Abstract
Dentin Matrix Protein 1 (DMP1) plays a regulatory role in dentin mineralization and can also function as a signaling molecule. MMP-2 (matrix metalloproteinase-2) is a predominant protease in the dentin matrix that plays a prominent role in tooth formation and a potential role during the carious process. The possibility that MMP-2 can cleave DMP1 to release biologically active peptides was investigated in this study. DMP1, both in the recombinant form and in its native state within the dentin matrix, was shown to be a substrate for MMP-2. Proteolytic processing of DMP1 by MMP-2 produced two major peptides, one that contains the C-terminal region of the protein known to carry both the ASARM (aspartic acid and serine rich domain) domain involved in biomineralization and the DNA binding site of DMP1. In vitro experiments with recombinant N- and C-terminal polypeptides mimicking the MMP-2 cleavage products of DMP1 demonstrated an effect of the C-polypeptide on the differentiation of dental pulp stem/progenitor cells to a putative odontoblast phenotype. In vivo implantation of this peptide in a rat injured pulp model induced a rapid formation of a homogeneous dentin bridge covered by a palisade of orientated cells expressing dentin sialoprotein (DSP) and DMP1, attesting an efficient repair process. These data suggest that a peptide generated through the proteolytic processing of DMP1 by MMP-2 can regulate the differentiation of mesenchymal cells during dentinogenesis and thus sustain reparative dentin formation in pathological situations such as carious decay. In addition, these data open a new therapeutic possibility of using this peptide to regenerate dentin after an injury.
Keywords: DMP1, MMP-2, dentin, cleavage product, pulp stem/progenitor cells, bioactivity, differentiation
Introduction
Dentin Matrix Protein 1 (DMP1) has been shown to play a central role in dentin mineralization (He and George 2004, He et al., 2003). Its carboxy-terminal domain rich in aspartic acid and serine (ASARM; residues 408 to 429) may be responsible for the nucleation of hydroxyapatite (Gajjeraman et al., 2007, Rowe et al., 2000). DMP1 was shown to be transported to the nuclear compartment and was implicated in signaling functions (Narayanan et al., 2003). It was shown to regulate the transcription of DSPP during early odontoblast differentiation by binding to the promoter of this gene through its carboxyl end (residues 420 to 489) (Narayanan et al., 2006).
Dentin and bone extracellular matrix (ECM) were shown to contain both the full length DMP1 as well as its processed N-terminal (N-ter) (37 kDa) and C-terminal (C-ter) (57 kDa) fragments (Huang et al., 2008, Qin et al., 2003). The bone morphogenetic protein-1 (BMP1) and its relative tolloid-like 1 (TLL1) metalloproteinases were proposed to be responsible for the physiological processing of DMP1 (Steiglitz et al., 2004). However, the role of matrix metalloproteinases (MMPs), in generating active DMP1 peptides in the extracellular matrix has not yet been evoked. Indeed, these proteases were shown to be contained within the dentin matrix and can be activated in pathological conditions (Chaussain-Miller et al., 2006, Tjaderhane et al., 1998).
It is now widely acknowledged that MMPs convert structural matrix proteins to signaling molecules (Page-McCaw et al., 2007) and may function in the differentiation program of mesenchymal stem cells (Mannello et al., 2006). Several MMPs have been identified in dentin and pulp (Boukpessi et al., 2008, Goldberg et al., 2003, Palosaari et al., 2003) and are thought to be implicated in the early stages of dentinogenesis. Among endogenous MMPs, MMP-2 has been proposed to be involved in dental ECM processing taking place at later stages of tooth development, and particularly in dentin sialophosphoprotein (DSPP) processing, possibly in association with MMP-20 (Bourd-Boittin et al., 2005, Yamakoshi et al., 2006), or in association with a self-processing mechanism of DSPP (Godovikova and Ritchie, 2007). MMP-2 was also isolated from mature human mineralized dentin matrix (Martin-De Las Heras et al., 2000) and zymographically identified in demineralized dentin (Mazzoni et al., 2007, van Strijp et al., 2003), playing a potential role in dentin matrix destruction during the carious process (Chaussain-Miller et al., 2006, Tjaderhane et al., 1998).
We therefore investigated the possibility that DMP1 in dentin matrix may be a substrate for MMP-2, as this enzyme was already shown to generate bioactive peptide from several matrix proteins in different models (Dean and Overall 2007, Dean et al., 2007). We hypothesized that DMP1 cleavage by MMP-2 under physiological conditions such as tooth development, or in pathological situations such as carious decay, may release bioactive peptides that conserve DMP1 biological activities or present specific new activity. Therefore, the purpose of this study was to determine whether DMP1 can be cleaved by MMP-2 and if the cleavage products can act as signaling molecules on the transformation of dental pulp cells into putative odontoblasts. An in vitro model using human dental pulp stem/progenitor cells (DPSCs), a heterogeneous pulp cell population and an in vivo rat pulp injury model were used to validate this hypothesis.
Materials and Methods
Expression and purification of recombinant DMP1 (r-DMP1) and its N- and C-DMP1 polypeptides
The r-DMP1 was expressed in E. coli and purified as published earlier (Srinivasan et al., 1999). The coding region of rat DMP1 cDNA was amplified using a set of sequence-specific sense and antisense primers with BamH1 and NotI recognition sites on the 5′ ends of both the primers.
The sense primer had the sequence: 5′ GCCGCCGGATCCCTCCCTGTCGCCAGATAC 3′ and the antisense primer: 5′ GCCGCCGCGGCCGCACTAGTAGCCATCTTGGCAATC 3′.
The amplified mature DMP1 cDNA was cloned into the BamHI and NotI sites of the prokaryotic expression vector PGEX-4T-3 (Pharmacia, Uppsala, Sweden) and transformed into the E. coli host JM 109 strain. Transformed colonies were grown in LB/ampicillin medium and the expression of fusion proteins was induced by adding 0.1mM isopropyl β-D-1-thiogalactopyranoside (IPTG). The fusion protein was purified from bacterial lysates by Sepharose 4B-Glutathione affinity and used for the proteolytic assay in this study. A similar procedure was used to express and purify recombinant N- and C-DMP1 polypeptides (Gajjeraman et al., 2007). The coding sequence of rat DMP1 was subamplified into two parts by PCR with a set of primers flanking the required sequence region:
N term-S, G GAT CCC ATG AAG ACT GTT ATC CTC CTT ACG;
N term-AS, GC GGC CGC TTC CTG GGA TCC CTC ACC AGC;
C-term-S, G GAT CCC AGC AGC AGC GAG TCT CAG GAA;
C-term-AS, GC GGC CGC GTA GCC ATC TTG GCA ATC ATT.
