than in the other proteins (8 amino acids compared to 103 amino acids for CYT-18; Figures 1 and 2). However, Aspergillus spp. mt TyrRSs have a larger Ins3 with a proline-rich region that is not present in other Pezizomycotina mt TyrRSs (Figure 2) (2). Because we had observed significant proteolysis of the full-length An mt TyrRS within the proline-rich region of Ins3 (data not shown), we designed a construct that begins within Ins3 downstream of this region. The coding sequence for this construct (residues 455–617) was cloned in the expression vector pET-11d, over-expressed in E. coli strain BL21(DE3), and purified as described in EXPERIMENTAL PROCEDURES. The construct produced large amounts of soluble protein that behaved as a monomer in size-exclusion chromatography and could be concentrated to over 100 mg/ml (data not shown).
Figure 2.
Structure-based sequence alignment of the Nc mt TyrRS (CYT-18) and An mt TyrRS with the Gs and Tt TyrRS C-terminal domains. The alignment begins with the last helix of the intermediate α-helical domain. Secondary structural elements in the An mt TyrRS C-terminal domain are shown at the top, and α-helices and β-strands observed in NMR or X-ray crystal structures (this work and (9, 11)) are highlighted in orange and beige, respectively, in the alignments. Residues in bold are conserved in two or more of the aligned proteins, and residues in italics are flexible in the An mt TyrRS C-terminal domain NMR structure based on 15N relaxation. The blue highlighted residue in the An mt TyrRS (P455) is the first residue in the NMR construct whose structure is described here, and red highlighted residues in CYT-18 are those that were EPD-Fe conjugation sites in site-directed hydroxyl radical cleavage experiments (see Figures 7, 8 below). Asterisks below the Tt TyrRS sequence indicate amino acid residues that contact tRNATyr in the co-crystal structure (11). The boundaries of the Pezizomycotina-specific insertions are redefined relative to those in (2) based on the structural alignment.
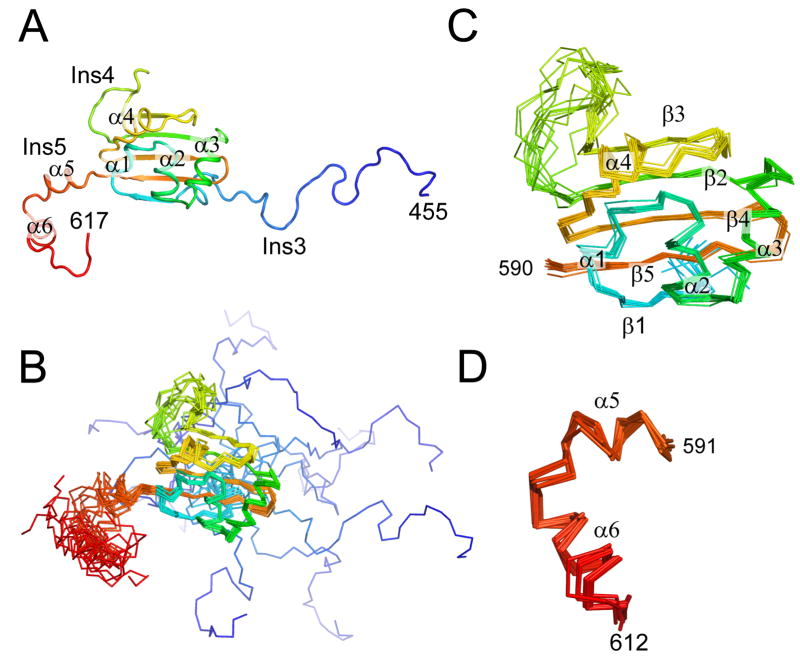
The An mt TyrRS C-terminal domain was well-suited for NMR studies, with resonance dispersion and line widths typical of a mostly globular polypeptide with a molecular weight of 17 kDa (Figure S1, S2). The structure was determined by restrained simulated annealing using NOE cross-peaks to provide inter-proton distance restraints. As expected from its homology to other TyrRSs, the An mt TyrRS C-terminal domain has an S4-like fold with two α-helices (α2 and α3 forming a helical hairpin and five β-strands forming a β-sheet with mixed parallel and antiparallel strands (Figure 3, S3). Structure-based sequence alignments (Figure 2) and superpositioning (Figure 4) shows that the arrangement of the β-sheet and α–helices comprising the S4 fold is similar to that of previously characterized bacterial TyrRSs.
Figure 3.
NMR structure of the An mt TyrRS. (A) Cartoon diagram of a representative structure from the NMR ensemble. (B) Superposition of 12 structures of the C-terminal domain of the A. nidulans mt TyrRS that satisfy the NMR-derived constraints equally well. The positions of the β-strands and α-helices that form the core of the S4 fold are well-defined by the NMR data, while the locations of the termini and the loop of Ins4 between β2 and β3 are less certain. (C) Superposition of residues 490–590 from the 12 structures showing that the core of the S4 fold is well-defined by the NMR constraints. (D) Superposition of α5 and α6 of Ins5 from the 12 structures showing that the positions of these two helices are well-defined relative to each other, although their positions relative to the rest of the domain are not as well-defined (panel B). The color is ramped blue to red from the N- to C-terminus.
Figure 4.
