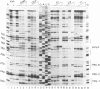
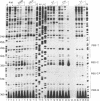
Abstract
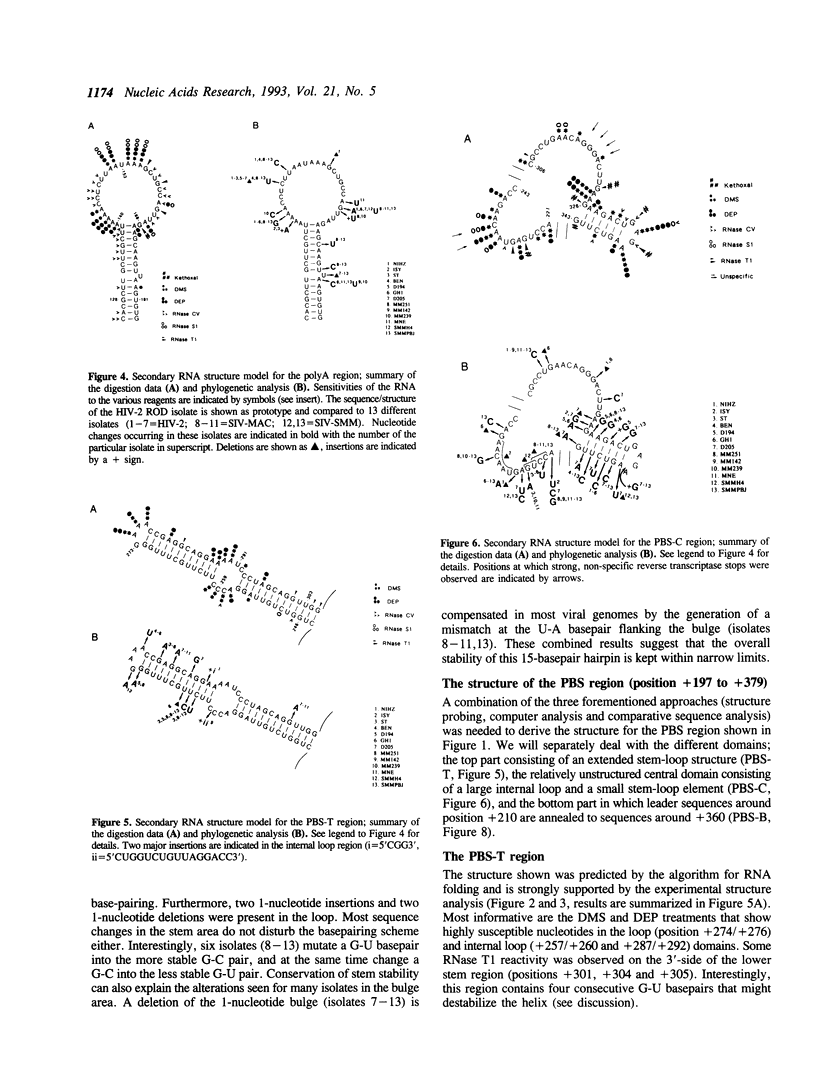
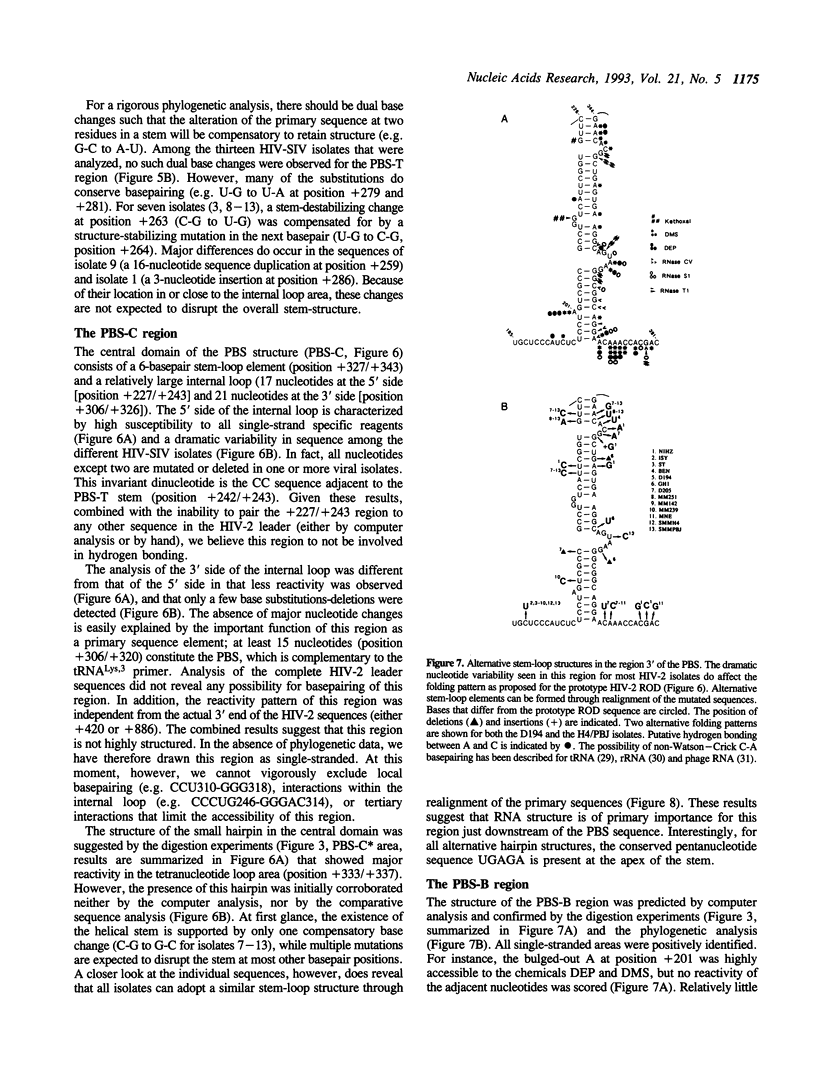
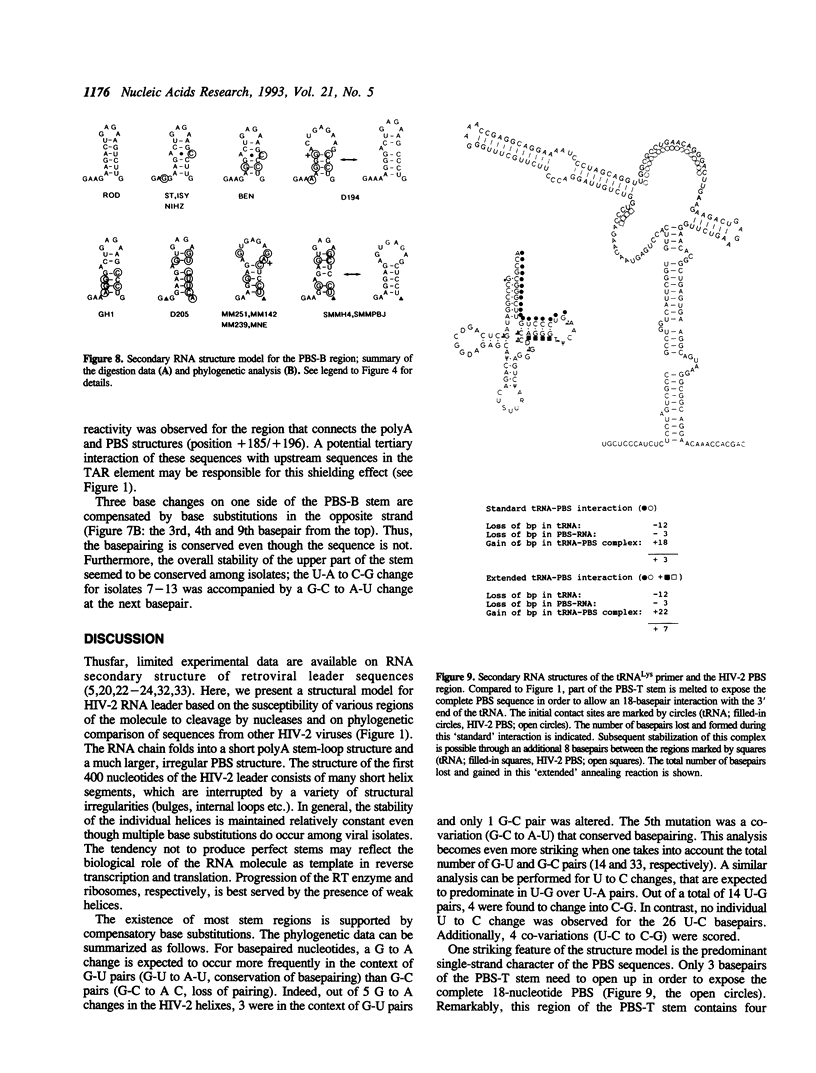
The initiation of reverse transcription of a retroviral RNA genome occurs by a tRNA primer bound near the 5' end of the genomic RNA at a position called the primer-binding site (PBS). To understand the molecular basis for this RNA-RNA interaction, the secondary structure of the leader RNA of the human immunodeficiency virus type 2 (HIV-2) RNA was analyzed. In vitro synthesized HIV-2 RNA was probed with various structure-specific enzymes and chemicals. A computer program was then used to predict the secondary structure consistent with these data. In addition, the nucleotide sequences of different HIV-2 isolates were used to screen for the occurrence of covariation among putative base pairs. The primary sequences have diverged rapidly in some HIV-2 isolates, however, some strikingly conserved secondary structure elements were identified. Most nucleotides in the leader region are involved in base pairing. An exception is the PBS sequence, of which 15 out of 18 nucleotides are exposed in an internal loop. These findings suggest that the overall structure of the HIV-2 genome has evolved to facilitate an optimal interaction with its tRNA primer.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abrahams J. P., van den Berg M., van Batenburg E., Pleij C. Prediction of RNA secondary structure, including pseudoknotting, by computer simulation. Nucleic Acids Res. 1990 May 25;18(10):3035–3044. doi: 10.1093/nar/18.10.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiyar A., Cobrinik D., Ge Z., Kung H. J., Leis J. Interaction between retroviral U5 RNA and the T psi C loop of the tRNA(Trp) primer is required for efficient initiation of reverse transcription. J Virol. 1992 Apr;66(4):2464–2472. doi: 10.1128/jvi.66.4.2464-2472.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alford R. L., Honda S., Lawrence C. B., Belmont J. W. RNA secondary structure analysis of the packaging signal for Moloney murine leukemia virus. Virology. 1991 Aug;183(2):611–619. doi: 10.1016/0042-6822(91)90990-s. [DOI] [PubMed] [Google Scholar]