The amplified fragments were sequence-verified and then inserted into the PGEX-4T3 vector and expressed in BL21-DE3 E. coli cells (Invitrogen, Carlsbad, CA, USA). The glutathione S-transferase fusion protein induced and purified by the standard procedure was cleaved by thrombin at 4 °C. The corresponding peptides were termed N-DMP1 (residues 1–334) and C-DMP1 (residues 334–489).
Total protein extraction from tooth germs
Tooth germs were dissected from 5 days post-natal Swiss mice (Charles River, Lyon, France). The protocol was approved by the Animal Care Committee of French Veterinary Services (DSV). According to the procedure of Bourd-Boittin et al. (2005), tooth germs were ground to a fine powder in the presence of liquid nitrogen, then homogenized in ice-cold extraction buffer (10 germs in 150 μl buffer: 50 mM Tris-HCl, pH 7.5, containing 5 mM CaCl2, 0.9% NaCl and 0.2 % Triton X-100), supplemented with 1/100 Protease Inhibitor Cocktail Set V EDTA free (Calbiochem, La Jolla, CA, USA). The resulting homogenate referred as HC was then briefly sonicated on ice (3 times 5 sec) and cleared by centrifugation at 10 000xg for 15 min at 4°C. Other extraction procedures also included, in addition to the Protease Inhibitor Cocktail, 1μM marimastat (British Biotechnology, Oxford, UK), or 0.005 M EDTA and the corresponding homogenates will be referred to as HMAR and HEDTA, respectively.
Proteolysis of native and r-DMP1 by recombinant MMP-2 (r-MMP-2)
r-MMP-2 (a gift from Dr R. Fridman, Wayne State University, Detroit, MI, USA) was first incubated with APMA (p-aminophenylmercuric acetate; 1 mM, Sigma, Saint Louis, MO, USA) for 1 h at room temperature to achieve activation. Activated MMP-2 (1 μg/ml) was added to 100 μg rat r-DMP1 in a final volume of 100 μl of 100 mM Tris buffer pH 7.2, containing 0.15 M NaCl and 5 mM CaCl2 (all the buffer reagents used are from Fisher Chemicals, Fair Lawn, NJ, USA). r-DMP1 incubated under the same conditions but without MMP-2 was used as a control. Following 0.5 to 6 h incubation periods at 37°C, 20 μg of DMP1 treated with or without MMP-2 were subjected to 12% SDS-PAGE. Gels were stained either with Coomassie blue R-250 (FisherBiotec, Wembley, Australia), or with Stains-All (Sigma). The apparent molecular weights of protein bands were estimated by comparison with Precision plus protein unstained standards for Coomassie and Precision plus protein kaleidoscope standards for Stains-All (Bio-Rad, Richmond, CA, USA). In parallel, the homogenate obtained from tooth germ (HC) was incubated with activated MMP-2 (3 μg/ml) for 1 and 3 h respectively, similar to the conditions described above incubated under the same conditions but for r-DMP1. HC without MMP-2 was used as a control.
Identification of cleaved DMP1 peptides by Western blot analysis
Western blot analysis was performed to identify intact DMP1 and its proteolytic cleavage products using tooth germ homogenates when incubated with MMP-2. 20 μg of homogenates were subjected to electrophoresis on 12% SDS-PAGE and transferred onto a nitrocellulose membrane (Bio-Rad). Membranes were incubated with a polyclonal anti-C-DMP1 antibody (C-857; 1: 2000) (a gift from Dr Chunlin Qin, Baylor College of Dentistry, Dallas, TX, USA) (Qin et al., 2003).
Effect of DMP1 polypeptides on the differentiation of dental pulp stem/progenitor cells (DPSCs)
Dental pulp stem/progenitor cells (DPSCs) were a gift from Dr S. Shi (Univ. of Southern California, Growth and Development, Los Angeles, CA, USA). These cells represent a heterogeneous population obtained from the enzyme treatment of human pulp containing a fraction of progenitor/stem cells displaying phenotypes consistent with different perivascular cell populations, and particularly positive for STRO-1 and CD146 antigens (Shi and Gronthos, 2003). If seeded onto dentin, some DPSCs convert into putative odontoblast cells with a polarized cell body and a cell process extending into the existing dentinal tubules (Huang et al., 2006). In addition to their dentinogenic potential, subpopulations of DPSCs also possess adipogenic and neurogenic differentiation capacities (Gronthos et al., 2002; for review see Huang et al., 2009). More recently, DPSCs were also found to undergo osteogenic, chondrogenic and myogenic differentiation in vitro (d’Aquino et al., 2007, Laino et al., 2005, Zhang et al., 2006).
The pulp was obtained at the Dental Clinic of the National Institute of Dental and Craniofacial Research, from normal third molar teeth in healthy young adults (19–29 yrs of age), who gave their informed consent according to approved guidelines set by the National Institutes of Health Office Human Subject Research (Gronthos et al., 2002). Approximately 105 cells were collected from each pulp sample.
The cells at P4 were grown in Dulbecco’s modified Eagle’s (DMEM)/F12 medium containing 10% fetal bovine serum (FBS), 1% penicillin/streptomycin and 1% amphotericin B. Cells were seeded in 6 well plates and grown to 70% confluence. 12–16 h before the beginning of the experiment the cells were cultured in DMEM/F-12 medium supplemented with 1% FBS. N- or C-DMP1 polypeptides were added for varying time periods 4, 24 and 48 h, at 0 and 0.25 μg/ml. Experiments were repeated in triplicates. For each time point and at each concentration, one well of cells were fixed with cold ethanol, stained for 10 min with Giemsa’s stain (Giemsa STAIN Solution; Sigma), rinsed extensively with double-deionized (DDI) water and the cell morphology was observed under an inverted microscope (DM IRB Leica Bannockburn, IL, USA) equipped with an QImaging Retiga Exi Fast 1394 camera and QCapture software (Qimaging, Surrey, BC, Canada).
The differentiation potential of the treated cells was investigated by analyzing the expression of DMP1 and DSP after extraction of total RNA from cultured cells 4, 24 and 48 h after addition of DMP1 polypeptides using RNeasy kit (Qiagen, Hilden, Germany). RT-PCR was performed with DNase I (Qiagen)-treated RNA. 3 μg of total RNA was reverse transcribed for 90 min at 50°C with Superscript III (Gibco, Invitrogen). cDNA was amplified with 0.1 g of specific primers in a reaction mixture (PCR supermix, Invitrogen) containing Taq DNA polymerase, Tris HCl, potassium chloride, magnesium chloride, and deoxynucleoside triphosphates. Quantitative real-time PCR (qPCR) analysis was carried out using ABI step one plus. Expression for DSP, DMP1 and GAPDH transcripts were analyzed by qPCR during its linear phase. The relative gene expression level was estimated by using the 2−ΔΔCT method where CT value = log-linear plot of PCR signal versus the cycle number. ΔCT= CT value of target gene-CT value of GAPDH. Results were expressed as mean ± SD of relative mRNA level compare to untreated cells for each time point. Primers were obtained from IDT DNA.