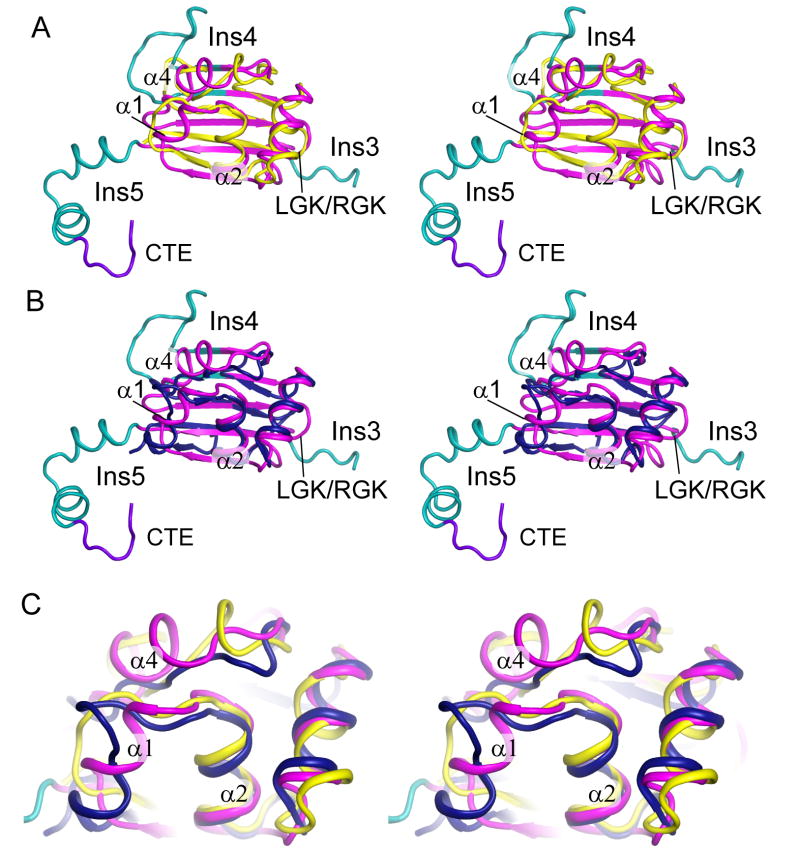
The An mt TyrRS C-terminal domain structure is similar to that of bacterial TyrRSs. (A) Stereoview superposition of the An mt TyrRS C-terminal domain NMR structure (magenta/cyan/purple) with that of the Gs TyrRS C-terminal domain (yellow; (8)). (B) Stereoview superposition of the An mt TyrRS C-terminal domain with the Tt TyrRS C-terminal domain (blue) from the co-crystal structure with tRNATyr (11). (C) Magnified view of α-helical bundle region of the An mt TyrRS C-terminal domain with superposed bacterial TyrRS structures. α1 differs from the shorter helical region in the Tt TyrRS and is not present in the Gs TyrRS, and α4 is not present in either of the bacterial TyrRSs.
C-terminal Domain Insertions and Novel α-Helical Bundle Structure
The solved structure includes a large portion of Ins3 (residues 455–489) and all of Ins4 and Ins5 (residues 533–552 and 590–610, respectively; Figure 3). The structural alignments reveal that Ins3 is an expansion of the linker region between the N- and C-terminal domains, which begins after the last α-helix of the intermediate domain and extends to first structural element (β1) of the S4 fold (Figure 2). Except for the proline-rich region characteristic of Aspergillus spp. mt TyrRSs, we could find no clear sequence conservation between the linker regions of different Pezizomycotina mt TyrRS or between the linker regions of Pezizomycotina mt TyrRSs and those of bacterial or other mt TyrRSs. The portion of the Ins3/linker present in the NMR construct is not well defined structurally by the NMR-derived distance constraints (Figure 3B), and 15N NMR relaxation data confirmed that these residues are flexible in solution (Figures S4, S5). Further, a longer An mt TyrRS C-terminal domain construct (residues 427–617) that included the last α-helix of the α-helical domain and all of the Ins3/linker exhibited N-terminal degradation (possibly at the same position as the full-length protein), but no substantial changes in 15N or 1H chemical shifts that would indicate the formation of additional stable secondary structure (data not shown). This structural flexibility of the Ins3/linker could reflect that contacts with other protein regions are needed to stabilize a higher-order structure. However, the lack of defined structure could also be an inherent and functionally important characteristic of the linker regions of TyrRSs. Consistent with this hypothesis, the linker region of the Tt TyrRS lacks regular secondary structure even when ordered in the co-crystal structure by binding of the C-terminal domain to tRNATyr (11).
Ins4 is shown by the NMR structure to be comprised of an expanded loop between β2 and β3 (residues 538–548) and short extensions of both β2 and β3 compared to the bacterial TyrRSs (Figures 2, 3, 4). Other fungal mt TyrRSs that do not function in splicing have smaller (2–6 residue) insertions in this region relative to bacterial TyrRSs, and the mt TyrRSs of several fungal species (Debraryomyces hansenii, Pichia stipitis, Candida albicans) may also have an extended β3, as judged by sequence similarity (2). 15N NMR relaxation measurements indicate that the Ins4 loop is flexible in solution.
Ins5 follows β5 and has two well-defined α–helices (α5 and α6) that are not present in non-splicing TyrRSs. The orientation of α5 and α6 relative to each other is defined by the NMR data (Figure 3D), but their orientation relative to the other elements of the C-terminal domain has greater uncertainty (Figure 3B). α6 extends beyond our previous definition of Ins5 based on sequence conservation in the Pezizomycotina mt TyrRSs (2). Redefining Ins5 to include all of α6 leaves the An mt TyrRS CTE at only 8 residues long, and these residues appear flexible in solution based on 15N NMR relaxation (Figure S4). Sequence conservation with other Pezizomycotina mt TyrRSs ends three amino acids into α6, just after W604 in the An mt TyrRS. The CTE downstream of α6 is highly variable in length and sequence between the different Pezizomycotina mt TyrRSs (2).
In addition to the Pezizomycotina-specific insertions, the An mt TyrRS C-terminal domain contains two α-helices, α1 and α4, that are either missing or abbreviated in bacterial TyrRS structures. α1 is composed of residues 497–502 and was originally identified as part of Ins3 (2). This helix is absent in the Gs TyrRS NMR structure and consists of only three residues in the Tt TyrRS X-ray crystal structure. α4 is composed of residues 560–567 and was originally identified as a part of Ins4 (2). The Gs TyrRS structure contains two short helical elements in this region, but both bacterial TyrRSs lack an extended helical structure. Hydrophobic side chain packing between α1, α4 and the conserved helix α2 forms a novel α-helical bundle that is not present in the bacterial TyrRS structures (Figure 4C). Modeling below suggests that this α-helical bundle could contribute to binding both tRNATyr and group I intron RNAs.