- Araya A., Sarih L., Litvak S. Reverse transcriptase mediated binding of primer tRNA to the viral genome. Nucleic Acids Res. 1979 Aug 24;6(12):3831–3843. doi: 10.1093/nar/6.12.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barat C., Lullien V., Schatz O., Keith G., Nugeyre M. T., Grüninger-Leitch F., Barré-Sinoussi F., LeGrice S. F., Darlix J. L. HIV-1 reverse transcriptase specifically interacts with the anticodon domain of its cognate primer tRNA. EMBO J. 1989 Nov;8(11):3279–3285. doi: 10.1002/j.1460-2075.1989.tb08488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B., Gatignol A., Silver J., Jeang K. T. Efficient trans-activation by the HIV-2 Tat protein requires a duplicated TAR RNA structure. Nucleic Acids Res. 1990 Apr 11;18(7):1839–1846. doi: 10.1093/nar/18.7.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B., Jeang K. T. trans activation of human immunodeficiency virus type 1 is sequence specific for both the single-stranded bulge and loop of the trans-acting-responsive hairpin: a quantitative analysis. J Virol. 1989 Dec;63(12):5501–5504. doi: 10.1128/jvi.63.12.5501-5504.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B., Silverman R. H., Jeang K. T. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell. 1989 Oct 20;59(2):273–282. doi: 10.1016/0092-8674(89)90289-4. [DOI] [PubMed] [Google Scholar]
- Berkhout B. Structural features in TAR RNA of human and simian immunodeficiency viruses: a phylogenetic analysis. Nucleic Acids Res. 1992 Jan 11;20(1):27–31. doi: 10.1093/nar/20.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobrinik D., Aiyar A., Ge Z., Katzman M., Huang H., Leis J. Overlapping retrovirus U5 sequence elements are required for efficient integration and initiation of reverse transcription. J Virol. 1991 Jul;65(7):3864–3872. doi: 10.1128/jvi.65.7.3864-3872.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobrinik D., Soskey L., Leis J. A retroviral RNA secondary structure required for efficient initiation of reverse transcription. J Virol. 