Effect of C-DMP1 polypeptide on dentin regeneration in a rat pulp injury model
Affi-gel agarose beads (Bio-Rad) were soaked in a solution of C-DMP1 polypeptide dissolved in phosphate-buffered saline (PBS) solution (1 μg/μl) for 2 h, and dried overnight at 37°C (Six et al., 2007). The beads were implanted in the pulp of the first upper jaw molar of 16 rats (6 weeks, OFA/SD, Charles River). In brief, after gingival electrosurgery, half-moon-shaped Class V 0.5 mm cavities were prepared on the mesial aspect of the first maxillary molars using a 0.2 mm diameter round bur (E0123, Dentsply Maillefer, Ballaigues, Switzerland). Pulp perforation was accomplished by pressure with the tip of a steel probe and 10 C-DMP1 soaked beads were implanted in the injury (Six et al., 2007). The protocol was approved by the Animal Care Committee of the French Veterinary Services (DSV). Co-lateral molars were implanted with buffer soaked beads as a control. Cavities were filled with glass ionomer cement (GC Fuji II, GC Corporation, Tokyo, Japan). Animals were anesthetized and sacrificed at 7, 15 and 30 days after implantation by an intracardiac perfusion with paraformaldehyde/glutaraldehyde solution buffered with sodium cacodylate 0.1 M at pH 7.2–7.4. Block sections including molars were dissected from the maxilla after a few minutes and immersed in the fixative solution for 24 h at 4°C. Tissue demineralization was performed in 4.13% EDTA for approximately 8 weeks. After tissue embedding in Paraplast, 8 μm thick sections were cut and either prepared for staining with hematoxylin-eosin and trichrome stain, or prepared for immunohistochemical staining. Sections were deparaffined and rehydrated before blocking with 10% normal goat serum for 45 min at room temperature. Sections were incubated overnight at 4°C with primary antibodies against DSP and DMP1 at 1:200 dilution (Hao et al., 2009). A negative control was included by using an equal concentration of rabbit pre-immune serum as primary antibody. Following incubation with primary antibody, the tissue sections were incubated with an anti-rabbit immunoglobulin (1:500) for 1 h at room temperature. The color reaction was developed using 3,3′-diaminobenzidine tetrahydrochloride (DAB, Vector Laboratories, Burlingame, CA, USA) for 1–5 min at room temperature. The tissue sections were then counterstained with hematoxylin.
Results
Native and r-DMP1 are substrates for MMP-2
Results in Fig. 1 show that recombinant r-DMP1 (96 kDa) can be cleaved by MMP-2. Proteolytic processing of the r-DMP1 at 37°C was visible by both Coomassie and ‘Stains-All’ staining of the SDS PAGE gels. While the cleavage of the 96 kDa band could be seen already at 1 h incubation with MMP-2, the decrease in intensity of this band became more apparent after 2 h incubation with the appearance of 2 main cleavage bands at approximately 50 kDa and 40 kDa, the intensity of which further increased between 3–6 h cleavage times.
Figure 1.
In-vitro cleavage assay A: SDS-PAGE stained with Coomassie blue (A) and Stains-All (B). APMA activated MMP-2 was added to recombinant rat DMP1 (96 kDa). Following 1 to 6 h incubation periods at 37°C, aliquots containing 20 μg were subjected to 12% SDS-PAGE analysis to reveal DMP1 peptides cleaved by MMP-2. The arrows indicate the bands that result from r-DMP1 cleavage (50 kDa and ~40 kDa).
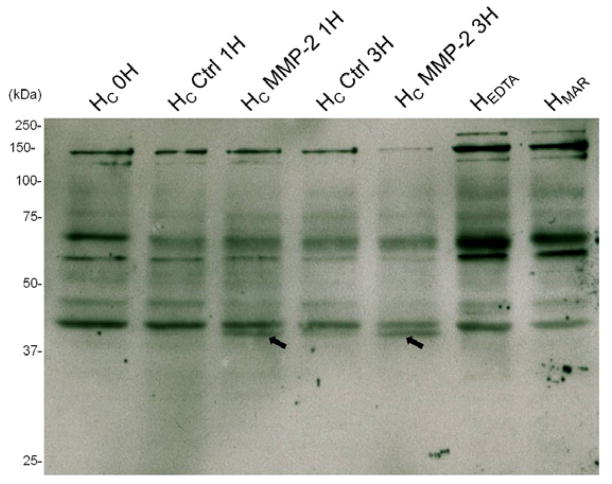
In order to verify that native DMP1, when incorporated within a complex extracellular matrix, can also be cleaved by MMP-2, we treated dental tissue with this enzyme. To this end, total proteins were gently extracted from tooth germs obtained from 5-day-old mice using a Triton containing buffer. Since the C-ter was shown to contain domains likely to be biologically active, we have looked more specifically for the release of peptides enclosing the C-ter region with a specific antibody against this region. The Western blot analysis using a C-ter DMP1 antibody confirmed the cleavage of matrix-bound DMP1 by MMP-2 (Fig. 2). In controls, (non treated Hc, HMAR, HEDTA), matrix-bound DMP1 was revealed as several bands: the ~150 kDa which probably corresponds to the full-length DMP1, the fragment observed at ~57 kDa may correspond to the previously described 57 kDa C-fragment of DMP1 and a band at ~42 kDa, which was not yet reported in the literature. The ~150 kDa and the ~57 kDa isoforms were more prominent in EDTA and Marimastat extracts (HEDTA and HMAR). The fact that without MMPs inhibitors, these bands decreased while the 42 kDa band became more intense suggests that during the experimental extraction procedure DMP1 was cleaved by endogenous dental MMPs to generate a 42 kDa cleavage product and supports the idea that DMP1 may be a potential MMP substrate in vivo.
Figure 2.
Identification of the cleavage products resulting from tooth germ extract incubated with MMP-2 by western blot analysis. In dentin homogenates, C-ter DMP1 antibody recognized a doublet at ~57 kDa, a band at ~42 kDa and a triplet at ~150 kDa. When Hc was treated with MMP-2, the ~150 kDa band progressively decreased between 1 and 3 h. A cleavage-product became apparent at ~40 kDa indicated by a black arrow. This ~40 kDa product was also faintly observed in control Hc at 3 h. Hc: dentin homogenate; HEDTA: dentin homogenate made in the presence of EDTA; HMAR: dentin homogenate made in presence of marimastat.