C-terminal Domain Motion in the Full-Length TyrRS
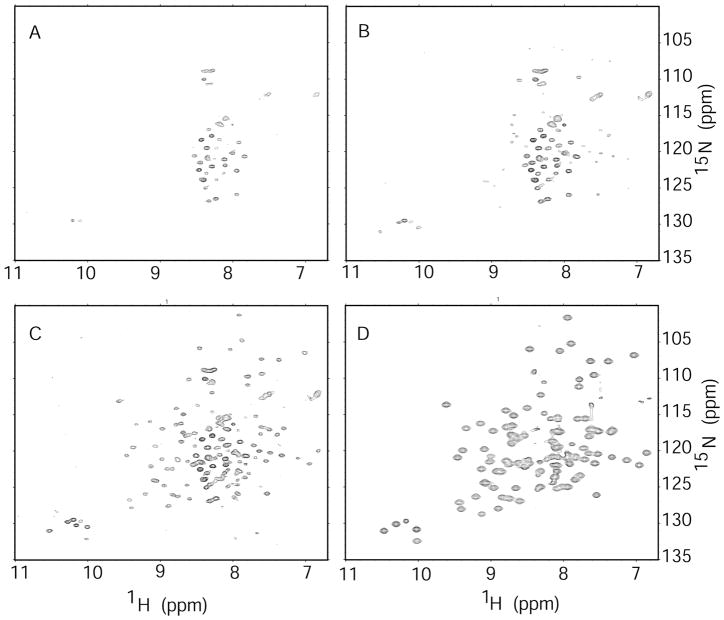
The flexible attachment of the C-terminal domain is potentially critical for its ability to interact with tRNATyr and group I introns RNAs, which bind on different sides of the catalytic domain. Here, we used 15N-1H two-dimensional NMR to investigate the extent to which the motions of the C-terminal domain are independent of other regions in the full-length TyrRS (Figure 5). Globular single-domain proteins with molecular weights over 60 kDa typically yield broad NMR signals due to relatively slow molecular tumbling times and rapid NMR transverse relaxation rates. However, smaller domains that are flexibly attached to larger domains can be observed if the domains tumble independently (29). Initially, the spectrum of the full-length protein lacked any resemblance to that of the isolated C-terminal domain. The relatively small number of visible peaks presumably arose from the amide groups of relatively mobile loops and termini (Figure 5). Over a period of 24 h, the peaks that are characteristic of the C-terminal domain began to appear in the spectrum (Figure 5B, C). SDS-PAGE showed that the full-length protein undergoes proteolysis during this period (data not shown), apparently releasing the C-terminal domain to move independently and resulting in observable NMR signals. The lack of signal prior to proteolysis suggests that the motion of the An mt TyrRS C-terminal domain is significantly restricted when attached to the remainder of the protein.
Figure 5.
A time series of two-dimensional 15N-1H spectra of the full-length An mt TyrRS. The resonances of the free C-terminal domain appear as the protein degrades. Spectrum A was acquired soon after sample preparation, spectrum B after ~10 h, and spectrum C after 24 h. Spectrum D is that of the purified C-terminal domain for comparison. The lack of observable resonances from the C-terminal domain within the intact full-length protein indicates that the tumbling of the C-terminal domain is restricted while it is part of the full-length protein.
The relatively restricted orientations of the two domains could be a novel structural adaptation that enables the An mt TyrRS to carry out its two RNA-binding functions. To examine this possibility further, the non-splicing Gs TyrRS protein was investigated using the same methods. The one-dimensional 1H NMR spectrum shows mostly broad peaks, consistent with a globular dimeric protein with the expected molecular weight of 94 kDa (Figure S6). The 15N-1H two dimensional correlated spectrum of the full-length Gs TyrRS shows only a few peaks with chemical shifts near those typical of disordered structure, which presumably arise from the flexible portions of the protein (Figure S7). There were no disperse NMR peaks observed that could be attributed to the C-terminal domain after any length of time, indicating that the motion of the C-terminal is restricted when in the context of the full-length TyrRS. Unlike the An mt TyrRS, the Gs TyrRS protein did not show significant degradation over several days. These results indicate that even in a non-splicing bacterial TyrRS, the motions of the C-terminal domain are restricted by attachment to the N-terminal domain.
Modeling of An mt TyrRS C-terminal Domain/tRNATyr Interactions
To identify potential tRNA-binding regions of the An mt TyrRS C-terminal domain, we superposed the NMR structure of the isolated domain on the corresponding region of the Thermus thermophilus (Tt) TyrRS-tRNATyr co-crystal structure (11) (Figure 6). The Tt TyrRS recognizes its cognate tRNA primarily via interactions between the α-helical hairpin and the tRNA’s anticodon stem and variable arms, and between the β-hairpin containing the RGK loop and the distal end of the anticodon stem and loop (11). Figure 6A shows that these regions of the An mt TyrRS S4 fold superpose well on the co-crystal structure, consistent with similar roles in tRNA recognition. The α-helical hairpin (α2 and α3 in the An mt TyrRS) is located between the variable and anticodon arms, with α3 positioned to contact both arms and α2 positioned to contact the variable arm. The β-hairpin is located beneath α3, with the LGK cognate of the RGK sequence positioned to contact the distal end of the anticodon stem and loop.
Figure 6.
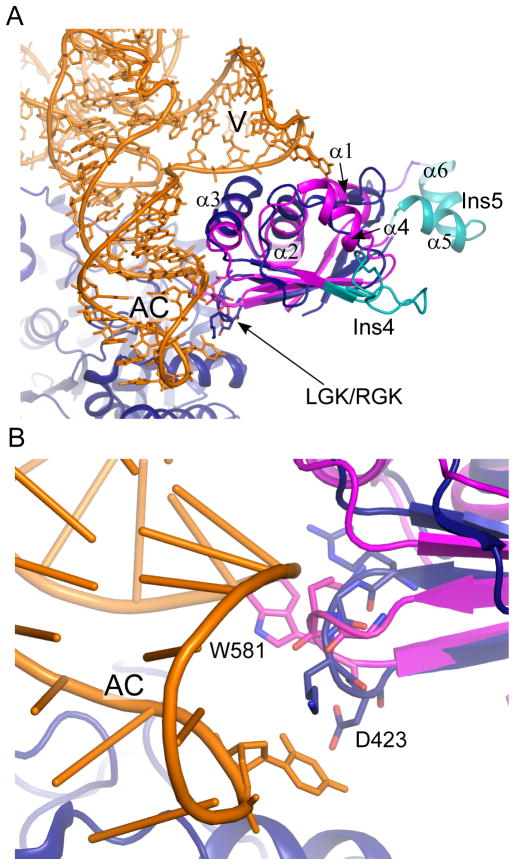
Model of tRNATyr binding by the An mt TyrRS C-terminal domain. (A) Superposition of the An mt TyrRS C-terminal domain (magenta/cyan) on the Tt TyrRS co-crystal structure (11) (blue, orange). Unordered residues preceding β1 in the An mt TyrRS structure have been removed for clarity. (B) The RGK β-hairpin contacts the anticodon arm of tRNATyr. Residues 420-RGKD-423 from Tt TyrRS and the equivalent residues (578-LGKW-581) from the An mt TyrRS are shown as sticks. D423 of the Tt TyrRS is equivalent to W581 of the An mt TyrRS. Ac, anticodon arm; V, variable arm.