1988 Oct;62(10):3622–3630. doi: 10.1128/jvi.62.10.3622-3630.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collett M. S., Leis J. P., Smith M. S., Faras A. J. Unwinding-like activity associated with avian retrovirus RNA-directed DNA polymerase. J Virol. 1978 May;26(2):498–509. doi: 10.1128/jvi.26.2.498-509.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell B., Swanstrom R., Goodman H. M., Bishop J. M. tRNATrp as primer for RNA-directed DNA polymerase: structural determinants of function. J Biol Chem. 1979 Mar 25;254(6):1866–1874. [PubMed] [Google Scholar]
- Darlix J. L., Zuker M., Spahr P. F. Structure-function relationship of Rous sarcoma virus leader RNA. Nucleic Acids Res. 1982 Sep 11;10(17):5183–5196. doi: 10.1093/nar/10.17.5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Holland E. C. HIV-1 tat trans-activation requires the loop sequence within tar. Nature. 1988 Jul 14;334(6178):165–167. doi: 10.1038/334165a0. [DOI] [PubMed] [Google Scholar]
- Gurevich V. V., Pokrovskaya I. D., Obukhova T. A., Zozulya S. A. Preparative in vitro mRNA synthesis using SP6 and T7 RNA polymerases. Anal Biochem. 1991 Jun;195(2):207–213. doi: 10.1016/0003-2697(91)90318-n. [DOI] [PubMed] [Google Scholar]
- Gutell R. R., Weiser B., Woese C. R., Noller H. F. Comparative anatomy of 16-S-like ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- Hackett P. B., Dalton M. W., Johnson D. P., Petersen R. B. Phylogenetic and physical analysis of the 5' leader RNA sequences of avian retroviruses. Nucleic Acids Res. 1991 Dec 25;19(24):6929–6934. doi: 10.1093/nar/19.24.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison G. P., Lever A. M. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. J Virol. 1992 Jul;66(7):4144–4153. doi: 10.1128/jvi.66.7.4144-4153.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseltine W. A., Kleid D. G., Panet A., Rothenberg E., Baltimore D. Ordered transcription of RNA tumor virus genomes. J Mol Biol. 1976 Sep 5;106(1):109–131. doi: 10.1016/0022-2836(76)90303-x. [DOI] [PubMed] [Google Scholar]