When the Hc extract was treated with exogenously added MMP-2, the intensity of the ~ 150 kDa form further decreased (more obvious at 3 h). At the same time, a cleavage product at ~40 kDa became apparent in the sample treated with MMP-2. It is interesting that after 3 h this cleavage product could also be seen in the control group, albeit very faintly. This may correspond to the cleavage of DMP1 by endogenous MMP-2 already reported to be present in the extracted dentin (Bourd-Boittin et al., 2005). The 42 kDa fragment was not cleaved by MMP-2 at these incubation periods.
Thus, both recombinant and matrix-bound DMP1 can be cleaved by MMP-2 to release a peptide of approximately 40 kDa in size which would therefore contain the C-ter region known to carry both the ASARM domain (residues 408 to 429) and the DNA binding site of DMP1 (residues 420 to 489). The MMP-2 cleavage products we obtained are in accord with at least two potential MMP-2 cleavage sites (Turk et al., 2001) that we identified in the rat and mouse DMP1 sequences (Table 1). Based on the sequence of the protein, the C-ter peptide should contain at least one glycosylation site and several phosphorylation sites, which may account for the slightly higher molecular weight band present in the extracts from the dentin matrix (Fig. 2) compared to that obtained from the recombinant protein (Fig. 1).
Table 1.
Predicted cleavage site for MMP-2 (Turk et al., 2001) mediated DMP1 cleavage (P for position in the cleavage-site motif, knowing that the cleavage occurs between P1 and P1′ residues). The number of occurrences of each residue that arise more than once is given for each position. In the protein sequence, the signal peptide appears in italics and the P1 and P1′ positions are underlined. These sites are also present within the mouse sequence (NP_058059).
| P5 | P4 | P3 | P2 | P1 | P1′ | P2′ | P3′ | |
|---|---|---|---|---|---|---|---|---|
| Rat DMP1 sequence (NP_987089) | ||||||||
| E59-L60GS | 1.3 | 4.2 | 2.2 | |||||
| AG78-M79DA | 1.3 | 1.4 | 2.8 | 2.2 | ||||
| E170-Y171R | 1.3 | 1.9 | 1.5 | |||||
| E188-F189 | 1.3 | 1.8 | ||||||
| G193-M194QS | 1.4 | 2.8 | 2.2 | |||||
| E240-F241 | 1.3 | 1.8 | ||||||
| DDRGE260-L261 | 1.4 | 1.3 | 4.2 | |||||
| S380-L381 | 1.9 | 4.2 | ||||||
| NESLS382-L383 | 1.3 | 1.9 | 4.2 | |||||
| G401-L402QS | 1.4 | 4.2 | 2.2 |
NP_987089.1 dentin matrix protein 1 [Rattus norvegicus]
MKTVILLTFLWGLSCALPVARYQNTESESSEERTGNLAQSPPPPMANSDHTDSSESGEELGSDRSQYRP AGGLSKSAGMDADKEEDEDDSGDDTFGDEDNGPGPEERQWGGPSRLDSDEDSADTTQSSEDSTSQ ENSAQDTPSDSKDHHSDEADSRPEAGDSTQDSESEEYRVGGGSEGESSHGDGSEFDDEGMQSDDPG STRSDRGHTRMSSAGIRSEESKGDHEPTSTQDSDDSQDVEFSSRKSFRRSRVSEEDDRGELADSNSR ETQSVSTEDFRSKEESRSETQEDTAETQSQEDSPEGQDPSSESSEEAGEPSQESSSESQEGVASESRGD NPDNTSQTGDQRDSESSEEDRLNTFSSSESQSTEEQGDSESNESLSLSEESQESAQDEDSSSQEGLQS QSASRESRSQESQSEQDSRSEENRDSDSQDSSRSKEESNSTGSTSSSEEDNHPKNIEADNRKLIVDAY HNKPIGDQDDNDCQDGY
C-DMP1 polypeptide promotes differentiation of DPSCs in-vitro
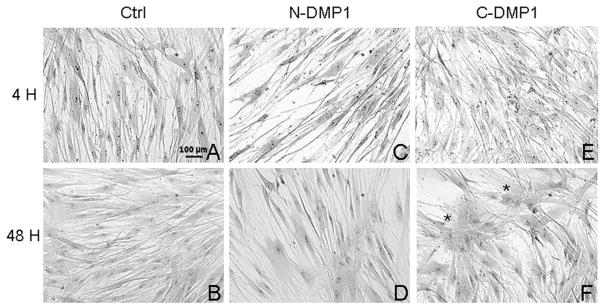
We next examined the functional role of C- and N-ter recombinant polypeptides of DMP1 closely matching those released by MMP-2 (Fig. 3), in dental pulp stem/progenitor cells (DPSCs) differentiation. DPSCs were cultured for 48 h in the presence of the DMP1 polypeptides and were then examined using light microscopy for changes in cell morphology. At 48 h, cells cultured with the C-DMP1 polypeptide formed cellular clusters whereas in the presence of the N-DMP1 polypeptide they retained their fibroblast-like morphology as observed with control cells (Fig. 4). We then analyzed the mRNA levels of DMP1 and DSPP, the expression of which indicates differentiation towards the odontoblast phenotype. We observed that the C-DMP1 polypeptide regulated the expression of DMP1, as shown by the progressive increased RNA levels observed between 4 and 48 h (Fig. 5). Similarly, C-DMP1 polypeptide also induced higher DSPP expression with time when compared with N-DMP1. Hence, the C-DMP1 polypeptide seems to activate cellular differentiation of DPSCs towards the odontoblast phenotype, inducing an organization of cells in clusters and an increased expression of odontoblast markers.
Figure 3.
Schematic representation of the recombinant N- and C- DMP1 polypeptides with respect to the possible cleavage sites of DMP1 by MMP-2. Potential cleavage sites for MMP-2 identified in the rat DMP1 sequence (Table 1) which agree with our experimental data (see Figs. 1 and 2) are indicated with arrows (E260-L261, , S380-L381 , S382-L383). The recombinant N-polypeptide (residues 1 to 334) and C-polypeptide (residues 334 to 489) are represented below. A = ASARM motif; DNAb = DNA binding region; Rec = recombinant.
Figure 4.
In-vitro functions of DMP1 polypeptides on dental pulp stem/progenitor cell (DPSC) morphology. At 4h, DPSCs displayed a fibroblastic morphology in the 3 groups. At 48 h DPSCs cultured with the C-DMP1 polypeptide organized themselves in clusters whereas cells cultured with the N-DMP1 polypeptide presented no particular organization and were similar to the control. A and B: control; C and D: cells cultured with 0.25 μg/ml N-DMP1; E and F: cells cultured with 0.25 μg/ml C-DMP1. Clusters are indicated by stars.