In the An mt TyrRS NMR structure, the RGK β-hairpin closely resembles that seen in the absence of bound tRNA in the Gs TyrRS solution structure (Figure 6B) (9). In the tRNA-bound state, seen in the Tt TyrRS co-crystal structure, the RGK β-hairpin contacts the anticodon arm stem and loop of tRNATyr with a non-phylogenetically conserved aspartic acid residue (D424) forming a hydrogen bond with the central base of the anticodon (11). In the An mt TyrRS, this aspartic acid is replaced by a solvent-exposed aromatic residue, W581, which was indicated by sequence harmony analysis to be functionally important (2) and could play a similar role in contacting the anticodon in the Pezizomycotina mt TyrRSs.
The model shows that positioning of the S4 fold of the An mt TyrRS to bind tRNATyr as in the bacterial enzymes places α1 and α4 of the novel α-helical bundle near the distal end of the tRNA’s variable arm (Figure 6B). The variable arms of all Pezizomycotina and most fungal mt tRNATyr are one to three nucleotides longer than those of bacterial tRNAsTyr (30), and it is thus possible that the α-helical bundle is a Pezizomycotina- or fungal-specific adaptation that enables extended binding to this longer variable arm.
Notably, the model also shows that when the S4 fold of the An mt TyrRS is positioned to bind tRNATyr, Ins4 and 5 are located on the opposite side of the C-terminal domain, positioned well away from the tRNA. We made no attempt to model the flexible Ins3/linker region, although part of it could be located similarly to the smaller flexible linker in the bacterial enzymes. The linker region of the Tt TyrRS does not contact tRNATyr in the co-crystal structure (11), but the An mt TyrRS Ins3/linker is considerably larger and we cannot exclude that it contributes to tRNA binding.
Modeling of An mt TyrRS C-terminal Domain/Group I Intron Interactions
The CYT-18/Δ424–669-Twort orf142-I2 ribozyme co-crystal structure showed that the group I intron RNA binds to the protein across the surface of the two subunits of the homodimer with most of the contacts made by one subunit (arbitrarily designated subunit B) and a smaller number of contacts made by the other subunit (subunit A) (18). By combining the NMR structure of the An mt TyrRS C-terminal domain with the co-crystal structure and distance restraints from biochemical data, we modeled how the C-terminal domain might contribute to binding group I introns.
Previous site-directed hydroxyl radical cleavage assays using Fe-EPD conjugated to single cysteine residues in CYT-18 identified two positions in the C-terminal domain (the native C-terminal cysteine, C494, and G493C) that cleaved the Nc ND1 intron at ten sites clustered in the P6–P6a and P3–P8 stacked helices and at two positions in P5 ((19) and Paukstelis, Coon, Lambowitz, unpublished results). These site-directed cleavages provide distance restraints with an upper limit of 25 Å from the cysteine residue to the cleaved nucleotide position (31). Using aligned amino acid positions between CYT-18 (G493, C494) and the An mt TyrRS (G529, A530) and equivalent nucleotide positions in the Twort and ND1 introns (based on their relative position in base paired regions), we determined orientations of the An mt TyrRS C-terminal domains that are consistent with these distance restraints in the context of the CYT-18/Δ424–669-Twort co-crystal structure.
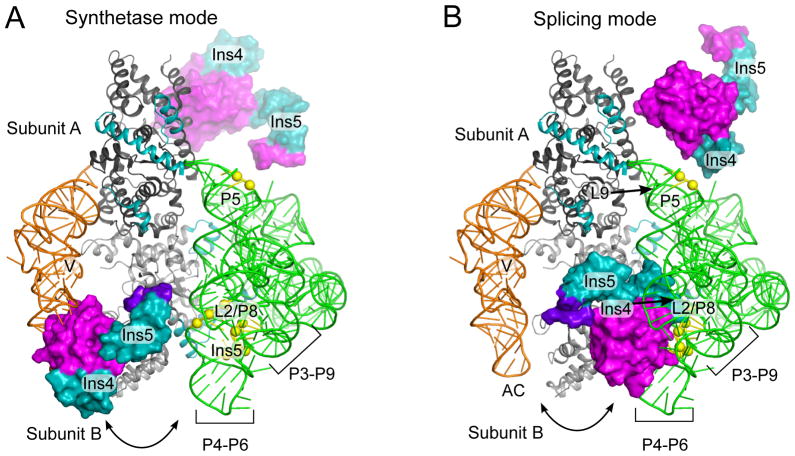
Manual rotations of the An mt TyrRS C-terminal domain starting in the tRNA-bound position resulted in orientations of the domain that met distance, spatial, and geometric restraints. One of these models is shown in Figures 7 and 8. A common feature of all the models was that the C-terminal domain must undergo a large positional shift to interact with the group I intron RNA and tRNATyr on opposite sides of the catalytic domain (Figure 7, Figure 8A,B). Globally, the models suggest that subunit B of the mt TyrRS acts as a clamp, with the N-terminal domain and Pezizomycotina-specific insertions H0 and Ins1 forming one side and the bottom of the clamp, and the C-terminal domain wrapping around to form another side (Figure 8B, C).
Figure 7.