- Haseltine W. A., Maxam A. M., Gilbert W. Rous sarcoma virus genome is terminally redundant: the 5' sequence. Proc Natl Acad Sci U S A. 1977 Mar;74(3):989–993. doi: 10.1073/pnas.74.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseltine W. A., Panet A., Smoler D., Baltimore D., Peters G., Harada F., Dahlberg J. E. Interaction of tryptophan tRNA and avian myeloblastosis virus reverse transcriptase: further characterization of the binding reaction. Biochemistry. 1977 Aug 9;16(16):3625–3632. doi: 10.1021/bi00635a019. [DOI] [PubMed] [Google Scholar]
- Jeang K. T., Chang Y., Berkhout B., Hammarskjöld M. L., Rekosh D. Regulation of HIV expression: mechanisms of action of Tat and Rev. AIDS. 1991;5 (Suppl 2):S3–14. [PubMed] [Google Scholar]
- Levin J. G., Seidman J. G. Selective packaging of host tRNA's by murine leukemia virus particles does not require genomic RNA. J Virol. 1979 Jan;29(1):328–335. doi: 10.1128/jvi.29.1.328-335.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muesing M. A., Smith D. H., Capon D. J. Regulation of mRNA accumulation by a human immunodeficiency virus trans-activator protein. Cell. 1987 Feb 27;48(4):691–701. doi: 10.1016/0092-8674(87)90247-9. [DOI] [PubMed] [Google Scholar]
- Nagashunmugam T., Velpandi A., Goldsmith C. S., Zaki S. R., Kalyanaraman V. S., Srinivasan A. Mutation in the primer binding site of the type 1 human immunodeficiency virus genome affects virus production and infectivity. Proc Natl Acad Sci U S A. 1992 May 1;89(9):4114–4118. doi: 10.1073/pnas.89.9.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters G. G., Hu J. Reverse transcriptase as the major determinant for selective packaging of tRNA's into Avian sarcoma virus particles. J Virol. 1980 Dec;36(3):692–700. doi: 10.1128/jvi.36.3.692-700.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prats A. C., Sarih L., Gabus C., Litvak S., Keith G., Darlix J. L. Small finger protein of avian and murine retroviruses has nucleic acid annealing activity and positions the replication primer tRNA onto genomic RNA. EMBO J. 1988 Jun;7(6):1777–1783. doi: 10.1002/j.1460-2075.1988.tb03008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhim H., Park J., Morrow C. D. Deletions in the tRNA(Lys) primer-binding site of human immunodeficiency virus type 1 identify essential regions for reverse transcription. J Virol. 