Figure 5.
Expression of DMP1 and DSPP by dental pulp stem/progenitor cells (DPSCs) treated with N- and C-DMP1 polypeptides. Treatment of DPSCs with C-DMP1 (left panel) increased the expression of DMP1; 10 fold DMP1 expression was observed at 48 h with C-DMP1 peptide, while it is <1 fold at 48 h with N-DMP1. DSPP expression, which is a marker for terminally differentiated odontoblasts, showed a faint expression in control untreated cells and a slightly increased expression in cells cultured with the N-DMP1 polypeptide. In contrast, cells cultured with the C-DMP1 polypeptide strongly expressed DSPP with a signal that increased with time (right panel). GAPDH was used as a control for the cDNA loading. Results were expressed as relative mRNA level for N- and C-DMP1 treated cells compared to untreated cells at each time point.
C-DMP1 polypeptide promotes pulp repair in an animal model of pulp injury
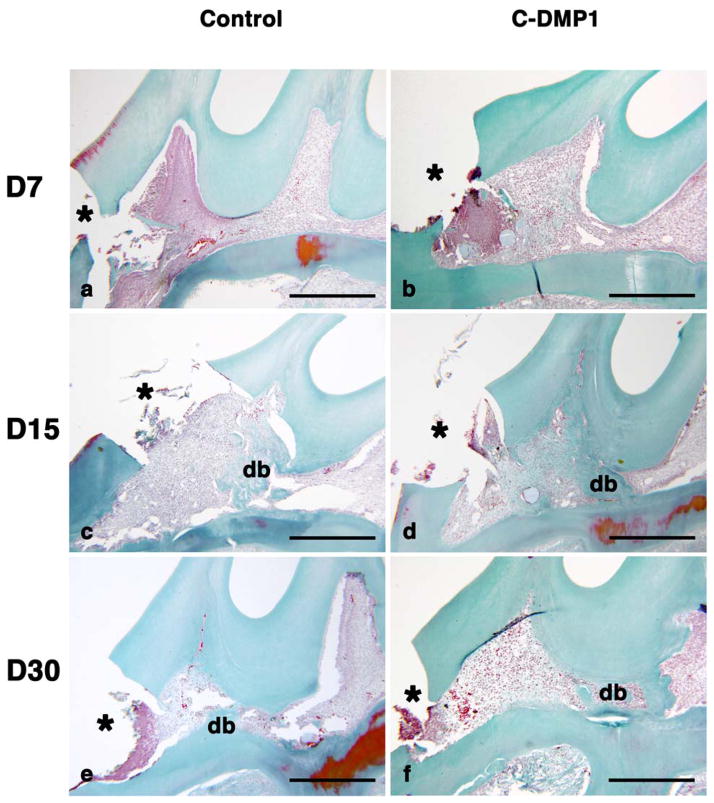
In order to explore the in vivo potential of C-DMP1 polypeptide to promote pulp cells differentiation, we implanted agarose beads soaked with this peptide in the injured pulp of young rats and followed the rate of healing in comparison to a control group treated with beads soaked in buffer only. In this model a reparative dentin bridge is formed to serve as a protective barrier to the dental pulp. In both control and C-DMP1 treated animals, an important inflammation process with a high cell infiltrate was observed at day 7 in the pulp horn where the pulp injury was performed (Fig. 6A). At day 15, a continuous well-defined reparative dentin bridge was observed in the C-DMP1 treated pulps, isolating the injured cusp from the rest of the pulp. In contrast, a poorly organized reparative structure was observed in the control group at the same location. At one month, the reparative bridges expanded in both groups, but still maintained the same histological differences as those observed at 15 days (Fig. 6).
Figure 6.
C-DMP1 polypeptide promotes pulp repair in an injured rat pulp model. Beads soaked with the polypeptide (right panel) or with buffer (left panel) were implanted in the injured pulp of young rats. Day7: At day 7, the pulp exposure is clearly visualized in the crown of the tooth. In both groups, an important inflammatory process is observed in the pulp horn as visualized by the high tissue cell infiltrate in this area (a, b). Day 15: A reparative dentin bridge is present in both groups at a distance from the inflammatory area, poorly organized in the control group (c) and well structured in the C-DMP1 implanted group (d). Day 30: The dentin bridges had expanded in both groups, in the control teeth still appearing as non homogenous with cell inclusion (e) but constituting an efficient protective barrier in the C-DMP1 treated pulp (f).
Scale bars: 0.5 mm
db: dentin bridge, *: pulp exposure
In Fig. 7, the reparative bridge region observed at day 15 is shown at a higher magnification. In the C-DMP1 treated group, the cells associated with the bridge and located at the opposite side of the injury, presented a polarized morphology and were organized as a palisade, with a basal location of the nucleus. These polarized cells were positively labeled for DSP and DMP1 at their secretory pole boarding the dentin bridge (Fig. 7). The corresponding cells in the control group were not polarized and showed a faint immunostaining for either DSP or DMP1. These results demonstrate that a peptide comprising the C-ter region of DMP1 induces a rapid formation of a homogenous reparative dentin bridge in an injured pulp model and therefore improves pulp healing.
Figure 7.
Characterization of the reparative dentin bridge at day 15. In control teeth (left panel), no particular cell organization is observed related to the dentin bridge (a); a faint DSP (c) or DMP1 (e) labeling could be observed associated to the cells (arrow-heads). In the C-DMP1 treated pulps (right panel), cells presenting a polarized morphology (arrows) were seen organized as a palisade behind the dentin bridge, at the opposite side of the injury (b). Their hematoxylin stained nucleus was located at the basal pole of the cell body whereas the apical pole was highly labelled for DSP (d) and DMP1 (f). Formation of an odontoblast process is observed in the forming matrix as indicated by large arrows.
Scale bars a – b: 100 μm; c–f: 50 μm
db: dentin bridge
Discussion
The main function attributed to MMPs is the degradation of the ECM (Visse and Nagase, 2003). However, it is now widely acknowledged that MMPs play an important role in signaling by generating peptides with specific biological activities (Page-McCaw et al., 2007). In this study, we have shown that DMP1, an important NCP of bone and dentin ECM, can be cleaved by MMP-2 to release a C-ter peptide, potentially able to promote the differentiation of dental pulp stem cells. MMP-2 was selected since it is a predominant protease in the dentin matrix (Mazzoni et al., 2007) with a relatively large substrate specificity and has been previously shown to produce active peptides from matrix proteins in other systems (Dean et al., 2007).