The An mt TyrRS C-terminal domain must undergo a large shift on the flexible linker to bind tRNATyr and a group I intron RNA on opposite sides of the catalytic domain. The figure shows the An mt TyrRS C-terminal domain (magenta, cyan) appended to the CYT-18/Δ424–669-Twort co-crystal structure (18) (grey, green) with tRNATyr (orange) from T. thermophilus co-crystal structure (11). (A) Model for tRNA-binding (synthetase) mode. The An mt TyrRS C-terminal domain is positioned to match that of the Tt TyrRS in the Tt TyrRS-tRNATyr co-crystal structure (11). (B) Model for group I intron-binding (splicing) mode. The An mt TyrRS C-terminal domain is positioned to satisfy distance restraints from site-directed hydroxyl radical cleavage experiments in which EPD-Fe conjugated at CYT-18 positions G423C and C424 (G529 and A530 in the An mt TyrRS; not visible in this view) cleaved the bound Nc ND1 intron at P3[5′] bp 3–4; P6[3′] bp 3; J6/6a[3′] nt 1; P6a[5′] bp 1–3 ; P6a[3′] bp 1; P8[3′] bp 4–5, and P5[5′] bp 3–4 ((19) and Paukstelis, Coon, and Lambowitz, unpublished data). The yellow spheres show the cleaved group I intron backbone phosphate positions in the Twort intron. Pezizomycotina-specific Ins4 and Ins5 are in cyan. Ins3 is not shown.
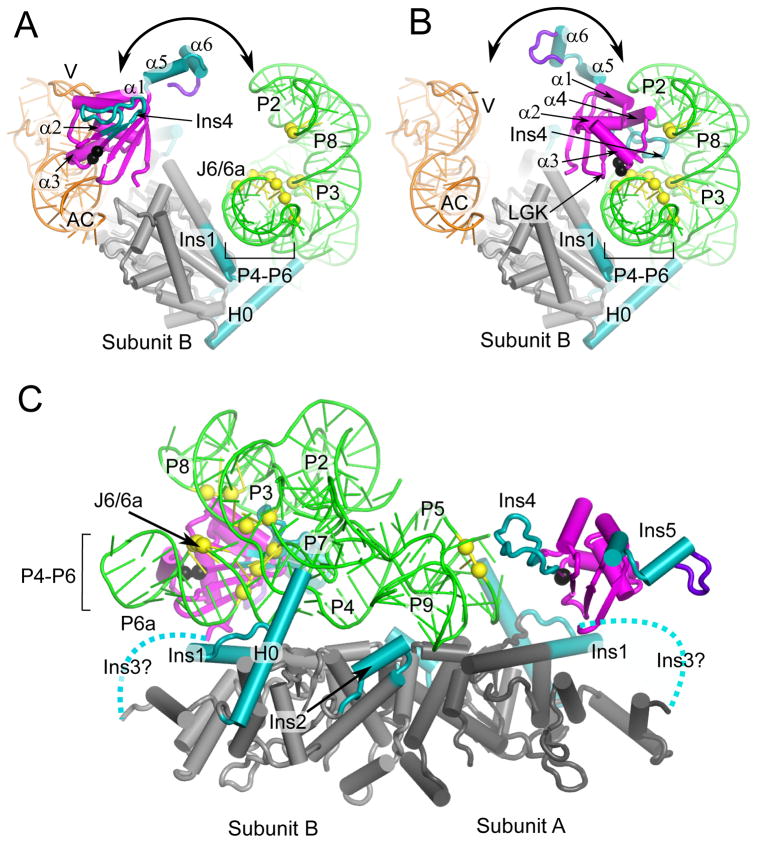
Figure 8.
The An mt TyrRS N- and C-terminal domain function together to clamp the two ends of the group I intron RNA. The models are the same as in Figure 7. (A) Model for subunit B in tRNA-binding mode viewed looking down the P4–P6 helical axis. (B) Model for subunit B in intron-binding mode viewed looking down the P4–P6 stacked helices. The view shows that the position of the C-terminal domain of subunit B satisfies distant restraints based on directed hydroxyl-radical cleavage experiments (see legend Figure 7) between G529 and A530 (black spheres) and Twort ribozyme phosphodiester backbone positions in P6a: 95–97; J6/6a-P6: 112–114; P3: 162, 163; P8: 178, 179 (yellow spheres). (C) Model for group I intron binding viewed perpendicular to P4–P6 domain axis showing C-terminal domains from both subunits. The view shows that the position of C-terminal domain of subunit A satisfies distance restraints between A530 (black sphere) and phosphodiester backbone cleavages at P5a positions 61 and 62 (yellow spheres). Dashed lines represent the Ins3/linker connecting the N- and C-terminal domains. The intron RNA peripheral structures P9.2, P7.1, P7.2 have been removed for clarity.
In the model shown, the An mt TyrRS residues corresponding to Fe-EPD conjugation sites (black spheres) are brought near the phosphodiester backbone residues corresponding to hydroxyl radical cleavage sites (yellow spheres). A consequence of satisfying the 25-Å distance restraints is that residues involved in tRNA recognition in the α-helical hairpin and RGK β-hairpin must be positioned away from the group I intron (Figure 8B). In the orientation shown, the C-terminal domain is located between P6–P6a, P2, and P8, with the loop of Ins4 positioned just below the L2/P8 tetraloop/receptor interaction and several of the loop residues near the phosphodiester backbone of the P4/P6 junction. Notably, α1 and α4 of the α-helical bundle, which may be involved in extended contacts with the variable arm of mt tRNATyr, are located close enough to potentially contact the intron RNA (α1 is near the P2 backbone and α4 is near the L2/P8 tertiary contact; Figure 8B). Thus, these elements may be Pezizomycotina adaptations that function in both tRNA and group I intron binding.
The two site-directed RNA cleavages in P5 of the ND1 introns from CYT-18 G493C and C494 (corresponding to An mt TyrRS G529 and A530) cannot be accounted for by binding of the C-terminal domain of subunit B and suggest that the C-terminal domain of subunit A also contributes to group I intron binding. Satisfying the distance restraints from CYT-18’s C494 position to the cleaved RNA nucleotides (P5-bp 3–4 in the ND1 intron) places the C-terminal domain of subunit A near P5, with the loop of Ins4 near P9 (Figure 8C). The proximity of subunit A’s C-terminal domain to the L9/P5 tetraloop/tetraloop-receptor interaction may explain the ability of the full-length protein to partially compensate for deletions in Ins2, which is involved in orientating and stabilizing P9 (18).