1991 Sep;65(9):4555–4564. doi: 10.1128/jvi.65.9.4555-4564.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallafranque-Andreola M. L., Robert D., Barr P. J., Fournier M., Litvak S., Sarih-Cottin L., Tarrago-Litvak L. Human immunodeficiency virus reverse transcriptase expressed in transformed yeast cells. Biochemical properties and interactions with bovine tRNALys. Eur J Biochem. 1989 Sep 15;184(2):367–374. doi: 10.1111/j.1432-1033.1989.tb15028.x. [DOI] [PubMed] [Google Scholar]
- Sawyer R. C., Hanafusa H. Comparison of the small RNAs of polymerase-deficient and polymerase-positive Rous sarcoma virus and another species of avian retrovirus. J Virol. 1979 Mar;29(3):863–871. doi: 10.1128/jvi.29.3.863-871.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skripkin E. A., Adhin M. R., de Smit M. H., van Duin J. Secondary structure of the central region of bacteriophage MS2 RNA. Conservation and biological significance. J Mol Biol. 1990 Jan 20;211(2):447–463. doi: 10.1016/0022-2836(90)90364-R. [DOI] [PubMed] [Google Scholar]
- Sobol R. W., Suhadolnik R. J., Kumar A., Lee B. J., Hatfield D. L., Wilson S. H. Localization of a polynucleotide binding region in the HIV-1 reverse transcriptase: implications for primer binding. Biochemistry. 1991 Nov 5;30(44):10623–10631. doi: 10.1021/bi00108a004. [DOI] [PubMed] [Google Scholar]
- Taylor J. M. An analysis of the role of tRNA species as primers for the transcription into DNA of RNA tumor virus genomes. Biochim Biophys Acta. 1977 Mar 21;473(1):57–71. doi: 10.1016/0304-419x(77)90007-5. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn M. H., Klug A. A model for the tertiary structure of mammalian mitochondrial transfer RNAs lacking the entire 'dihydrouridine' loop and stem. EMBO J. 1983;2(8):1309–1321. doi: 10.1002/j.1460-2075.1983.tb01586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]