We identified several potential MMP-2 cleavage sites in the DMP1 protein sequence, based on the fact that gelatinases present a deep hydrophobic S1′ pocket in their catalytic site and interact favorably with hydrophobic residues in P1′, leucine being the most favorable candidate (Turk et al., 2001). Considering these potential cleavage sites and the results obtained with SDS-PAGE and Western blot analysis, it is most likely that the initial cleavage of DMP1 by MMP-2 occurs between S382 and L383 or E260 and L261. Site-directed mutations in the recombinant protein would allow to definitively identify the cleavage site. As other potential cleavage sites in DMP1 sequence are located at the amino and the carboxyl end of DMP1, it is possible that secondary proteolysis may occur with extended incubation periods with MMP-2.
Published reports have demonstrated the presence of the 37 kDa N- and 57 kDa C-fragments of DMP1 in rat and bovine dentin matrix (Huang et al., 2008, Maciejewska et al., 2009). These fragments were thought to be the only forms of DMP1 present in the matrix although the full-length protein was recently detected in the matrix, albeit at a low level (Huang et al., 2008). In our study, we showed a high level of full-length protein in the developing tooth using a gentle extraction buffer containing Triton. It may be that the extraction techniques combining guanidine-HCl with chromatographic separation of a mineralized tissue is too harsh to recover the proteins in their native forms within the matrix, accounting for the low level of the native form of DMP1 detected in previous studies (Huang et al., 2008, Qin et al., 2003). The addition of the total metalloproteinase inhibitor EDTA or the more specific MMP inhibitor marimastat during the extraction of dentin appeared to protect the full-length DMP1 and the C-DMP1 57 kDa fragment from further proteolysis, as the bands were more intense in their presence. This suggests that native isoforms of DMP1 can be potentially degraded in vivo by endogenous MMPs present in the dentin matrix, generating a 42 kDa peptide which contains the biologically active C-ter region of the protein. In addition, treatment of the extracts with exogenous MMP-2 further released a 40 kda peptide. Taken together these data demonstrate that DMP1 is a substrate for MMP-2 but also for other MMPs, such as MMP-3 shown to be present within the dentin matrix (Boukpessi et al., 2008).
The biological activity of the N and C fragments of DMP1 were shown to be different, based on their localization patterns (Lu et al., 2009); the N-DMP1 fragment is believed to be a mineralization inhibitor as it is localized in unmineralized matrices (predentin, proliferation and hypertrophic zones in the growth plate of bone), whereas the C-DMP1 fragment was suggested to be a nucleator as it is localized within mineralized matrices (Maciejewska et al., 2009). As DMP1 can be cleaved by MMP-2, it was of interest to investigate whether its cleavage products retain functional activity. In a first approach, we demonstrated that the recombinant C-DMP1 polypeptide, mimicking the limited cleavage products obtained by MMP-2, had a profound effect on the differentiation of dental pulp stem/progenitor cells, not observed with the N- polypeptide. Differentiation into putative odontoblast was revealed by the rapid organization of most of the cells into clusters and the increased expression of odontoblast differentiation markers, namely DMP1 and DSPP (Hao et al., 2009, Hao et al., 2004, Paula-Silva et al., 2009). However, as DPSCs are a heterogeneous cell population, further investigations using sorted pulp stem cells will allow us to investigate the mechanism induced by DMP1 derived peptides on pulp cell differentiation.
It is worth noting that the DNA binding region of DMP1 on the DSPP promoter which we had previously mapped (Narayanan et al., 2006) is also located within the C-ter DMP1, between residues 420 and 489. Therefore, the hypothesis that a peptide derived from a cleavage of DMP1 within the matrix, enclosing this region, could induce odontoblast differentiation by binding the DSPP promoter require further investigations. We recently demonstrated that DMP1 binding to GRP-78 receptor was an important mechanism by which DMP1 was internalized and transported to the nucleus during bone and tooth development (Ravindran et al., 2008).
We previously reported that recombinant DMP1 can induce the cytodifferentiation of pulp cells after an insult or trauma of the tooth in a rat model (Almushayt et al., 2006). In this study, we demonstrated that the C-DMP1 polypeptide also enhanced pulp-healing capacity when implanted in a rat pulp-injury model that mimics an injury/carious process in humans tooth. A dense and continuous reparative dentin bridge was observed within two weeks suggesting accelerated pulp healing. The cells boarding this bridge presented a palisade organization and expressed DSP and DMP1, demonstrating the ability of C-DMP1 polypeptide to promote pulp cell differentiation toward a putative odontoblastic phenotype, and supporting our in vitro data obtained with pulp stem/progenitor cells.
BMP1, or its close relative TLL1, were suggested to be responsible for the physiological processing of DMP1 producing active fragments detected in the dentin matrix (Qin et al., 2003, Steiglitz et al., 2004). However, the cleavage of DMP1 by exogenous MMP-2 demonstrated in this study and its protection by marimastat against degradation by endogenous MMPs, suggest that MMPs, including MMP-2, can contribute to the release of DMP1 active peptides in the ECM and is likely to play a role in a more pathological situation (Chaussain-Miller et al., 2006). It is thus tempting to extrapolate that during the carious process, cleavage of DMP1 by MMPs would be activated by the acidic environment leading to the release of active fragments which could promote pulp healing. Future work, including the synthesis of DMP1 polypeptides identical to those produced by MMP-2 cleavage, will be necessary to better define the precise function of the cleaved products.
Conclusions
We demonstrated in this study that DMP1 possesses endogenous biological activity that can be released by proteolytic cleavage. MMPs, which are able to cleave DMP1 into peptides of various molecular sizes, represent good candidates for regulating the function of ECM molecules during dentin development or during pulp repair after an injury such as carious decay. In addition, in vivo confirmation of the capacity of the C-ter peptide to promote cell differentiation opens a therapeutic interest of using this peptide to regenerate dentin after an injury. Overall, these data support the current approach that ECM constitutes a tank of healing molecules.
Acknowledgments
We thank Dr. Bao-Shiang Lee (Laboratory of Proteomics, UIC, USA) for his assistance, Mrs Dominique Septier and Pr Jean Louis Saffar (EA 2496) for their assistance with histology and Mr Benoit Letang (EA 2496) for his assistance with animal experiments. This work was supported by National Institutes of Health Grants DE 11657 (AG) and DE 13836 (AG), and by grants from University Paris Descartes and AP-HP, France (CC).