Surprisingly, Ins5, which is the most highly conserved Pezizomycotina-specific insertion, is positioned away from the RNA substrate in the models for both the tRNA-bound and intron-bound states. However, the variability in position of the Ins5 helices in the NMR structure (Figure 3) could reflect that it is flexibly attached to the remainder of the C-terminal domain and could thus bind the intron RNA by assuming an orientation that was not sampled in the solution structure.
Attempts to further model C-terminal domain interactions by using similar site-directed hydroxyl radical cleavage data for the Nc mt LSU intron mapped onto the Twort intron structure were unsuccessful, likely due at least in part to the disparate sizes and structures of these introns. The Nc mt LSU and ND1 introns have five C-terminal domain cleavage sites in common (P6a[5′] bp 1–3, J6/6a[3′] nt 1, and P6[3′] bp 3), but the Nc mt LSU intron had additional cleavages from K540C in P6-P6a, which were not seen in the ND1 intron; from G493C in P2, which is not present in the ND1 intron; and from G493C and G497C in P4, which were consistently outliers from the 25-Å distance restraint. These additional cleavages as well as previous RNA footprinting data (5) raise the possibility that CYT-18’s C-terminal domain interacts somewhat differently with different group I introns, including more extensive interactions with the P4–P6 domain of the Nc mt LSU intron.
DISCUSSION
Here, we determined an NMR structure of the C-terminal domain of the group I intron splicing An mt TyrRS, and we used this structure to construct models for tRNATyr and group I intron binding by the C-terminal domains of Pezizomycotina mt TyrRSs. The structure shows that the An mt TyrRS C-terminal domain has an S4-like fold similar to those of bacterial TyrRSs, but with additional features, including three Pezizomycotina-specific insertions, Ins3–5, and a novel α-helical bundle, appended to this conserved structural framework. The regions of the S4 fold that bind tRNATyr in bacterial TyrRSs are conserved in the An mt TyrRS, and modeling suggests that they function similarly in binding of the anticodon and long variable arms of mt tRNATyr. The modeling of group I intron interactions to fit previous directed hydroxyl-radical cleavage data with CYT-18 suggests that the C-terminal domains of both subunits of the homodimeric protein interact with different ends of the intron RNA and that the C-terminal domain must undergo a large shift on the flexible linker to interact with tRNATyr and group I intron RNAs on different sides of the catalytic domain. The modeling also indicates that Ins4 and possibly Ins5 may function in group I intron splicing, while the α-helical bundle may bind both tRNATyr and the intron RNA. The previous finding that a group I intron RNA competitively inhibits tRNATyr aminoacylation by CYT-18 (4), which was difficult to explain by the non-overlapping RNA-binding sites in the N-terminal domain, is readily accounted for by the mutually exclusive binding of the C-terminal domain to tRNATyr and group I intron RNAs.
The NMR structure provides the first detailed view of the Pezizomycotina-specific insertions. Ins3 is seen to be an expansion of the unstructured linker between the α-helical and C-terminal domains; Ins4 consists largely of an expanded loop between β2 and β3; and Ins5 consists of two α-helices (α5 and α6) that are not found in non-splicing TyrRSs but are conserved in sequence among Pezizomycotina mt TyrRSs (2). These C-terminal α-helices are the last structured elements preceding the CTE, which varies in length and sequence among Pezizomycotina mt TyrRSs and is dispensable for protein function in vitro (32). As was found for the N-terminal domain insertions (2), the C-terminal domain insertions in Pezizomycotina mt TyrRSs appear to have evolved from unstructured loops or termini in non-splicing TyrRS. Adaptive mutations in unstructured loops or termini that do not affect the overall protein fold may be a general mechanism by which proteins can progressively acquire new functions (see Discussion ref. 2).
Modeling of tRNATyr interactions by fitting the An mt TyrRS C-terminal domain to the Tt TyrRS-tRNATyr co-crystal structure suggests that the α-helical hairpin of the S4 fold binds the tRNA’s variable and anticodon arms and the β-hairpin containing the RGK/LGK motif binds the anticodon, as in bacterial TyrRSs. This positioning of the S4 fold to bind tRNATyr places two distinctive helices (α1 and α4) of the novel α-helical bundle in position to extend binding to the distal end of the variable arm, possibly reflecting co-evolution with the variable arm, which is 1–3 nt longer in Pezizomycotina and most other fungal mt tRNAsTyr than in bacterial tRNAsTyr. By contrast, Ins4 and Ins5 are located on the opposite side of the S4 fold away from the tRNA. The location of the unstructured Ins3/linker region was not modeled, but may in part correspond to that of the smaller linker region of bacterial TyrRSs, which does not bind tRNATyr (11).
Models for group I intron RNA binding were constructed by starting with the CYT-18/Twort co-crystal structure and positioning the An mt TyrRS C-terminal domain to fit previous site-directed hydroxyl radical cleavages obtained with CYT-18. Although the number of cleavages was insufficient to precisely model the C-terminal domain locations, the models indicate that the C-terminal domain of one subunit (subunit B) is located between P6-P6a, P2 and P8 and acts together with the N-terminal domain of that subunit to “clamp” the intron’s P4–P6 domain. To account for cleavages in P5 on the opposite end of the intron RNA, the C-terminal domain of the other subunit (Subunit A) must be located in proximity to the L9-P5 tertiary interaction. These locations explain previous findings that CYT-18’s C-terminal domain can suppress mutations that weaken both the L2-P8 and L9-P5 tertiary interactions (15) and that small deletions in Ins2, which helps orient P9, have more severe effects in the C-terminally truncated than in the full-length protein (17). The C-terminal domain can also compensate for mutations in several other regions of the intron that may indirectly affect the relative orientation of the core domains or positioning of P2 or P8 (15).
Although additional biochemical data are needed to draw firm conclusions about the function of the Pezizomycotina-specific insertions, in order to satisfy the distance restraints, Ins4 of subunit B must be positioned toward the intron near the L2/P8 interaction, while Ins5 of this subunit may have sufficient flexibility to contact the intron near L2. Notably, the models suggest that the α-helical bundle of subunit B, which may bind the variable arm of tRNATyr, could potentially bind P2 and/or P8 of the intron RNA. Thus, the α-helical bundle may be a Pezizomycotina TyrRS adaptation that binds both the intron RNA and tRNATyr.