List of Abbreviations
- ASARM
aspartic acid and serine rich domain
- C-ter
C-terminal
- DDI
double deionized
- DMEM
Dulbecco’s modified Eagle’s medium
- DMP1
dentin matrix protein 1
- DPSCs
dental pulp stem cells
- DSP
dentin sialoprotein
- DSPP
dentin sialoprotein phosphoprotein
- DSV
Animal Care Committee of the French Veterinary Services
- DSPP
sentin sialophosphoprotein
- ECM
extracellular matrix
- FBS
fetal bovine serum
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- IPTG
isopropyl b-D-1-thiogalactopyranoside
- MMPs
matrix metalloproteinases
- NCPs
noncollagenous proteins
- N-ter
N-terminal
- PCR
polymerase chain reaction
- SDS-PAGE
SDS-polyacrylamide gel
- TIMPs
tissue inhibitors of metalloproteinases
References
- Almushayt A, Narayanan K, Zaki AE, George A. Dentin matrix protein 1 induces cytodifferentiation of dental pulp stem cells into odontoblasts. Gene Ther. 2006;13:611–620. doi: 10.1038/sj.gt.3302687. [DOI] [PubMed] [Google Scholar]
- Boukpessi T, Menashi S, Camoin L, Tencate JM, Goldberg M, Chaussain-Miller C. The effect of stromelysin-1 (MMP-3) on non-collagenous extracellular matrix proteins of demineralized dentin and the adhesive properties of restorative resins. Biomaterials. 2008;29:4367–4373. doi: 10.1016/j.biomaterials.2008.07.035. [DOI] [PubMed] [Google Scholar]
- Bourd-Boittin K, Fridman R, Fanchon S, Septier D, Goldberg M, Menashi S. Matrix metalloproteinase inhibition impairs the processing, formation and mineralization of dental tissues during mouse molar development. Exp Cell Res. 2005;304:493–505. doi: 10.1016/j.yexcr.2004.11.024. [DOI] [PubMed] [Google Scholar]
- Chaussain-Miller C, Fioretti F, Goldberg M, Menashi S. The role of matrix metalloproteinases (MMPs) in human caries. J Dent Res. 2006;85:22–32. doi: 10.1177/154405910608500104. [DOI] [PubMed] [Google Scholar]
- d’Aquino R, Graziano A, Sampaolesi M, Laino G, Pirozzi G, De Rosa A, Papaccio G. Human postnatal dental pulp cells co-differentiate into osteoblasts and endotheliocytes: a pivotal synergy leading to adult bone tissue formation. Cell Death Differ. 2007;14:1162–1171. doi: 10.1038/sj.cdd.4402121. [DOI] [PubMed] [Google Scholar]
- Dean RA, Overall CM. Proteomics discovery of metalloproteinase substrates in the cellular context by iTRAQ labeling reveals a diverse MMP-2 substrate degradome. Mol Cell Proteomics. 2007;6:611–623. doi: 10.1074/mcp.M600341-MCP200. [DOI] [PubMed] [Google Scholar]
- Dean RA, Butler GS, Hamma-Kourbali Y, Delbe J, Brigstock DR, Courty J, Overall CM. Identification of candidate angiogenic inhibitors processed by matrix metalloproteinase 2 (MMP-2) in cell-based proteomic screens: disruption of vascular endothelial growth factor (VEGF)/heparin affin regulatory peptide (pleiotrophin) and VEGF/Connective tissue growth factor angiogenic inhibitory complexes by MMP-2 proteolysis. Mol Cell Biol. 2007;27:8454–8465. doi: 10.1128/MCB.00821-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajjeraman S, Narayanan K, Hao J, Qin C, George A. Matrix macromolecules in hard tissues control the nucleation and hierarchical assembly of hydroxyapatite. J Biol Chem. 2007;282:1193–1204. doi: 10.1074/jbc.M604732200. [DOI] [PubMed] [Google Scholar]
- Godovikova V, Ritchie HH. Dynamic processing of recombinant dentin sialoprotein-phosphophoryn protein. J Biol Chem. 2007;282:31341–31348. doi: 10.1074/jbc.M702605200. [DOI] [PubMed] [Google Scholar]
- Goldberg M, Septier D, Bourd K, Hall R, George A, Goldberg H, Menashi S. Immunohistochemical localization of MMP-2, MMP-9, TIMP-1, and TIMP-2 in the forming rat incisor. Connect Tissue Res. 2003;44:143–153. doi: 10.1080/03008200390223927. [DOI] [PubMed] [Google Scholar]
- Gronthos S, Brahim J, Li W, Fisher LW, Cherman N, Boyde A, DenBesten P, Robey PG, Shi S. Stem cell properties of human dental pulp stem cells. J Dent Res. 2002;81:531–535. doi: 10.1177/154405910208100806. [DOI] [PubMed] [Google Scholar]
- Hao J, Ramachandran A, George A. Temporal and spatial localization of the dentin matrix proteins during dentin biomineralization. J Histochem Cytochem. 2009;57:227–237. doi: 10.1369/jhc.2008.952119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J, Zou B, Narayanan K, George A. Differential expression patterns of the dentin matrix proteins during mineralized tissue formation. Bone. 2004;34:921–932. doi: 10.1016/j.bone.2004.01.020. [DOI] [PubMed] [Google Scholar]
- He G, George A. Dentin matrix protein 1 immobilized on type I collagen fibrils facilitates apatite deposition in vitro. J Biol Chem. 2004;279:11649–11656. doi: 10.1074/jbc.M309296200. [DOI] [PubMed] [Google Scholar]
- He G, Dahl T, Veis A, George A. Dentin matrix protein 1 initiates hydroxyapatite formation in vitro. Connect Tissue Res. 2003;44:240–245. [PubMed] [Google Scholar]
- Huang GT, Shagramanova K, Chan SW. Formation of odontoblast-like cells from cultured human dental pulp cells on dentin in vitro. J Endod. 2006;32:1066–1073. doi: 10.1016/j.joen.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Huang B, Maciejewska I, Sun Y, Peng T, Qin D, Lu Y, Bonewald L, Butler WT, Feng J, Qin C. Identification of full-length dentin matrix protein 1 in dentin and bone. Calcif Tissue Int. 2008;82:401–410. doi: 10.1007/s00223-008-9140-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang GT, Gronthos S, Shi S. Mesenchymal stem cells derived from dental tissues vs. those from other sources: their biology and role in regenerative medicine. J Dent Res. 2009;88:792–806. doi: 10.1177/0022034509340867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laino G, d’Aquino R, Graziano A, Lanza V, Carinci F, Naro F, Pirozzi G, Papaccio G. A new population of human adult dental pulp stem cells: a useful source of living autologous fibrous bone tissue (LAB) J Bone Miner Res. 2005;20:1394–1402. doi: 10.1359/JBMR.050325. [DOI] [PubMed] [Google Scholar]
- Lu Y, Qin C, Xie Y, Bonewald LF, Feng JQ. Studies of the DMP1 57-kDa functional domain both in vivo and in vitro. Cells Tissues Organs. 2009;189:175–185. doi: 10.1159/000151727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejewska I, Cowan C, Svoboda K, Butler WT, D’Souza R, Qin C. The NH2-terminal and COOH-terminal fragments of dentin matrix protein 1 (DMP1) localize differently in the compartments of dentin and growth plate of bone. J Histochem Cytochem. 2009;57:155–166. doi: 10.1369/jhc.2008.952630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannello F, Tonti GA, Bagnara GP, Papa S. Role and function of matrix metalloproteinases in the differentiation and biological characterization of mesenchymal stem cells. Stem Cells. 2006;24:475–481. doi: 10.1634/stemcells.2005-0333. [DOI] [PubMed] [Google Scholar]
- Martin-De Las Heras S, Valenzuela A, Overall CM. The matrix metalloproteinase gelatinase A in human dentine. Arch Oral Biol. 2000;45:757–765. doi: 10.1016/s0003-9969(00)00052-2. [DOI] [PubMed] [Google Scholar]
- Mazzoni A, Mannello F, Tay FR, Tonti GA, Papa S, Mazzotti G, Di Lenarda R, Pashley DH, Breschi L. Zymographic analysis and characterization of MMP-2 and -9 forms in human sound dentin. J Dent Res. 2007;86:436–440. doi: 10.1177/154405910708600509. [DOI] [PubMed] [Google Scholar]
- Narayanan K, Ramachandran A, Hao J, He G, Park KW, Cho M, George A. Dual functional roles of dentin matrix protein 1. Implications in biomineralization and gene transcription by activation of intracellular Ca2+ store. J Biol Chem. 2003;278:17500–17508. doi: 10.1074/jbc.M212700200. [DOI] [PubMed] [Google Scholar]
- Narayanan K, Gajjeraman S, Ramachandran A, Hao J, George A. Dentin matrix protein 1 regulates dentin sialophosphoprotein gene transcription during early odontoblast differentiation. J Biol Chem. 2006;281:19064–19071. doi: 10.1074/jbc.M600714200. [DOI] [PubMed] [Google Scholar]
- Page-McCaw A, Ewald AJ, Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Biol. 2007;8:221–233. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palosaari H, Pennington CJ, Larmas M, Edwards DR, Tjaderhane L, Salo T. Expression profile of matrix metalloproteinases (MMPs) and tissue inhibitors of MMPs in mature human odontoblasts and pulp tissue. Eur J Oral Sci. 2003;111:117–127. doi: 10.1034/j.1600-0722.2003.00026.x. [DOI] [PubMed] [Google Scholar]
- Paula-Silva FW, Ghosh A, Silva LA, Kapila YL. TNF-alpha promotes an odontoblastic phenotype in dental pulp cells. J Dent Res. 2009;88:339–344. doi: 10.1177/0022034509334070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C, Brunn JC, Cook RG, Orkiszewski RS, Malone JP, Veis A, Butler WT. Evidence for the proteolytic processing of dentin matrix protein 1. Identification and characterization of processed fragments and cleavage sites. J Biol Chem. 2003;278:34700–34708. doi: 10.1074/jbc.M305315200. [DOI] [PubMed] [Google Scholar]
- Ravindran S, Narayanan K, Eapen AS, Hao J, Ramachandran A, Blond S, George A. Endoplasmic reticulum chaperone protein GRP-78 mediates endocytosis of dentin matrix protein 1. J Biol Chem. 2008;283:29658–29670. doi: 10.1074/jbc.M800786200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe PS, de Zoysa PA, Dong R, Wang HR, White KE, Econs MJ, Oudet CL. MEPE, a new gene expressed in bone marrow and tumors causing osteomalacia. Genomics. 2000;67:54–68. doi: 10.1006/geno.2000.6235. [DOI] [PubMed] [Google Scholar]
- Shi S, Gronthos S. Perivascular niche of postnatal mesenchymal stem cells in human bone marrow and dental pulp. J Bone Miner Res. 2003;18:696–704. doi: 10.1359/jbmr.2003.18.4.696. [DOI] [PubMed] [Google Scholar]
- Six N, Septier D, Chaussain-Miller C, Blacher R, DenBesten P, Goldberg M. Dentonin, a MEPE fragment, initiates pulp-healing response to injury. J Dent Res. 2007;86:780–785. doi: 10.1177/154405910708600818. [DOI] [PubMed] [Google Scholar]
- Srinivasan R, Chen B, Gorski JP, George A. Recombinant expression and characterization of dentin matrix protein 1. Connect Tissue Res. 1999;40:251–258. doi: 10.3109/03008209909000703. [DOI] [PubMed] [Google Scholar]
- Steiglitz BM, Ayala M, Narayanan K, George A, Greenspan DS. Bone morphogenetic protein-1/Tolloid-like proteinases process dentin matrix protein-1. J Biol Chem. 2004;279:980–986. doi: 10.1074/jbc.M310179200. [DOI] [PubMed] [Google Scholar]
- Tjaderhane L, Larjava H, Sorsa T, Uitto VJ, Larmas M, Salo T. The activation and function of host matrix metalloproteinases in dentin matrix breakdown in caries lesions. J Dent Res. 1998;77:1622–1629. doi: 10.1177/00220345980770081001. [DOI] [PubMed] [Google Scholar]
- Turk BE, Huang LL, Piro ET, Cantley LC. Determination of protease cleavage site motifs using mixture-based oriented peptide libraries. Nat Biotechnol. 2001;19:661–667. doi: 10.1038/90273. [DOI] [PubMed] [Google Scholar]
- van Strijp AJ, Jansen DC, DeGroot J, ten Cate JM, Everts V. Host-derived proteinases and degradation of dentine collagen in situ. Caries Res. 2003;37:58–65. doi: 10.1159/000068223. [DOI] [PubMed] [Google Scholar]
- Visse R, Nagase H. Matrix metalloproteinases and tissue inhibitors of metalloproteinases: structure, function, and biochemistry. Circ Res. 2003;92:827–839. doi: 10.1161/01.RES.0000070112.80711.3D. [DOI] [PubMed] [Google Scholar]
- Yamakoshi Y, Hu JC, Iwata T, Kobayashi K, Fukae M, Simmer JP. Dentin sialophosphoprotein is processed by MMP-2 and MMP-20 in vitro and in vivo. J Biol Chem. 2006;281:38235–38243. doi: 10.1074/jbc.M607767200. [DOI] [PubMed] [Google Scholar]
- Zhang W, Walboomers XF, Shi S, Fan M, Jansen JA. Multilineage differentiation potential of stem cells derived from human dental pulp after cryopreservation. Tissue Eng. 2006;12:2813–2823. doi: 10.1089/ten.2006.12.2813. [DOI] [PubMed] [Google Scholar]