The C-terminal domains of bacterial and mt TyrRSs are commonly described as being attached to the remainder of the protein via a “flexible peptide” (9), and this flexible attachment had been invoked to explain why the C-terminal domain is disordered in crystals of full-length TyrRSs (11). Thus, we were surprised to find that 15N-1H two-dimensional NMR experiments with the full-length An mt TyrRS and Gs TyrRS showed that the C-terminal domains of these enzymes do not tumble independently in solution, but instead exist in one or more preferred orientations relative to the remainder of the protein. These preferred orientations may reflect properties of the linker that restrain the C-terminal domain in a position that facilitates tRNA binding and/or transient interactions with the N-terminal domains that prevent independent tumbling on the time-scale of the NMR experiments. Our modeling indicates that the Pezizomycotinia mt TyrRS C-terminal domains must exist in at least two orientations to bind tRNATyr and group I intron RNAs on either side of the catalytic domain, and it is tempting to speculate that the longer Ins3/linker region in Pezizomycotina mt TyrRSs evolved to favor these two orientations.
Finally, our results underscore that the evolution of Pezizomycotina mt TyrRSs to function in splicing was a multistep process in which both the N- and C-terminal domains underwent multiple structural adaptations that contribute to binding group I intron RNAs. We speculated previously that the initial interaction between the mt TyrRS and a group I intron RNA involved the N-terminal catalytic domain, possibly with the region around Ins1, which is present in non-splicing fungal mt TyrRSs and binds the P4–P6 stacked helices of group I introns (2). The present results suggest an alternate scenario in which the initial interaction between the mt TyrRS and the group I intron was with the pre-existing RNA-binding site in the C-terminal domain, perhaps involving a region that recognizes similar structural features in the group I intron and tRNATyr. The flexible attachment of CYT-18’s C-terminal domain may have facilitated this interaction by enabling it to sample different binding orientations (11). With time, this initial C-terminal domain interaction with the group I intron RNA would become fixed by structural mutations in the intron RNA, resulting in dependence upon protein binding for RNA splicing. Such dependence would ensure continued proximity of the protein and RNA and provide the opportunity for more extensive interactions involving other protein and RNA regions. Both the N- and C-terminal domain first scenarios are compatible with the hypothesis of pre-adaptive/neutral evolution of biological complexity in which fortuitous interactions become fixed by mutations that result in structural dependence and are ratcheted into ever more complex macromolecular machines (33, 34).
Supplementary Material
Acknowledgments
We thank Mark Del Campo and Georg Mohr for comments on the manuscript, and Chun Geng for technical assistance.
Abbreviations
- An
Aspergillus nidulans
- Cp
Coccidioides posadasii
- EPD-Fe
(EDTA-2-aminoethyl)-2-pyridyl disulfide-Fe(III)
- Gs
GeoBacillus stearothermophilus
- Hc
Histoplasma capsulatum
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- LSU
large subunit rRNA
- MBP
maltose-binding protein
- mt
mitochondrial
- Nc
Neurospora crassa
- Pa
Podospora anserina
- TEV
tobacco etch virus
- Tt
Thermus thermophilus
- TyrRS
tyrosyl-tRNA synthetase
Footnotes
This work was supported by NIH grant GM37951 to A.M.L.
Supporting Information Available. Figures S1, S2, S4, S6, and S7 show NMR spectra of the An mt TyrRS C-terminal domain. Figure S3 shows a schematic diagram of the mixed parallel-antiparallel β-sheet. Figure S5 shows relaxation rates and order parameters determinations. Table S1 is a summary of refinement and structure statistics. This material is available free of charge on the Internet at http://pubs.acs.org.
References
- 1.Akins RA, Lambowitz AM. A protein required for splicing group I introns in Neurospora mitochondria is mitochondrial tyrosyl-tRNA synthetase or a derivative thereof. Cell. 1987;50:331–345. doi: 10.1016/0092-8674(87)90488-0. [DOI] [PubMed] [Google Scholar]
- 2.Paukstelis PJ, Lambowitz AM. Identification and evolution of fungal mitochondrial tyrosyl-tRNA synthetases with group I intron splicing activity. Proc Natl Acad Sci USA. 2008;105:6010–6015. doi: 10.1073/pnas.0801722105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohr G, Zhang A, Gianelos JA, Belfort M, Lambowitz AM. The Neurospora CYT-18 protein suppresses defects in the phage T4 td intron by stabilizing the catalytically active structure of the intron core. Cell. 1992;69:483–494. doi: 10.1016/0092-8674(92)90449-m. [DOI] [PubMed] [Google Scholar]
- 4.Guo Q, Lambowitz AM. A tyrosyl-tRNA synthetase binds specifically to the group I intron catalytic core. Genes Dev. 1992;6:1357–1372. doi: 10.1101/gad.6.8.1357. [DOI] [PubMed] [Google Scholar]
- 5.Caprara MG, Mohr G, Lambowitz AM. A tyrosyl-tRNA synthetase protein induces tertiary folding of the group I intron catalytic core. J Mol Biol. 1996;257:512–531. doi: 10.1006/jmbi.1996.0182. [DOI] [PubMed] [Google Scholar]
- 6.Vicens Q, Cech TR. Atomic level architecture of group I introns revealed. Trends Biochem Sci. 2006;31:41–51. doi: 10.1016/j.tibs.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 7.Chadee AB, Bhaskaran H, Russell R. Protein roles in group I intron RNA folding: the tyrosyl-tRNA synthetase CYT-18 stabilizes the native state relative to a long-lived misfolded structure without compromising folding kinetics. J Mol Biol. 2010;395:656–670. doi: 10.1016/j.jmb.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guez V, Nair S, Chaffotte A, Bedouelle H. The anticodon-binding domain of tyrosyl-tRNA synthetase: state of folding and origin of the crystallographic disorder. Biochemistry. 2000;39:1739–1747. doi: 10.1021/bi992382v. [DOI] [PubMed] [Google Scholar]
- 9.Guijarro JI, Pintar A, Prochnicka-Chalufour A, Guez V, Gilquin B, Bedouelle H, Delepierre M. Structure and dynamics of the anticodon arm binding domain of Bacillus stearothermophilus tyrosyl-tRNA synthetase. Structure. 2002;10:311–317. doi: 10.1016/s0969-2126(02)00699-8. [DOI] [PubMed] [Google Scholar]
- 10.Carter P, Bedouelle H, Winter G. Construction of heterodimer tyrosyl-tRNA synthetase shows tRNATyr interacts with both subunits. Proc Natl Acad Sci USA. 1986;83:1189–1192. doi: 10.1073/pnas.83.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yaremchuk A, Kriklivyi I, Tukalo M, Cusack S. Class I tyrosyl-tRNA synthetase has a class II mode of cognate tRNA recognition. EMBO J. 2002;21:3829–3840. doi: 10.1093/emboj/cdf373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ward WH, Fersht AR. Asymmetry of tyrosyl-tRNA synthetase in solution. Biochemistry. 1988;27:1041–1049. doi: 10.1021/bi00403a029. [DOI] [PubMed] [Google Scholar]
- 13.Saldanha RJ, Patel SS, Surendran R, Lee JC, Lambowitz AM. Involvement of Neurospora mitochondrial tyrosyl-tRNA synthetase in RNA splicing. A new method for purifying the protein and characterization of physical and enzymatic properties pertinent to splicing. Biochemistry. 1995;34:1275–1287. doi: 10.1021/bi00004a022. [DOI] [PubMed] [Google Scholar]
- 14.Mohr G, Rennard R, Cherniack AD, Stryker J, Lambowitz AM. Function of the Neurospora crassa mitochondrial tyrosyl-tRNA synthetase in RNA splicing. Role of the idiosyncratic N-terminal extension and different modes of interaction with different group I introns. J Mol Biol. 2001;307:75–92. doi: 10.1006/jmbi.2000.4460. [DOI] [PubMed] [Google Scholar]
- 15.Chen X, Mohr G, Lambowitz AM. The Neurospora crassa CYT-18 protein C-terminal RNA-binding domain helps stabilize interdomain tertiary interactions in group I introns. RNA. 2004;10:634–644. doi: 10.1261/rna.5212604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cherniack AD, Garriga G, Kittle JD, Jr, Akins RA, Lambowitz AM. Function of Neurospora mitochondrial tyrosyl-tRNA synthetase in RNA splicing requires an idiosyncratic domain not found in other synthetases. Cell. 1990;62:745–755. doi: 10.1016/0092-8674(90)90119-y. [DOI] [PubMed] [Google Scholar]
- 17.Paukstelis PJ, Coon R, Madabusi L, Nowakowski J, Monzingo A, Robertus J, Lambowitz AM. A tyrosyl-tRNA synthetase adapted to function in group I intron splicing by acquiring a new RNA binding surface. Mol Cell. 2005;17:417–428. doi: 10.1016/j.molcel.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 18.Paukstelis PJ, Chen J-H, Chase E, Lambowitz AM, Golden BL. Structure of a tyrosyl-tRNA synthetase splicing factor bound to a group I intron RNA. Nature. 2008;451:94–97. doi: 10.1038/nature06413. [DOI] [PubMed] [Google Scholar]
- 19.Myers CA, Kuhla B, Cusack S, Lambowitz AM. tRNA-like recognition of group I introns by a tyrosyl-tRNA synthetase. Proc Natl Acad Sci USA. 2002;99:2630–2635. doi: 10.1073/pnas.052596299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vicens Q, Paukstelis PJ, Westhof E, Lambowitz AM, Cech TR. Toward predicting self-splicing and protein-facilitated splicing of group I introns. RNA. 2008;14:2013–2029. doi: 10.1261/rna.1027208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 22.Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 23.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 24.Berjanskii MV, Neal S, Wishart DS. PREDITOR: a web server for predicting protein torsion angle restraints. Nucl Acids Res. 2006;34:W63–69. doi: 10.1093/nar/gkl341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 26.Palmer AG, Rance M, Wright PE. Intramolecular motions of a zinc finger DNA-binding domain from Xfin characterized by proton-detected natural abundance carbon-13 heteronuclear NMR spectroscopy. J Amer Chem Soc. 1991;113:4371–4380. [Google Scholar]
- 27.Mandel AM, Akke M, Palmer AG. Backbone dynamics of Escherichia coli ribonuclease HI: correlations with structure and function in an active enzyme. J Mol Biol. 1995;246:144–163. doi: 10.1006/jmbi.1994.0073. [DOI] [PubMed] [Google Scholar]
- 28.Delano W. The PyMOL molecular graphics system, version 1.2r3pre. Schrödinger, LLC; [Google Scholar]
- 29.Laursen BS. The N-terminal domain (IF2N) of bacterial translation initiation factor IF2 is connected to the conserved C-terminal domains by a flexible linker. Prot Sci. 2004;13:230–239. doi: 10.1110/ps.03337604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jühling F, Mörl M, Hartmann RK, Sprinzl M, Stadler PF, Pütz J. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucl Acids Res. 2009;37:D159–162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall KB, Fox RO. Directed cleavage of RNA with protein-tethered EDTA-Fe. Methods. 1999;18:78–84. doi: 10.1006/meth.1999.0759. [DOI] [PubMed] [Google Scholar]
- 32.Kittle JD, Jr, Mohr G, Gianelos JA, Wang H, Lambowitz AM. The Neurospora mitochondrial tyrosyl-tRNA synthetase is sufficient for group I intron splicing in vitro and uses the carboxy-terminal tRNA-binding domain along with other regions. Genes Dev. 1991;5:1009–1021. doi: 10.1101/gad.5.6.1009. [DOI] [PubMed] [Google Scholar]
- 33.Lynch M. The frailty of adaptive hypotheses for the origins of organismal complexity. Proc Natl Acad Sci USA. 2007;104:8597–8604. doi: 10.1073/pnas.0702207104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gray MW, Lukeš J, Archibald JM, Keeling PJ, Doolittle WF. Irremediable Complexity? Science. 2010;330:920–921. doi: 10.1126/science.1198594. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.